Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(3):571-582. doi:10.7150/ijms.90611 This issue Cite
Review
DARS-AS1: A Vital Oncogenic LncRNA Regulator with Potential for Cancer Prognosis and Therapy
1. Department of Gastrointestinal Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330008, Jiangxi, China.
2. Department of Spleen and Stomach Diseases, Jiujiang Hospital of Traditional Chinese Medicine, Jiujiang 332000, Jiangxi, China.
3. Department of Neurosurgery, Yingtan People's Hospital, Yingtan 335000, Jiangxi, China.
#These two authors contributed equally to this work.
Received 2023-9-28; Accepted 2024-1-1; Published 2024-1-20
Abstract
DARS-AS1, short for Aspartyl-tRNA synthetase antisense RNA 1, has emerged as a pivotal player in cancers. Upregulation of this lncRNA is a recurrent phenomenon observed across various cancer types, where it predominantly assumes oncogenic roles, exerting influence on multiple facets of tumor cell biology. This aberrant expression of DARS-AS1 has triggered extensive research investigations, aiming to unravel its roles and clinical values in cancer. In this review, we elucidate the significant correlation between dysregulated DARS-AS1 expression and adverse survival prognoses in cancer patients, drawing from existing literature and pan-cancer analyses from The Cancer Genome Atlas (TCGA). Additionally, we provide comprehensive insights into the diverse functions of DARS-AS1 in various cancers. Our review encompasses the elucidation of the molecular mechanisms, ceRNA networks, functional mediators, and signaling pathways, as well as its involvement in therapy resistance, coupled with the latest advancements in DARS-AS1-related cancer research. These recent updates enrich our comprehensive comprehension of the pivotal role played by DARS-AS1 in cancer, thereby paving the way for future applications of DARS-AS1-targeted strategies in tumor prognosis evaluation and therapeutic interventions. This review furnishes valuable insights to advance the ongoing efforts in combating cancer effectively.
Keywords: LncRNA, DARS-AS1, Tumor biomarker, Biological Functions, Regulatory mechanism
Introduction
Initially, long non-coding RNAs (lncRNAs) were considered as transcriptional noise or "junk RNA" since they lacked the functionality of encoding proteins [1-3]. However, as scientific research has advanced, it has become evident that lncRNAs play crucial roles in gene regulation [4-8], involving processes such as transcription, splicing, translation, and chromatin structure modifications. LncRNAs have been found to play key roles in various diseases [9-13], particularly in cancer [14-16]. Moreover, lncRNAs have the potential to serve as valuable biomarkers for progression and prognosis assessment, as well as promising therapeutic targets [17-22]. With the development and widespread use of high-throughput sequencing technologies, an increasing number of lncRNAs have been discovered [23-26].
LncRNAs are classified into several categories, such as antisense, sense, intronic, and intergenic, each possessing unique characteristics [27]. Focusing on antisense lncRNAs, these transcripts are uniquely synthesized from the opposite DNA strand of genes that may encode proteins or serve non-coding functions, leading to their critical role in the onset and development of tumors, which has recently garnered attention [28-31]. At the molecular level, they execute regulatory functions through a series of mechanisms, encompassing epigenetic modification, transcriptional control, post-transcriptional regulation, and impacts on translation [16, 28, 32]. Additionally, due to their nucleotide sequence complementarity, antisense lncRNAs possess a distinctive regulatory capability, specifically targeting and interacting with their corresponding sense genes [28], further highlighting the complexity of their involvement in cellular and pathological processes, particularly within the realm of cancer.
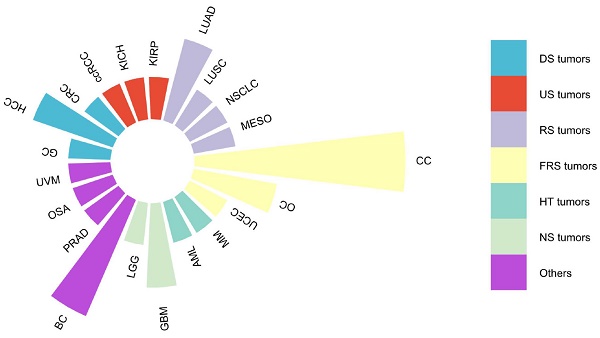
DARS-AS1, or Aspartyl-tRNA synthetase antisense RNA 1, serves as a prime example of an lncRNA, situated on chromosome 2 at q21.3 of the human genome. Spanning 22,367 nucleotides and comprising five exons (source: https://www.ncbi.nlm.nih.gov/gene/101928243), DARS-AS1 gives rise to a remarkable 34 splice variants, ranging in size from 311 base pairs for DARS1-AS1-228 to 1634 base pairs for DARS1-AS1-233(source: https://www.ensembl.org/Homo_sapiens/Gene/Summary?g = ENSG00000231890; r = 2:135985124-136022593). As an antisense lncRNA, DARS-AS1 has emerged as a recognized oncogenic entity, implicated in a wide array of cancer types as supported by extensive research publications [33-51], as depicted in Figure 1. These encompass cancers originating from various organ systems, including the digestive, urinary, respiratory, female reproductive, hematological, and neurological systems, among others. Notably, elevated DARS-AS1 expression has been linked to unfavorable clinicopathological characteristics and diminished overall survival outcomes. DARS-AS1 plays a pivotal part in tumor-related processes such as cell proliferation, migration, invasion, and autophagy, and potentially influences the sensitivity of tumor cells to radiotherapy or chemotherapy. These effects are mainly mediated through a complex ceRNA network and signaling pathways, which regulate tumorigenesis and disease progression. Here, we provide a concise review based on existing literature and The Cancer Genome Atlas (TCGA) analysis, mainly around examining the clinical significance of DARS-AS1, its impact on regulating cellular processes and related mechanisms, and its potential as a promising therapeutic target.
Malignancies associated with lncRNA DARS-AS1 in existing literature studies. This figure summarizes the association between various malignancies and lncRNA DARS-AS1 reported in the existing literature. One publication studied the link between lncRNA DARS-AS1 and GC, CRC, ccRCC, KICH, KIRP, LUSC, NSCLC, MESO, UCEC, MM, AML, LGG, PRAD, OSA, and UVM. Two publications studied the link between lncRNA DARS-AS1 and HCC, LUAD, OC, and GBM. Three publications studied the role of lncRNA DARS-AS1 in BC. And five publications studied the effects of lncRNA DARS-AS1 in CC. Abbreviations: CC: Cervical cancer, OC: Ovarian cancer, UCEC: Uterine corpus endometrial carcinoma, MM: Myeloma, AML: Acute myeloid leukemia, GBM: Glioblastoma, LGG: Lower grade glioma, BC: Breast cancer, PRAD: Prostate adenocarcinoma, OSA: Osteosarcoma, UVM: Uveal melanoma, GC: Gastric cancer, HCC: Hepatocellular carcinoma, CRC: Colorectal cancer, ccRCC: Clear cell renal cell carcinoma, KICH: Kidney chromophobe, KIRP: Kidney renal papillary cell carcinoma, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, NSCLC: Non-small cell lung cancer, MESO: Mesothelioma. DS tumors: Digestive system tumors, US tumors: Urinary system tumors, RS tumors: Respiratory system tumors, FRS tumors: Female reproductive system tumors, HT tumors: Hematological tumors, NS tumors: Neurological system tumors.
Expression of DARS-AS1 in tumor tissues
DARS-AS1, an emerging tumor marker, exhibits significant upregulation in a diverse array of cancer types based on research involving in-house tissue specimens and analyses of TCGA data (Table 1). These cancer types span various bodily systems, including the neurological system (such as brain lower-grade glioma and glioblastoma), the respiratory system (including non-small cell lung cancer, and mesothelioma), the digestive system (comprising gastric cancer, liver cancer, and colorectal cancer), the urinary system (featuring renal cell carcinoma), the female reproductive system (encompassing cervical and ovarian cancers), and the hematological system (including myeloma and acute myeloid leukemia). Additionally, DARS-AS1 demonstrates upregulation in other tumor types, such as bladder cancer, prostate adenocarcinoma, uveal melanoma, breast cancer, and osteosarcoma. These findings suggest that DARS-AS1 may hold significance as a potential biomarker or therapeutic target across a wide spectrum of cancers, warranting further investigation into its specific roles and clinical applications.
LncRNA DARS-AS1 is associated with tumor-related clinical features
Several investigations have explored the relationship between DARS-AS1 expression and clinicopathological characteristics across a spectrum of five tumor types (Table 1). In gastric cancer [51], DARS-AS1 exhibits a significant positive correlation with T category, N category, TNM stage. In hepatocellular carcinoma [35], overexpression DARS-AS1 indicates larger tumor size and advanced TNM stage. In clear cell renal cell carcinoma [36] and triple-negative breast cancer [40], high expression of DARS-AS1 is indicative of higher clinical-stage. In acute myeloid leukemia [34], patients with high DARS-AS1 expression exhibit a significantly higher leukocyte count in peripheral blood, while their hemoglobin levels and platelet counts are notably lower compared to those with low DARS-AS1 expression.
LncRNA DARS-AS1 is a valuable prognostic marker
LncRNA DARS-AS1 serves as a valuable prognostic indicator. The abnormal expression of DARS-AS1 has been closely associated with overall survival (OS) in cancer patients. As summarized in Table 1, drawing from both the in-house survival data and TCGA analyses conducted in these studies for OS, high levels of DARS-AS1 expression are predictive of poorer OS in a range of cancer types, including gastric cancer, hepatocellular carcinoma, lung adenocarcinoma, triple-negative breast cancer, cervical cancer, acute myeloid leukemia, UVM, KICH, KIRP, MESO, GBM, and LGG.
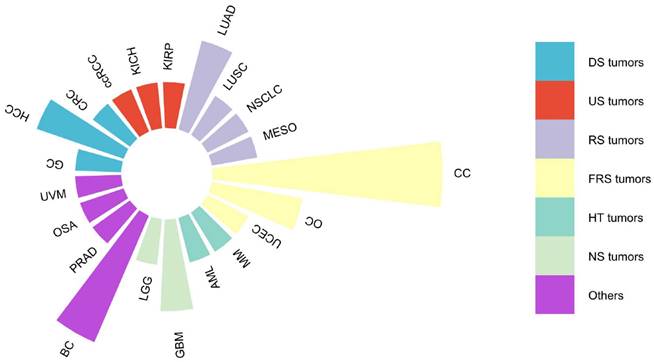
In addition to the effect on OS, we also performed a comprehensive assessment of the prognostic relevance of DARS-AS1 in a pan-cancer analysis using TCGA, considering disease-specific survival (DSS), disease-free interval (DFI) and progression-free interval (PFI).
Correlations between the expression levels of lncRNA DARS-AS1 in tumor tissues, clinical characteristics, and prognostic outcomes.
| Tumor type | Expression in tumor tissues | Clinical features | Prognosis | Methods for survival analysis | Indicator for poor survival | Ref. |
|---|---|---|---|---|---|---|
| Gastric cancer | Up-regulated | T category, N category, TNM stage | OS | K-M plot; Multivariate analysis | High expression | [51] |
| Hepatocellular carcinoma | Up-regulated | TNM stage, Tumor size | OS | K-M plot; Multivariate analysis | High expression | [35] |
| Clear cell renal cell carcinoma | Up-regulated | Clinical stage | - | - | - | [36] |
| Lung adenocarcinoma | Up-regulated | - | OS | K-M plot | High expression | [42] |
| Non-small cell lung cancer | Up-regulated | - | - | - | - | [39] |
| Triple-negative breast cancer | Up-regulated | Clinical stage | OS | K-M plot | High expression | [40] |
| Cervical cancer | Up-regulated | - | - | - | - | [33] |
| Cervical cancer | Up-regulated | - | - | - | - | [43] |
| Cervical Cancer | Up-regulated | - | OS | K-M plot | High expression | [49] |
| Cervical Cancer | Up-regulated | - | - | - | - | [50] |
| Cervical cancer | Up-regulated | - | OS | K-M plot | High expression | [37] |
| Ovarian cancer | Up-regulated | - | - | - | - | [38] |
| Ovarian cancer | Up-regulated | - | - | - | - | [48] |
| Acute myeloid leukemia | Up-regulated | White blood cells, Hemoglobin, Platelet count | OS | K-M plot | High expression | [34] |
| Osteosarcoma | Up-regulated | - | - | - | - | [45] |
| Colorectal cancer | Up-regulated | - | - | - | - | [46] |
| Hepatocellular carcinoma | Up-regulated | - | - | - | - | [46] |
| UVM, KICH, KIRP, MESO, GBM, and LGG (TCGA datasets) | - | - | OS | K-M plot | High expression | [46] |
| BLCA, KIRC, PRAD, LUSC, UCEC, LUAD, LIHC, KIRP and COAD (TCGA datasets) | Up-regulated | - | - | - | - | [46] |
| Glioblastoma | Up-regulated | - | OS | K-M plot; Multivariate analysis | High expression | [47] |
OS: Overall Survival, K-M plot: Kaplan-Meier plot, UVM: Uveal melanoma, KICH: Kidney chromophobe, KIRP: Kidney renal papillary cell carcinoma, MESO: Mesothelioma, GBM: Glioblastoma, LGG: lower grade glioma, BLCA: Bladder urothelial carcinoma, PRAD: Prostate adenocarcinoma, LUSC: Lung squamous cell carcinoma, UCEC: Uterine corpus endometrial carcinoma, LUAD: Lung adenocarcinoma, LIHC: Liver hepatocellular carcinoma, COAD: Colon adenocarcinoma, TCGA: The Cancer Genome Atlas. "-": Indicates missing or not applicable data.
In terms of DSS (Figure 2A), elevated expression of DARS-AS1 indicated unfavorable prognosis in eight tumor types, namely TCGA-GM (HR = 1.87(1.62,2.16), TCGA-KICH (HR=3.52(1.80,6.86), TCGA-KIRP (HR=1.71(1.23,2.37), TCGA-HNSC (HR = 1.30(1.06,1.59), TCGA-PRAD (HR=5.41(1.33,21.99), TCGA-ACC (HR=1.42(1.08,1.87),TCGA-LGG (HR = 1.25(1.02,1.53), and TCGA-UVM (HR=1.72(1.03,2.86), where high expression was indicative of poorer outcomes.
In terms of DFI (Figure 2B), increased DARS-AS1 expression was linked to a poor prognosis in three tumor types, TCGA-PAAD (HR = 1.96(1.127,3.41), TCGA-SARC (HR = 1.22(1.004,1.48) and TCGA-ACC (HR = 1.54(1.008,2.34) where high expression was associated with worse outcomes.
Regarding PFI (Figure 2C), it was observed that high expression of DARS-AS1 led to poorer prognosis in five tumor types, namely TCGA-GM (HR = 1.52(1.35,1.70), TCGA-KIRP (HR = 1.69(1.34,2.15), TCGA-KICH (HR = 2.20(1.43,3.40), TCGA-HNSC (HR = 1.29(1.09,1.53), and TCGA-ACC (HR = 1.37(1.11,1.68), highlighting the adverse impact of elevated DARS-AS1 expression on prognosis.
Functions of DARS-AS1 in human tumors
Extensive research has been conducted on the role of DARS-AS1 in twelve different types of tumors, utilizing in vivo and/or in vitro experiments (Table 2). DARS-AS1 expression has been consistently observed to be up-regulated in numerous tumor cell lines. The subcellular localization of this lncRNA was reported within the cytoplasm in six different types of tumor cells, including clear cell renal cell carcinoma, cervical cancer, acute myeloid leukemia, colorectal cancer, and glioblastoma cells. DARS-AS1 plays a pivotal oncogenic role in tumor development, impacting a series of biological processes (Figure 3). It promotes epithelial-mesenchymal transition (EMT), augments cell proliferation, enhances cell viability, facilitates migration and invasion, induces autophagy, contributes to therapy resistance, facilitates tumor growth, and metastasis. Conversely, DARS-AS1 inhibits apoptosis and hinders cell cycle arrest in tumor cells. These findings emphasize the profound importance of DARS-AS1 as a critical oncogenic regulator influencing various aspects of tumor progression.
Significant prognostic significance of lncRNA DARS-AS1 for DSS/DFI/PFI in pan-cancer analysis using TCGA. (A) High expression levels of lncRNA DARS-AS1 are associated with poorer DSS in GM, KICH, KIRP, HNSC, PRAD, ACC, LGG, and UVM. (B) Elevated expression of lncRNA DARS-AS1 is linked to inferior DFI specifically in PRAD, SARC, and ACC. (C) High expression levels of lncRNA DARS-AS1 are indicative of poorer PFI in GM, KIRP, KICH, HNSC, and ACC. Abbreviations: DSS: Disease-specific survival; DFI: Disease-free interval; PFI: Progression-free interval; GM: Glioma; KICH: Kidney chromophobe; KIRP: Kidney renal papillary cell carcinoma; HNSC: Head and neck squamous cell carcinoma; PRAD: Prostate adenocarcinoma; ACC: Adrenocortical carcinoma; LGG: Lower grade glioma; UVM: Uveal melanoma; SARC: Sarcoma.
Functions and regulatory mechanisms of lncRNA DARS-AS1 in different cancers.
| Cancer Type | Utilized Cell Lines | Expression Levels in Cancer Cell Lines | Subcellular localization | Animal models | Regulatory Pathways | Functional Role of LncRNA DARS-AS1 | Effects in vitro | Effects in vivo | Signaling pathway | Resistance | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gastric cancer | AGS, KATO-III, HGC-27, NCI-N87, MKN45, SNU-1, GES-1 | Up- regulated | - | - | DARS-AS1/miR-330-3p/NAT10 | ceRNA | Proliferation, migration, invasion | - | - | - | [51] |
| Hepatocellular carcinoma | THLE-3, Huh-7, HCCLM3, HLE, MHCC97, HCCLM6 | Up- regulated | - | BALB/c mice (male, 6-8 weeks old) | DARS-AS1/miR3200-5p/CKAP2 | ceRNA | Proliferation, apoptosis, migration, invasion, EMT | Tumor growth and metastasis | FAK-ERK pathway | - | [35] |
| Clear cell renal cell carcinoma | HK-2, ClearCa1, HH332, Caki-1, KMRC-2, KN-41 | Up- regulated | Mainly in cytoplasm | - | DARS-AS1/miR-194-5p/DARS | ceRNA | Proliferation, apoptosis | - | - | - | [36] |
| Lung adenocarcinoma | NCI-H23, A549, HCC827, PC-9, C422L, HBE | Up- regulated | - | Orthotopic xenografts and subcutaneous tumor transplantation | DARS-AS1/miR-188-5p/KLF12 | ceRNA | Proliferation, invasion, migration, apoptosis | Tumor growth and metastasis | PI3K/AKT pathway | - | [42] |
| Non-small cell lung cancer | SPca1, H1299, Pc-9, H358, 16HBe | Up- regulated | - | - | DARS-AS1/miR-532-3p | ceRNA | Cell viability, proliferation, invasion, migration | - | - | - | [39] |
| Triple-negative breast cancer | MDA-MB-231, MDA-MB-468, BT549,MCF-10A | Up- regulated | - | Xenograft model (Female BALB/c nude mice) | DARS-AS1/miR-129-2-3p/CDK1 | ceRNA | Cell viability, proliferation, migration, invasion, EMT | Tumor growth and metastasis | NF-κB/STAT3 pathway | - | [40] |
| Triple-negative breast cancer | MDA-MB-231, MDA-MB-468, BT549, MDA-MB-231/ADR, MCF-10A | - | - | Orthotopic models (6 weeks old female BALB/c nude mice) | DARS-AS1/TGF-β/Smad3 | Molecular mediator | Proliferation, migration, invasion, apoptosis, autophagy, cell viability | Tumor growth | TGF-β pathway | Chemoresistance (Doxorubicin) | [41] |
| Cervical cancer | HeLa, C33A, MS751, SiHa, ME-180, CaSki, Ect1/E6E7 | Up- regulated | Mainly in cytoplasm | - | DARS-AS1/miR-628-5p/JAG1 | ceRNA | Proliferation cell apoptosis | - | Notch pathway | - | [33] |
| Cervical cancer | SiHa, CaSki, C33A, DoTc24510, HeLa, End1/E6E7 | Up- regulated | Both in nucleus and cytoplasm | - | HIF1α/DARS-AS1/ DARS/ATG5/ATG3 | Modular scaffold | Autophagy | - | - | - | [43] |
| Cervical Cancer | SiHa, HeLa | - | - | - | DARS-AS1/miR-188-5p/HMGB1 | ceRNA | Proliferation, apoptosis, invasion | - | - | - | [49] |
| Cervical Cancer | SiHa, Hela | - | Mainly in cytoplasm | subcutaneous xenograft model (BALB/c nude mice, 4-6-weeks old) | DARS-AS1/ IGF2BP3 | Modular scaffold | Proliferation, apoptosis, invasion, cell cycle progression | Tumor growth | - | - | [50] |
| Cervical cancer | SiHa, CaSki, C-33A, Ect1/ E6E7 | Up- regulated | - | - | DARS-AS1/ ATP1B2 | Molecular mediator | Cell viability, proliferation, invasion, migration, cell cycle progression | - | cGMP-PKG pathway | - | [37] |
| Ovarian cancer | Caov-3, A2780, SKOV3, CoC1, IOSE80 | Up- regulated | - | - | DARS-AS1/miR-194-5p/TSPAN1 /ITGA2 | ceRNA | Cell viability, migration, invasion, apoptosis | - | - | - | [38] |
| Ovarian cancer | A2780, HeyA8, SKOV3 | Up- regulated | - | - | DARS-AS1/miR-194-5p/RBX1 /TP53 | ceRNA | Proliferation, apoptosis, invasion, migration | - | - | - | [48] |
| Myeloma | RPMI 8226, LP-1, U266, H929 | Up- regulated | - | NOD-SCID mice | HIF-1/DARS-AS1/ RBM39 | Protein decoy | Proliferation, apoptosis | Tumor growth | mTOR pathway | Chemoresistance (Bortezomib) | [44] |
| Acute myeloid leukemia | HS-5, BF-24, MV4-11, U937, HL-60 | Up- regulated | In the cytoplasm | Six-week-old male NOD/SCID mice | DARS-AS1/miR-425/TGFB1 | ceRNA | Proliferation | Tumor growth | TGF-β pathway | / | [34] |
| Osteosarcoma | U2OS, SOSP-9607, Saos-2, MG-63, hFOB | Up- regulated | - | - | DARS-AS1/miR-532-3p/CCR7 | ceRNA | Proliferation, invasion | - | - | - | [45] |
| Colorectal cancer | SW620 HCT116 | - | Mainly in cytoplasm | Xenograft mouse models | DARS-AS1/ PACT/ PKR | Protein decoy | Cell proliferation, apoptosis | Tumor growth | PACT-PKR pathway | - | [46] |
| Breast cancer | MBA-MD-231 | - | - | - | DARS-AS1/ PACT / PKR | Protein decoy | Cell proliferation, apoptosis | - | PACT-PKR pathway | - | [46] |
| Hepatocellular carcinoma | HepG2 | - | - | - | DARS-AS1/ PACT / PKR | Protein decoy | Cell proliferation, apoptosis | - | PACT-PKR pathway | - | [46] |
| Glioblastoma | U251, U87, LN229, GSC11, GSC17, GSC20, GSC262, GSC272, GSC295, Hs683, SW1783, NHAs, ReNcell | Up- regulated | Mainly in cytoplasm | Orthotopic glioblastoma models (athymic nude mice,6-8-week-old female) | DARS1-AS1/YBX1/E2F1/CCND1 | Modular scaffold | Proliferation, self-renewal, DSB repair, cell cycle progression | Tumor growth, Survival | - | Radioresistance | [47] |
ceRNA: Competing endogenous RNA, DSB: Double-strand break, "-": Indicates missing or not applicable data.
Diverse roles of lncRNA DARS-AS1 in twelve distinct human tumors through in vitro and/or in vivo experiments. LncRNA DARS-AS1 exhibits oncogenic effects in gastric, liver, kidney, lung, breast, cervical, ovarian, and colorectal cancers, as well as myeloma, acute myeloid leukemia, osteosarcoma, and glioblastoma. Abbreviations: EMT: Epithelial-mesenchymal transition; DSB repair: double-strand break repair.

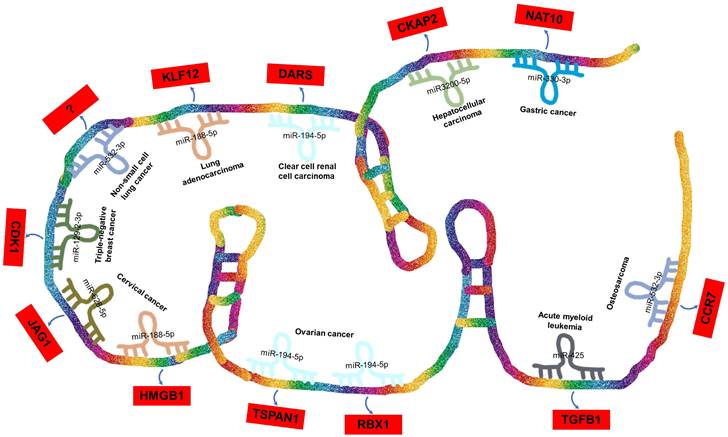
ceRNA Networks Showcasing lncRNA DARS-AS1 Interactions Across Diverse Human Cancers. LncRNA DARS-AS1 upregulates target gene expression by sponging miRNAs, including miR-330-3p, miR-3200-5p, miR-194-5p, miR-188-5p, miR-532-3p, and miR-129-2. This modulates the progression of various tumors, including gastric, liver, kidney, lung, breast, cervical, and ovarian cancers, alongside acute myeloid leukemia and osteosarcoma. Abbreviations: GC: Gastric cancer; HCC: Hepatocellular carcinoma; ccRCC: Clear cell renal cell carcinoma; LUAD: Lung adenocarcinoma; NSCLC: Non-small cell lung cancer; TNBC: Triple-negative breast cancer; CC: Cervical cancer; OC: Ovarian cancer; AML: Acute myeloid leukemia; OSA: Osteosarcoma; NAT10: N-Acetyltransferase 10; CKAP2: Cytoskeleton Associated Protein 2; DARS: Aspartyl-tRNA synthetase; KLF12:KLF Transcription Factor 12; CDK1:Cyclin Dependent Kinase 1; JAG1:Jagged Canonical Notch Ligand 1; HMGB1:High mobility group box 1; TSPAN1: Traspanin 1; RBX1:Ring-Box 1; TGFB1:Transforming growth factor beta 1; CCR7: C-C chemokine receptor type 7.
ceRNA network involving lncRNA DARS-AS1 in tumor progression
In recent years, there has been a notable surge in the exploration of competing endogenous RNAs (ceRNAs) networks [52-56]. These ceRNAs function as defensive shields, safeguarding mRNAs from the inhibitory actions of miRNAs [57]. Within the cancer research, the ceRNA regulatory network orchestrated by lncRNAs plays a pivotal and indispensable role [58-62].
In the case of DARS-AS1, its ceRNA network encompasses eight miRNAs across nine different types of cancers, as illustrated in Figure 4. These miRNAs include miR-330-3p in gastric cancer [51], miR-3200-5p in hepatocellular carcinoma [35], miR-194-5p in clear cell renal cell carcinoma [36] and ovarian cancer [38, 48], miR-188-5p in lung adenocarcinoma [42] and cervical cancer [49], miR-532-3p in non-small cell lung cancer [39] and osteosarcoma [45], miR-129-2-3p in triple-negative breast cancer [40], miR-628-5p in cervical cancer [33], and miR-425 in childhood acute myeloid leukemia [34].
BBOX1-AS1 exerts its regulatory influence across various types of tumors by competitively binding to a diverse range of miRNAs. Furthermore, it contributes to the progression of distinct tumor types by influencing common miRNAs. Intriguingly, in the context of a single tumor, BBOX1-AS1 can engage with multiple miRNAs to impact tumor development. For instance, in cervical cancer [33, 49], BBOX1-AS1 engages in two distinct ceRNA mechanisms, thereby promoting both the proliferation and apoptosis of cervical cancer cells. These mechanisms involve the targeting of miR-628-5p/JAG1 [33] and miR-188-5p/HMGB1 [49].
LncRNA DARS-AS1 as Functional Modules in Tumors
DARS-AS1 is a versatile regulator of gene expression, operating through ceRNA networks, as described above. However, it also exerts its influence in gene regulation by functioning as a modular scaffold, protein decoy, and molecular mediator impacting downstream targets.
When functioning as a molecular mediator, in triple-negative breast cancer [41], DARS-AS1 overexpression significantly upregulated the levels of TGF-β, p-Smad3, ATG5, and the conversion from LC3-I to LC3-II. Conversely, silencing DARS-AS1 reversed these effects. Silencing DARS-AS1 enhanced the sensitivity of triple-negative breast cancer (TNBC) cells to doxorubicin by suppressing autophagy induced by the TGF-β/Smad3 signaling pathway, thereby strengthening the synergistic antitumor effects. In cervical cancer [37], ATP1B2 was identified as a target mRNA of DARS-AS1, and it showed a negative correlation with DARS-AS1 expression. DARS-AS1/ATP1B2 partially regulated malignant behaviors through the cGMP-PKG signaling pathway.
When serving as a modular scaffold, in cervical cancer [43], DARS-AS1 enhanced DARS mRNA stability and translation by recruiting METTL3 and METTL14. Moreover, DARS-AS1 positively regulated IGF2BP3 expression by stabilizing IGF2BP3 mRNA. In glioblastoma [47], DARS1-AS1 interacted with YBX1 to promote the binding and stability of target mRNA. This established a mixed transcriptional/post-transcriptional feed-forward loop, enhancing the expression of key regulators of the G1-S transition, including E2F1 and CCND1. DARS1-AS1/YBX1 also increased the stability of FOXM1 mRNA, a master transcription factor regulating GSC self-renewal and DSB repair.
When acting as a protein decoy, in colorectal cancer [46], DARS-AS1 directly bound to PACT. This interaction inhibited the association between PACT and PKR, preventing the phosphorylation of the PKR downstream substrate eIF2α, ultimately inhibiting apoptotic cell death. In breast cancer and HCC [46], DARS-AS1 promoted cancer cell proliferation and inhibited apoptosis by inhibiting the function of PACT. In myeloma [44], DARS-AS1 exerts its function by binding RNA-binding motif protein 39 (RBM39), which impedes the interaction between RBM39 and its E3 ubiquitin ligase RNF147 and prevents RBM39 from degradation.
The intricate interplay of ceRNA networks and DARS-AS1's interactions with both mRNA and protein constituents endow it with a multifaceted role as a gene expression regulator. The perturbation of DARS-AS1 in cancer underscores its significance as a plausible therapeutic target, underscoring the urgency for in-depth exploration of its exact functionalities and molecular associations.
Signaling pathways influenced by lncRNA DARS-AS1
Accumulating scientific findings support the pivotal role of lncRNAs in orchestrating various signaling pathways [63-67], offering fresh perspectives for the development of targeted therapies. Presently, DARS-AS1 has been unequivocally established as an important participant in the regulation of multiple cancer-related signaling pathways, as outlined in Figure 5. These pathways encompass the FAK/ERK, PI3K/AKT, NF-κB/STAT3, TGF-β/Smad3, Notch, cGMP-PKG, mTOR, and PACT-PKR pathways. The involvement of DARS-AS1 in these intricate signaling networks implies its broader influence on the behavior of cancer cells and their responses to therapy.
In hepatocellular carcinoma [35], DARS-AS1 up-regulates CKAP2 by binding to miR-3200-5p, thereby activating the FAK-ERK pathway and promoting HCC proliferation and metastasis, and DARS-AS1 is also reported to promote HCC cell proliferation and inhibits apoptosis through inhibiting the function of PACT [46]. In lung adenocarcinoma [42], it was found that DARS-AS1 significantly enhances the malignant properties of LUAD cells. This effect was achieved through the activation of the PI3K/AKT pathway, triggering the EMT process, and the up-regulation of Cyclin D1 and Bcl-2 proteins, both recognized contributors to cell growth and survival. In breast cancer, DARS-AS1 promote the TNBC tumorigenesis by activation of the NF-κB/STAT3 signaling pathway [40], and increased the resistance of TNBC cells to doxorubicin by promote TGF-β/Smad3 signaling pathway-induced autophagy [41]. In addition, DARS-AS1 inhibits breast cancer cell proliferation, which is, at least partially, through repressing PACT-mediated PKR activation [46]. In cervical cancer, DARS-AS1 exhibits pro-tumorigenic effects by activating the Notch pathway [33], and cGMP-PKG pathway [37], exacerbating the tumorigenesis of cervical cancer. In myeloma, Tong et al. [44] revealed that hypoxia-induced lncRNA DARS-AS1 upregulates RBM39 protein expression via the ubiquitin-proteasome pathway, and further promotes mTOR signaling pathway to promote myeloma malignancy. In colorectal cancer [46], lncRNA DARS-AS1 is directly involved in the inhibition of the PACT-PKR pathway and promotes the proliferation and inhibit cancer cell apoptosis, and promote tumor growth in vivo.
Treatment resistance mediated by lncRNA DARS-AS1
Treatment resistance is a significant concern in cancer [68-71], and there is abundant evidence that lncRNAs play a role in regulating the sensitivity of tumor cells to drug and radiotherapy [72-77], thereby affecting tumor recurrence and metastasis.
Doxorubicin is the primary chemotherapy drug utilized to enhance the survival of triple-negative breast cancer patients [78-80]. However, it often leads to strong drug resistance during its usage. Liu et al. [41] reported that silencing DARS-AS1 decreases doxorubicin resistance by suppressing autophagy via inhibition of the TGF-β/Smad3 signaling pathway, and combination of DARS-AS1 siRNA and DOX significantly inhibited tumorigenesis and growth of TNBC cells, which indicated that combination of DARS-AS1 siRNA and doxorubicin can be used as new therapeutic agents for TNBC.
In the context of myeloma [44], lncRNA DARS-AS1 is directly upregulated by hypoxia inducible factor-1. And overexpression of DARS-AS1 resulted in a decreased responsiveness of myeloma cells to bortezomib. Additionally, Zheng et al. [47] investigated the impact of BBOX1-AS1 on glioblastoma tumorigenesis/radioresistance by multiomics analyses, and revealed that DARS1-AS1 depletion impaired the homologous recombination (HR)-mediated double-strand break (DSB) repair and enhanced the radiosensitivity of glioblastoma cells.
Signaling pathways regulated by lncRNA DARS-AS1. LncRNA DARS-AS1 promotes the tumor progression by activating multiple pathways, including FAK/ERK, PI3K/AKT, NF-κB/STAT3, TGF-β/Smad3, Notch, cGMP-PKG, and mTOR, as well as inactivating the PACT-PKR pathway. The red box indicates activation, the gray box indicates inactivation. Abbreviations: FAK/ERK: Focal adhesion kinase/Extracellular signal-regulated kinase; PI3K/AKT: Phosphoinositide 3-Kinase/Serine-threonine protein kinase; NF-κB/STAT3: Nuclear factor-kappa B/Signal transducer and activator of transcription 3; TGF-β/Smad3: Transforming growth factor beta/Smad family member 3; cGMP-PKG:Cyclic guanosine monophosphate/Protein kinase G; mTOR: Mammalian target of rapamycin; PACT-PKR: Protein activator of the interferon-induced protein kinase/Protein kinase R.
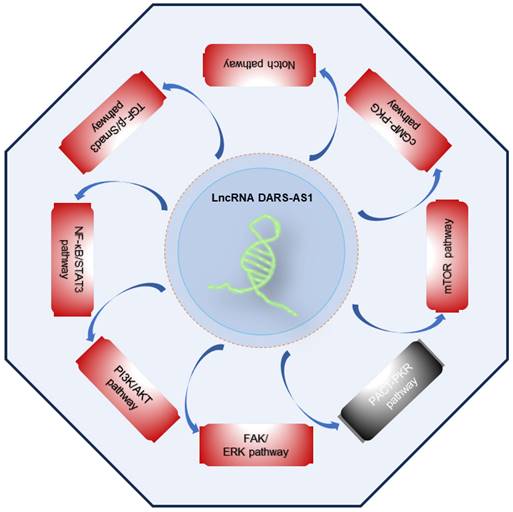
Summary of the molecular mechanisms of lncRNA DARS-AS1 as a central regulator of cellular processes in cancer. This figure provides an overview of how lncRNA DARS-AS1 serves as a central regulator of cellular processes in cancer. It influences gene expression through ceRNA networks and functions as a versatile entity. DARS-AS1 acts as a modular scaffold, protein decoy, and molecular mediator, thereby impacting downstream targets in gene regulation.
Overall, exploring the molecular mechanisms of the DARS1-AS1-associated regulatory axis in drug and radiotherapy resistance could provide valuable insights for the development of targeted therapies and improved clinical outcomes for cancer patients.
Future perspectives
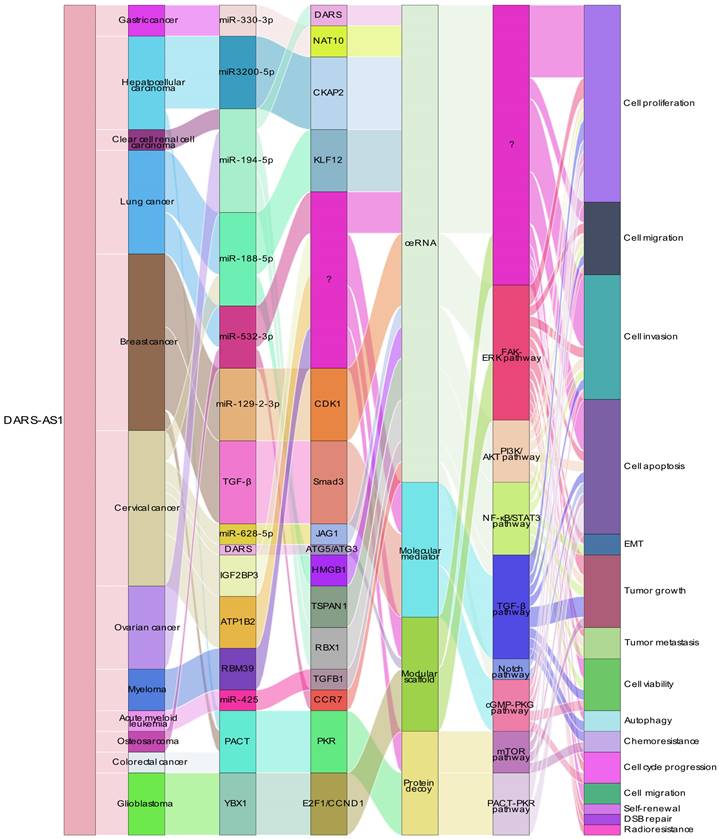
In recent years, DARS1-AS1, as an emerging oncogenic lncRNA, has been consistently found to be upregulated in various cancer types. Notably, DARS1-AS1 demonstrates significant clinical relevance in prognostic predictions. Studies have revealed that DARS1-AS1 plays a regulatory role in tumor development by influencing key molecules and genes involved in critical tumor-related biological processes, as shown in Figure 6. These findings highlight the potential therapeutic implications of targeting DARS1-AS1 in cancer treatment.
In terms of mechanisms, DARS-AS1 plays a crucial role as a multifaceted functional module, encompassing activities such as ceRNA, modular scaffold, protein decoy, and molecular mediator functions. It intricately influences gene expression and modulates signaling pathways, thereby regulating biological processes associated with tumorigenesis and progression. As a ceRNA, lncRNA DARS-AS1 binds to microRNA, leading to the upregulation of target genes such as NAT10, DARS, CDK1, JAG1, and HMGB1, and thus exhibits a broader regulatory effect on the development of different tumors. Notably, lncRNA DARS-AS1 is intricately involved in many signaling pathways, such as FAK/ERK, PI3K/AKT, NF-κB/STAT3, TGF-β/Smad3, Notch, cGMP-PKG, mTOR. Among these pathways, lncRNA DARS-AS1 plays an activating role, affecting key processes in cancer initiation and progression. By engaging with these signaling pathways, lncRNA DARS-AS1 plays a central role in shaping the dynamics of cancer cells, promoting their survival and enhancing their invasive and metastatic potential. And lncRNA DARS-AS1 inactivates PACT-PKR pathway, promotes proliferation and inhibits apoptotic cell death in multiple cancer cells. Remarkably, diminished levels of DARS1-AS1 expression demonstrated heightened susceptibility to both drug and radiation therapies, suggesting a potential avenue for augmenting the efficacy of current cancer treatment modalities. Overall, the intricate array of functions attributed to DARS1-AS1, encompassing its role as a ceRNA, functional modules, and its influence on pivotal signaling pathways, positions it as a promising therapeutic target for pioneering cancer interventions designed to combat metastasis and enhance treatment responses.
Despite the progress made, our understanding of DARS1-AS1 remains incomplete. Further studies are needed to assess DARS1-AS1 expression in hematopoietic cancers and to determine its impact on tumor progression and prognosis in larger study populations of different tumor types. In addition, no studies have explored the diagnostic potential of DARS1-AS1. It is imperative to evaluate the diagnostic value of DARS1-AS1 for cancer. In particular, the clinical value of detection of DARS1-AS1 in liquid tissue in the early diagnosis of tumors deserves further study. Furthermore, it is essential to acquire a more profound understanding of the specific regulatory mechanisms of DARS1-AS1 in various tumor types. DARS1-AS1 might be intricately connected to additional signaling pathways and could possess a broader ceRNA network. In vitro and in vivo studies are needed to elucidate the mechanisms through which DARS1-AS1 contributes to therapy resistance across diverse tumor types.
Conclusion
In a nutshell, DARS-AS1 is an oncogenic lncRNA consistently overexpressed in various cancers, strongly linked to poor patient outcomes. Research in lab and live settings reveals its cancer-promoting role in tumor processes through ceRNA networks and signaling pathways. DARS-AS1 holds promise as a cancer biomarker and therapeutic target, but more research and clinical validation are needed to fully understand its mechanisms and potential applications in cancer management.
Author contributions
HLL and YW designed and supervised the study. JS and KJX wrote the first draft. HLL and YW edited the manuscript. JS, KJX, HLL, and YW collected reference materials and produced tables and graphical visualizations. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Frontiers in genetics. 2015;6:2
2. Palazzo AF, Koonin EV. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell. 2020;183:1151-61
3. Lee H, Zhang Z, Krause HM. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends in genetics: TIG. 2019;35:892-902
4. Kazemzadeh M, Safaralizadeh R, Orang AV. LncRNAs: emerging players in gene regulation and disease pathogenesis. Journal of genetics. 2015;94:771-84
5. Chen X, Sun Y, Cai R, Wang G, Shu X, Pang W. Long noncoding RNA: multiple players in gene expression. BMB reports. 2018;51:280-9
6. Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging. 2014;6:992-1009
7. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome biology. 2017;18:206
8. Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends in genetics: TIG. 2018;34:142-57
9. Zhang X, Hong R, Chen W, Xu M, Wang L. The role of long noncoding RNA in major human disease. Bioorganic chemistry. 2019;92:103214
10. Han X, Mo J, Yang Y, Wang Y, Lu H. Crucial Roles of LncRNAs-Mediated Autophagy in Breast Cancer. Int J Med Sci. 2022;19:1082-92
11. Suwal A, Hao JL, Liu XF, Zhou DD, Pant OP, Gao Y. et al. NONRATT021972 long-noncoding RNA: A promising lncRNA in diabetes-related diseases. Int J Med Sci. 2019;16:902-8
12. Zhu K, Gong Z, Li P, Jiang X, Zeng Z, Xiong W. et al. A review of linc00673 as a novel lncRNA for tumor regulation. Int J Med Sci. 2021;18:398-405
13. Abdulle LE, Hao JL, Pant OP, Liu XF, Zhou DD, Gao Y. et al. MALAT1 as a Diagnostic and Therapeutic Target in Diabetes-Related Complications: A Promising Long-Noncoding RNA. Int J Med Sci. 2019;16:548-55
14. Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics, proteomics & bioinformatics. 2016;14:42-54
15. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. The international journal of biochemistry & cell biology. 2013;45:1895-910
16. Sun W, Yang Y, Xu C, Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer genetics. 2017;216-217:105-10
17. Golla U, Sesham K, Dallavalasa S, Manda NK, Unnam S, Sanapala AK. et al. ABHD11-AS1: An Emerging Long Non-Coding RNA (lncRNA) with Clinical Significance in Human Malignancies. Non-coding RNA. 2022;8:21
18. Liu FT, Zhu PQ, Ou YX, Lin QS, Qiu C, Luo HL. Long non-coding RNA-LET can indicate metastasis and a poor prognosis: a meta-analysis. Minerva medica. 2016;107:101-7
19. Wang S, Chen Z, Gu J, Chen X, Wang Z. The Role of lncRNA PCAT6 in Cancers. Frontiers in oncology. 2021;11:701495
20. Liu FT, Zhu PQ, Luo HL, Zhang Y, Qiu C. Prognostic value of long non-coding RNA UCA1 in human solid tumors. Oncotarget. 2016;7:57991-8000
21. Hussen BM, Azimi T, Abak A, Hidayat HJ, Taheri M, Ghafouri-Fard S. Role of lncRNA BANCR in Human Cancers: An Updated Review. Frontiers in cell and developmental biology. 2021;9:689992
22. Liu FT, Zhu PQ, Luo HL, Zhang Y, Hao TF, Xia GF. et al. Long noncoding RNA ANRIL: a potential novel prognostic marker in cancer: a meta-analysis. Minerva medica. 2016;107:77-83
23. Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X. et al. Global identification of human transcribed sequences with genome tiling arrays. Science (New York, NY). 2004;306:2242-6
24. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N. et al. The transcriptional landscape of the mammalian genome. Science (New York, NY). 2005;309:1559-63
25. Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y. et al. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Frontiers in oncology. 2020;10:598817
26. Tao S, Hou Y, Diao L, Hu Y, Xu W, Xie S. et al. Long noncoding RNA study: Genome-wide approaches. Genes & diseases. 2023;10:2491-510
27. Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA biology. 2013;10:925-33
28. Liu B, Xiang W, Liu J, Tang J, Wang J, Liu B. et al. The regulatory role of antisense lncRNAs in cancer. Cancer cell international. 2021;21:459
29. Mosca N, Russo A, Potenza N. Making Sense of Antisense lncRNAs in Hepatocellular Carcinoma. International journal of molecular sciences. 2023;24:8886
30. Jiang B, Yuan Y, Yi T, Dang W. The Roles of Antisense Long Noncoding RNAs in Tumorigenesis and Development through Cis-Regulation of Neighbouring Genes. Biomolecules. 2023;13:684
31. Zhao S, Zhang X, Chen S, Zhang S. Natural antisense transcripts in the biological hallmarks of cancer: powerful regulators hidden in the dark. Journal of experimental & clinical cancer research: CR. 2020;39:187
32. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nature reviews Molecular cell biology. 2021;22:96-118
33. Chen Y, Wu Q, Lin J, Wei J. DARS-AS1 accelerates the proliferation of cervical cancer cells via miR-628-5p/JAG1 axis to activate Notch pathway. Cancer cell international. 2020;20:535
34. Dou B, Jiang Z, Chen X, Wang C, Wu J, An J. et al. Oncogenic Long Noncoding RNA DARS-AS1 in Childhood Acute Myeloid Leukemia by Binding to microRNA-425. Technology in cancer research & treatment. 2020;19:1533033820965580
35. Feng Y, Wei G, Zhang L, Zhou H, Wang W, Guo P. et al. LncRNA DARS-AS1 aggravates the growth and metastasis of hepatocellular carcinoma via regulating the miR-3200-5p-Cytoskeleton associated protein 2 (CKAP2) axis. Bioengineered. 2021;12:8217-32
36. Jiao M, Guo H, Chen Y, Li L, Zhang L. DARS-AS1 promotes clear cell renal cell carcinoma by sequestering miR-194-5p to up-regulate DARS. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;128:110323
37. Kong X, Wang JS, Yang H. Upregulation of lncRNA DARS-AS1 accelerates tumor malignancy in cervical cancer by activating cGMP-PKG pathway. Journal of biochemical and molecular toxicology. 2021;35:1-11
38. Li J, Gao H, Chen B, Li L, Wang Q, Gao Z. lncRNA DARS-AS1 Modulates TSPAN1-Mediated ITGA2 Hypomethylation by Interaction with miR-194-5p Thus Promoting Ovarian Cancer Progression. Stem cells international. 2022;2022:4041550
39. Liu D, Liu H, Jiang Z, Chen M, Gao S. Long non-coding RNA DARS-AS1 promotes tumorigenesis of non-small cell lung cancer via targeting miR-532-3p. Minerva medica. 2021;112:408-9
40. Liu X, Zhang G, Yu T, He J, Liu J, Chai X. et al. Exosomes deliver lncRNA DARS-AS1 siRNA to inhibit chronic unpredictable mild stress-induced TNBC metastasis. Cancer letters. 2022;543:215781
41. Liu X, Zhang G, Yu T, Liu J, Chai X, Yin D. et al. CL4-modified exosomes deliver lncRNA DARS-AS1 siRNA to suppress triple-negative breast cancer progression and attenuate doxorubicin resistance by inhibiting autophagy. International journal of biological macromolecules. 2023;250:126147
42. Liu Y, Liang L, Ji L, Zhang F, Chen D, Duan S. et al. Potentiated lung adenocarcinoma (LUAD) cell growth, migration and invasion by lncRNA DARS-AS1 via miR-188-5p/ KLF12 axis. Aging. 2021;13:23376-92
43. Shen W, Zhu M, Wang Q, Zhou X, Wang J, Wang T. et al. DARS-AS1 recruits METTL3/METTL14 to bind and enhance DARS mRNA m(6)A modification and translation for cytoprotective autophagy in cervical cancer. RNA biology. 2022;19:751-63
44. Tong J, Xu X, Zhang Z, Ma C, Xiang R, Liu J. et al. Hypoxia-induced long non-coding RNA DARS-AS1 regulates RBM39 stability to promote myeloma malignancy. Haematologica. 2020;105:1630-40
45. Xue Y, Liu H, Nie G, Ren X. lncRNA DARS-AS1 Promoted Osteosarcoma Progression through Regulating miR-532-3p/CCR7. Disease markers. 2022;2022:4660217
46. Yang L, Lin K, Zhu L, Wang H, Teng S, Huang L. et al. Long non-coding RNA DARS-AS1 promotes tumor progression by directly suppressing PACT-mediated cellular stress. Communications biology. 2022;5:822
47. Zheng C, Wei Y, Zhang Q, Sun M, Wang Y, Hou J. et al. Multiomics analyses reveal DARS1-AS1/YBX1-controlled posttranscriptional circuits promoting glioblastoma tumorigenesis/radioresistance. Science advances. 2023;9:eadf3984
48. Zhou M, Cheng H, Fu Y, Zhang J. Long noncoding RNA DARS-AS1 regulates TP53 ubiquitination and affects ovarian cancer progression by modulation miR-194-5p/RBX1 axis. Journal of biochemical and molecular toxicology. 2021;35:e22865
49. Zhu J, Han S. DARS-AS1 Knockdown Inhibits the Growth of Cervical Cancer Cells via Downregulating HMGB1 via Sponging miR-188-5p. Technology in cancer research & treatment. 2020;19:1533033820971669
50. Zhu J, Han S. Downregulation of LncRNA DARS-AS1 Inhibits the Tumorigenesis of Cervical Cancer via Inhibition of IGF2BP3. OncoTargets and therapy. 2021;14:1331-40
51. Du C, Han X, Zhang Y, Guo F, Yuan H, Wang F. et al. DARS-AS1 modulates cell proliferation and migration of gastric cancer cells by regulating miR-330-3p/NAT10 axis. Open medicine (Warsaw, Poland). 2022;17:2036-45
52. Dai Q, Li J, Zhou K, Liang T. Competing endogenous RNA: A novel posttranscriptional regulatory dimension associated with the progression of cancer. Oncology letters. 2015;10:2683-90
53. Tehrani SS, Ebrahimi R, Al EAA, Panahi G, Meshkani R, Younesi S. et al. Competing Endogenous RNAs (CeRNAs): Novel Network in Neurological Disorders. Current medicinal chemistry. 2021;28:5983-6010
54. Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. Journal of hematology & oncology. 2015;8:30
55. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-52
56. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids research. 2014;42:D92-7
57. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
58. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. International journal of molecular sciences. 2019;20:5758
59. Yan P, Tang L, Liu L, Tu G. Identification of candidate RNA signatures in triple-negative breast cancer by the construction of a competing endogenous RNA network with integrative analyses of Gene Expression Omnibus and The Cancer Genome Atlas data. Oncology letters. 2020;19:1915-27
60. Zhang Y, Tian Y. Comprehensive analysis of lncRNA-mediated ceRNA regulatory networks and key genes associated with papillary thyroid cancer coexistent with Hashimoto's thyroiditis. BMC Endocr Disord. 2022;22:252
61. Xu J, Xu J, Liu X, Jiang J. The role of lncRNA-mediated ceRNA regulatory networks in pancreatic cancer. Cell death discovery. 2022;8:287
62. Li K, Yao T, Wang Z. lncRNA-mediated ceRNA network in bladder cancer. Non-coding RNA research. 2023;8:135-45
63. Mahdi Khanifar M, Zafari Z, Sheykhhasan M. Crosstalk between long non-coding RNAs and p53 signaling pathway in colorectal cancer: A review study. Pathology, research and practice. 2023;249:154756
64. Maharati A, Moghbeli M. Long non-coding RNAs as the critical regulators of PI3K/AKT, TGF-β, and MAPK signaling pathways during breast tumor progression. Journal of translational medicine. 2023;21:556
65. Hussain MS, Afzal O, Gupta G, Altamimi ASA, Almalki WH, Alzarea SI. et al. Long non-coding RNAs in lung cancer: Unraveling the molecular modulators of MAPK signaling. Pathology, research and practice. 2023;249:154738
66. Ashrafizadeh M, Mohan CD, Rangappa S, Zarrabi A, Hushmandi K, Kumar AP. et al. Noncoding RNAs as regulators of STAT3 pathway in gastrointestinal cancers: Roles in cancer progression and therapeutic response. Medicinal research reviews. 2023;43:1263-321
67. Almalki WH. LncRNAs and PTEN/PI3K signaling: A symphony of regulation in cancer biology. Pathology, research and practice. 2023;249:154764
68. Wang X, Zhang H, Chen X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resistance. 2019;2:141
69. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nature Reviews Cancer. 2013;13:714-26
70. Cabrera-Licona A, Pérez-Añorve IX, Flores-Fortis M, del Moral-Hernández O, González-de la Rosa CH, Suárez-Sánchez R. et al. Deciphering the epigenetic network in cancer radioresistance. Radiotherapy Oncology. 2021;159:48-59
71. Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nature Reviews Cancer. 2008;8:545-54
72. Zhang X, Xie K, Zhou H, Wu Y, Li C, Liu Y. et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Molecular cancer. 2020;19:1-26
73. Xiao J, He X. Involvement of non-coding RNAs in chemo-and radioresistance of nasopharyngeal carcinoma. Cancer Management Research. 2021:8781-94
74. Zhu J, Chen S, Yang B, Mao W, Yang X, Cai J. Molecular mechanisms of lncRNAs in regulating cancer cell radiosensitivity. Bioscience reports. 2019;39:BSR20190590
75. Chi H-C, Tsai C-Y, Tsai M-M, Yeh C-T, Lin K-H. Roles of long noncoding RNAs in recurrence and metastasis of radiotherapy-resistant cancer stem cells. International journal of molecular sciences. 2017;18:1903
76. Peng Y, Tang D, Zhao M, Kajiyama H, Kikkawa F, Kondo Y. Long non-coding RNA: a recently accentuated molecule in chemoresistance in cancer. Cancer metastasis reviews. 2020;39:825-35
77. Lin X, Kong D, Chen Z-S. Chemo-Radiation-Resistance in Cancer Therapy. Frontiers in pharmacology. 2022;13:904063
78. Schneeweiss A, Michel LL, Möbus V, Tesch H, Klare P, Hahnen E. et al. Survival analysis of the randomised phase III GeparOcto trial comparing neoadjuvant chemotherapy of intense dose-dense epirubicin, paclitaxel, cyclophosphamide versus weekly paclitaxel, liposomal doxorubicin (plus carboplatin in triple-negative breast cancer) for patients with high-risk early breast cancer. European journal of cancer (Oxford, England: 1990). 2022;160:100-11
79. Shepherd JH, Ballman K, Polley MC, Campbell JD, Fan C, Selitsky S. et al. CALGB 40603 (Alliance): Long-Term Outcomes and Genomic Correlates of Response and Survival After Neoadjuvant Chemotherapy With or Without Carboplatin and Bevacizumab in Triple-Negative Breast Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2022;40:1323-34
80. Symonds L, Jenkins I, Linden HM, Kurland B, Gralow JR, Gadi VVK. et al. A Phase II Study Evaluating the Safety and Efficacy of Sunitinib Malate in Combination With Weekly Paclitaxel Followed by Doxorubicin and Daily Oral Cyclophosphamide Plus G-CSF as Neoadjuvant Chemotherapy for Locally Advanced or Inflammatory Breast Cancer. Clinical breast cancer. 2022;22:32-42
Author contact
![]() Corresponding authors: Hongliang Luo, email: ndefy13028edu.cn, Tel: +86-13097280001; Yang Wang, email: iggsnrtcom, Tel: +86-18070390681.
Corresponding authors: Hongliang Luo, email: ndefy13028edu.cn, Tel: +86-13097280001; Yang Wang, email: iggsnrtcom, Tel: +86-18070390681.

 Global reach, higher impact
Global reach, higher impact