Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(12):1304-1311. doi:10.7150/ijms.25580 This issue Cite
Research Paper
The Role of YB1 in Renal Cell Carcinoma Cell Adhesion
1. Department of Urology, The Second Hospital of Tianjin Medical University, Tianjin Institute of Urology and Department of Microbiology, School of Medical Laboratory, Tianjin Medical University, Tianjin 300070, China
2. Department of Clinical Laboratory, Tianjin Medical University General Hospital, Tianjin Medical University, Tianjin 300052, China
3. Research Center of Molecular Biology, Inner Mongolia Medical University, Hohhot 010059, China
4. Department of Genetics, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China
5. School of Microelectronics, Tianjin University, Tianjin 300072, China
# These authors contribute equally to this work.
Received 2018-2-16; Accepted 2018-6-28; Published 2018-8-6
Abstract
Background: Y-box binding protein 1 (YB1) is a multifunctional protein involved in many processes related to cancer progression and metastasis.
Methods: In this study, we constructed YB1 knockdown stable renal cell carcinoma (RCC) cell line 786-0. The gene expression profile of 786-0 was performed by DNA microarray analysis to identify genes that were regulated by YB1. Real-time PCR and western blotting were used to test the genes and proteins expression. Transforming growth factor-β (TGF-β) activity was detected by dual-luciferase reporter assay. Cell adhesion assay was used to determine RCC cell adhesion ability.
Results: Pathway analysis revealed that YB1 knockdown influenced cell adhesion molecules (CAMs). We further verified four genes (CLDN4, NRXN3, ITGB8, and VCAN) related to CAMs by real-time PCR, and confirmed that YB1 regulated the expression of ITGB8 in RCC. Functional assays demonstrated that knockdown of YB1 significantly inhibited the cell adhesion of 786-0 cells in vitro. In addition, YB1 affected TGF-β activation.
Conclusion: Our study demonstrated that YB1 modulated the adhesion ability of renal cell carcinoma cells by regulating ITGB8 and TGF-β.
Keywords: cell adhesion, ITGB8, renal cell carcinoma, YB1
Introduction
Renal cell carcinoma (RCC) is highly malignant neoplasms and originated from the proximal tubular epithelial cells [1]. About 20%-30% of patients have occurred metastasis when they are first diagnosed [2]. Furthermore, RCC is insensitive to conventional radiotherapy and chemotherapy. Recently, surgical intervention followed by immunotherapy is emerging as a therapeutic option for RCC with metastasis [3]. However, it is still failed to demonstrate a clear benefit of these therapeutic strategies for the metastasis RCC patients. Therefore, the understanding of underlying molecular mechanisms of RCC progression is urgent.
Our previous studies on RCC have shown that the elevated nuclear Y-box binding protein 1 (YB1) expression is closely related to the tumor growth and aggressive cancer phenotype, leading to poor prognosis of RCC patients [4-6]. Actually, YB1 is a DNA/RNA-binding protein that can bind to the targets promoter in which contains an inverted CCAAT box (Y-box sequence) and regulates transcription and translation [7]. The cold shock domain (CSD) of YB1 binds to both DNA and RNA and involves in numerous biological processes such as cell proliferation, RNA splicing, DNA transcription and translation, DNA repair, drug resistance, stress response to extracellular signals, etc [8]. In addition to RCC, abnormally expression of YB1 is also frequently detected in different kinds of cancers, including prostate cancer [9], ovarian cancer [10], and breast cancer [11]. YB1 is closely related to the progression and prognosis of these cancers [12]. Mechanistically, YB1 indirectly enhances TGF-β signaling cascades via Smad2 phospho-activation in breast cancer [13]. YB1 has also been shown to regulate AR-V7 expression, and YB1 inhibition augmented the anticancer effect of enzalutamide [14]. However, the molecular mechanisms of how YB1 contributes to RCC progression still remain unclear.
In this study, to better understand the oncogenic potential of YB1, we performed gene expression profiles analysis following YB1 knockdown. Through validation of the microarray data, we confirmed that YB1 regulated the expression of ITGB8 in RCC.
Materials and Methods
Cell culture
Renal cell carcinoma cell line 786-0 was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). 786-0 cells were cultured with DMEM (Corning, NY, USA) supplemented with 10% fetal bovine serum (Biological Industries, Kibbutz Beit Haemek, Israel) and 1× penicillin/streptomycin (Gibco, NY, USA) at 37 ℃ in the presence of 5% CO2.
Plasmid construction and cell transfection
The pLKO.1-scr and pLKO.1-shYB1 recombinant lentiviruses were produced by co-transfection of HEK 293T cells with the shRNA lentivirus expression plasmids, lentivirus psAX2 packaging plasmid and pMD2G envelope plasmid for 48 h using lipofectamine2000 (Invitrogen, Carlsbad, CA, USA). The shRNA sequences targeting YB1 were sense 5'-CCGGCACCAGACTGACTGCCATAAAGACTCGAGTCTTTATGGCAGTCTTTATGGCAGTCTGGTGTTTTTG-3' and anti-sense 5'-AATTCAAAAACACCACCAGACTGCCATAAAGACTCGAGTCTTTATGGCAGTCTGGTG-3'. Virus particles were harvested 48 h after transfection. 786-0 cells were infected with recombinant lentivirus-transducing units plus 5 μg/ml polybrene. Two days after infection, cells were treated with 2 μg/ml puromycin (Sangon Biotech, Shanghai, Co., LTD) to select cells stably expressing the shRNAs.
Microarray
Triplicate samples of YB1 shRNA knockdown and respective control transfected 786-0 cells were extracted for RNA and prepared for microarray profiling. Samples were sent to Jingtai Bio-tech company (Shanghai, China) for mRNA isolation, quality control, chip hybridization, and microarray data analysis, the samples were purified according to the manufacture's instructions (QIAGEN, Valencia, CA), cDNA was synthesized with SuperScript II (Invitrogen), and then purified with RNeasy Mini Kit (QIAGEN). Labeled with biotin and hybridized at 45 ℃ for 16 h to Afymetrix GeneChip Human Gene 1.0 ST arrays (Afymetrix, Santa Clara, CA, USA). All arrays were washed and scanned using a GeneChip Scanner 3000 (Afymetrix) at correct pixel value (3 µm) and wavelength (570 nm), and data were collected and analysised. Genes expressed differentially with at least 2-fold change with p < 0.05 in either direction were considered as up or down regulated.
Quantitative real-time PCR
Total RNA were extracted from 786-0 using Trizol reagent (Ambion, Austin, TX, USA) and reversely transcribed cDNA using FastQuant RT Kit (TIANGEN, Beijing, China) according to the manufacturer's protocols. RNA and cDNA concentration and purity were measured using a NanoDrop 2000c (Thermo Fisher Scientifc, Waltham, MA, USA). Quantitative real-time PCR reactions were performed using the 7500-fast PCR Systems (Applied Biosystems, Foster City, CA). The primer sequences used for PCR were listed in Table 1. The following PCR parameters were used for each primer set: denaturing at 95 ℃ for 15 min, followed by 45 cycles of 94 ℃ for 15 s, annealing temperature of 56 ℃ for 30 s and extension at 72 ℃ for 30 s. Assay performance was validated by assessing amplification efficiencies by means of calibration curves, and ensuring that the plot of log input amount versus ∆Cq has a slope < |0.1|. At least three separated experiments were performed and each sample was assayed in triplicate. A mean value of the triplicates was used for the determination of relative mRNA levels by the comparative Cq method with GAPDH as the reference gene and using the formula 2-∆∆Cq.
Western blotting
Cells were harvested and lysed in SDS lysis buffer with added 1×protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany), and the concentration of total protein was determined by a BCA protein assay kit (Thermo). Equal amounts of each protein were subjected to SDS-PAGE and transferred to a PVDF membrane (Millipore, Bedford, MA, USA). After blocking with 5% skim milk (Transduction Laboratories, BD Biosciences, San Jose, CA, USA), the membrane was incubated with a specific primary antibodies β-actin (Affinity Biosciences, Cincinnati, OH, USA, T0022), YB1 (Santa Cruz Biotechnology, CA, sc-101198), or ITGB8 (Affinity, DF2545), at 4 °C overnight. Then the membrane was washed with TBS containing 0.1% Tween-20 and labeled with horse radish peroxidase conjugated, secondary anti-rabbit (Affinity, S0001) or -mouse antibody (Affinity, S0002). Finally, the membrane was developed by ECL Western Blotting Detection Reagents (Millipore).
Primer sequences.
| Primer name | Sequence 5′to 3′ |
|---|---|
| YB1 | Forward GGGTGCAGGAGAACAAGGTA |
| Reverse TCTTCATTGCCGTCCTCTCT | |
| GAPDH | Forward TGCACCACCAACTGCTTAGC |
| Reverse GGCATGGACTGTGGTCATGAG | |
| CLDN4 | Forward ATGCAGTGCAAGGTGTACGA |
| Reverse CTTTCATCCTCCAGGCAGTT | |
| ITGB8 | Forward TGTGTGCTGGGCATGGAGAGTGT |
| Reverse CAGTGCTGGGCTGCTGCTGAA | |
| NRXN3 | Forward TGCTGAATGTTCAAGTGATGATG |
| Reverse GTGCTTTGTAGCCACCTTCG | |
| VCAN | Forward GTAACCCATGCGCTACATAAAGT |
| Reverse GGCAAAGTAGGCATCGTTGAAA |
Cell adhesion Assay
Fibronectin (10 μg/ml, Sigma-Aldrich, St. Louis, MO, USA) was used to coat 35 mm culture dish, overnight at 4 ℃, washed 3 times with ddH2O. Cells were resuspended in serum-free medium (1×105/ml) and then plated in 35 mm dishes. After 5 min, 15 min and 30 min of incubation, the non-adherent cell containing media was aspirated off and each well was washed gently with cold PBS. Cells were fixed with 4% PFA (Solarbio, Beijing, China), then stained with crystal violet and counted under a microscope (Olympus, Tokyo, Japan) with five random fields.
Conventional cell adhesion assay
96-well plates were coated with Fibronectin at 4 ℃ overnight and then blocked with 2% BSA (Solarbio, Beijing, China) for 1 h at 37 ℃ before seeding cells. Cells were seeded at a density of 2×105/ml and the reactions were blocked with cold PBS at 15 min and 30 min respectively. Non-adherent cells were removed by washing with PBS, and adherent cells were determined by Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan). Absorbance was measured at 450 nm using a microplate spectrophotometer.
Dual-luciferase reporter assay
786-0 cells were co-transfected with p3TP or pSBE4 reporter plasmid and the internal control Renilla reporter plasmid (pRL-TK) using Lipofectamine2000 in 48-well plates. After 48 h, cells were washed with PBS and cellular proteins were lysed by 1× passive lysis buffer (PLB). Then the cellular proteins were analyzed for luciferase activity according to the standard protocol (Dual-luciferase Reporter Assay System Promega, Madison, WI, USA).
Statistical Analysis
Data are presented as mean ± standard deviations. Differences in mean values between two groups were analyzed by t-test. One-way ANOVA test was used to analyze the differences between parent, negative control, and YB1 knockdown groups (SPSS 17.0 software). Differences with *p < 0.05 were considered as statistically significant.
Results
Differential gene expression after YB1 knockdown in 786-0 cells
In an effort to characterize the function of YB1 in renal cell carcinoma, 786-0 cells were transfected with lentivirus-mediated YB1 knockdown. Real-time PCR and western blotting were applied to determine the expression of YB1 in 786-0, 786-0-scr and 786-0-shYB1 cells. The mRNA and protein levels were significantly decreased after YB1 knockdown compared with parent cell and negative controls (Figure 1A and 1B). We thus employed these cells to examine the effect on other genes expression.
Total RNA extracted from 786-0-scr and 786-0-shYB1 cells were subjected to microarray analysis. After normalizing the gene expression, we assessed the profile of gene expression in 786-0-scr and 786-0-shYB1 stable cells and set the threshold of differential expression at 2-fold and obtained a number of genes related to the YB1 expression. From the microarray data analysis, 196 genes were significantly down-regulated and 198 genes were up-regulated in 786-0-shYB1 cells compared with 786-0-scr cells with a 2-fold change (Figure 1C). Top ten significantly down-regulated and up-regulated genes were listed at Table 2. The volcano plot showed the distribution of differentially expressed genes according to fold-change and significance (Figure 1D). The horizontal grey line represented the p value cut-off (0.05), and the vertical grey line indicated the fold change cut-off. We further used pathway analysis to determine the pathway in which the differentially expressed genes involved. Pathway analysis based on the KEGG pathway database clearly revealed that YB1 knockdown affected many pathways. The down-regulated pathways focused on cell adhesion molecules (CAMs), axon guidance, sphingolipid metabolism, chemical carcinogenesis, and tryptophan metabolism (Figure 1E). CAMs are a group of transmembrane glycoproteins located on the cell surface, which mediates the cell-cell and cell-extracellular matrix adhesion [15]. The CAMs was the top modulated canonical pathway following YB1 knockdown. Therefore, we chose to further investigate the function of YB1 in cell adhesion.
Screening differentially expressed genes after YB1 knockdown. A. YB1 mRNA expression levels were detected by real-time PCR after lentivirus transfection. B. Confirmation of YB1 knockdown efficiency by western blotting: the level of YB1 protein was significantly decreased in 786-0-shYB1 cells compared with control cells. C. Clustering heat-map showing the significantly affected genes in 786-0 cells after YB1 knockdown. Red represents upregulated genes, while green represents downregulated genes. D. Volcano plot showing the differentially expressed genes between the experimental and control groups. Each dot represents one gene. Genes up-regulated with more than 2 fold change with a p<0.05 are depicted in red dot and those down-regulated with identical fold change and p-value are in green dot. E. Pathway analysis. The pathways that were down-regulated in the 786-0-shYB1 cells compared with the control cells. F. The cell adhesion molecules pathway diagram derived from KEGG pathway analysis. The blue color represents the genes were down-regulated on the microarray data. (** indicate p < 0.01, NS: no significant difference)
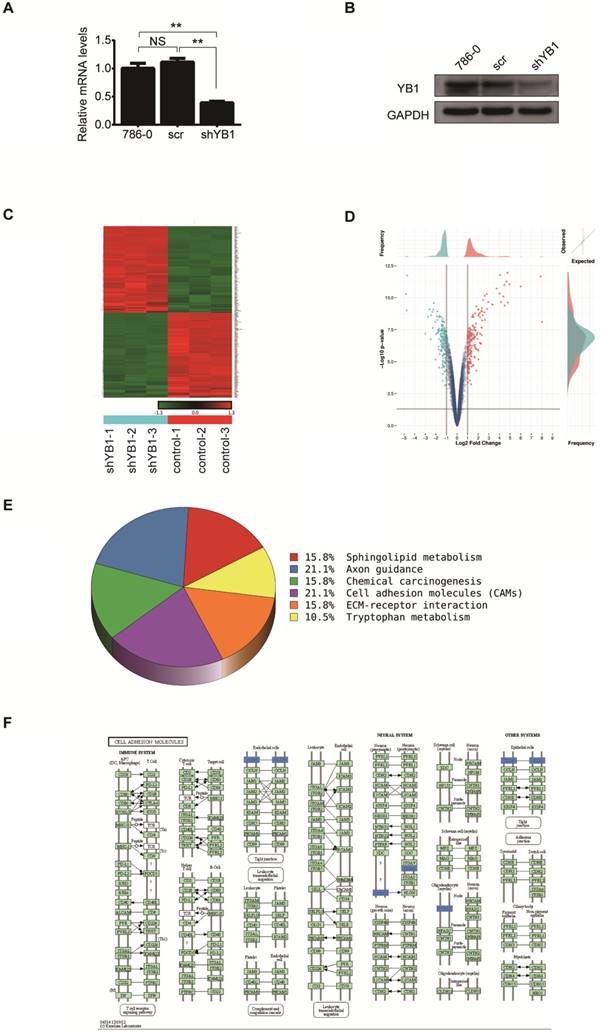
The down-regulated and up-regulated genes after YB1 knockdown
| down-regulated genes | up-regulated genes | ||||
|---|---|---|---|---|---|
| Gene name | Fold Change | FDR | Gene name | Fold Change | FDR |
| SHISA2 | -28.00 | 2.00E-07 | IGF2 | 30.88 | 6.60E-08 |
| HSPA1A | -26.88 | 1.51E-06 | IL8 | 27.46 | 5.98E-07 |
| SEMA6A | -25.79 | 2.48E-08 | CXCL1 | 27.28 | 2.48E-08 |
| CXADR | -7.90 | 2.25E-05 | IGF2 | 25.04 | 4.96E-08 |
| VAV3 | -6.59 | 2.91E-06 | IL8 | 22.44 | 8.52E-07 |
| ITGB8 | -5.47 | 2.58E-06 | C3 | 20.76 | 2.63E-07 |
| FXYD2 | -5.01 | 1.68E-06 | CXCL1 | 17.66 | 5.93E-07 |
| STMN3 | -4.69 | 1.38E-06 | CXCL2 | 16.62 | 2.48E-08 |
| CXADR | -4.59 | 6.25E-07 | CSF2 | 13.25 | 9.70E-08 |
| GMFB | -4.17 | 5.82E-06 | KYNU | 10.10 | 7.49E-07 |
Differentially expressed genes were enriched in top 6 pathways.
| Pathway ID | Definition | Genes |
|---|---|---|
| hsa00600 | Sphingolipid metabolism | GAL3ST1//KDSR//SGMS1 |
| hsa04360 | Axon guidance | EPHB1//SEMA5A//SEMA6A//SEMA6D |
| hsa05204 | Chemical carcinogenesis | ARNT//CYP1B1//GSTP1 |
| hsa04514 | Cell adhesion molecules (CAMs) | CLDN4//ITGB8//NRXN3//VCAN |
| hsa04512 | ECM-receptor interaction | ITGB8//RELN//SPP1 |
| hsa00380 | Tryptophan metabolism | CYP1B1//KMO |
YB1 regulated ITGB8 expression
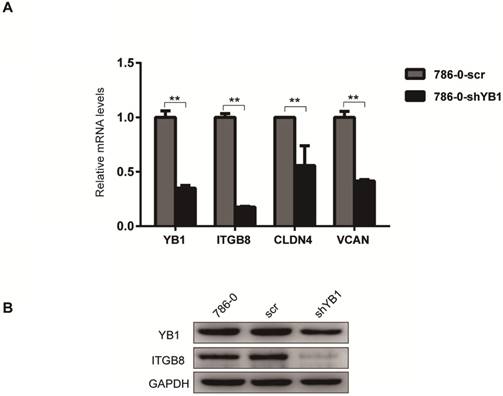
To further confirm the reliability of the microarray data, four significantly different expressed genes related to CAMs (Table 3), including ITGB8, CLDN4, VCAN, and NRXN3 were verified by real-time PCR analysis. The results showed that ITGB8, CLDN4 and VCAN were down-regulated after YB1 knockdown (Figure 2A). Thus, we focused on ITGB8 since its expression was the most significantly down-regulated in the 786-0-shYB1 cells. Western blotting showed that YB1 knockdown also resulted in a reduction of ITGB8 protein expression (Figure 2B), confirming the results seen at the gene level.
ITGB8 was significantly down-regulated in 786-0-shYB1 cells. A. The reliability of the microarray data was further verified by real-time PCR analysis. B. Western blotting analysis showing ITGB8 and YB1 protein levels in 786-0-shYB1 and control cells. (** indicate p < 0.01)
YB1 mediated RCC adhesion
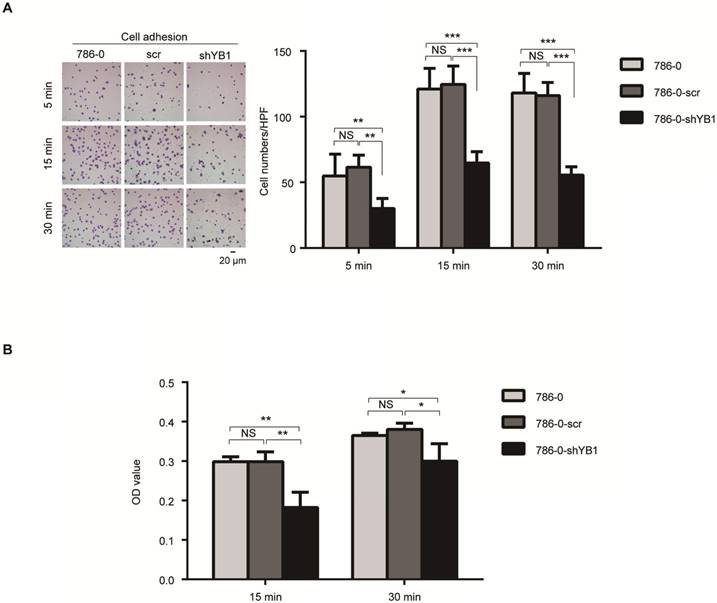
Integrins have been known to play an important role in cancer cell adhesion, a key step during metastasis [16]. These findings prompted us to test the role of YB1 in RCC-mediated adhesion. Fibronectin, the basement membrane components, plays a pivotal role in cell adhesion. We took advantage of this property to perform two kinds of adhesion assays. After knockdown of YB1, the adhesion ability of 786-0-shYB1 cells decreased obviously when compared with 786-0 and 786-0-scr cells (Figure 3A). Conventional cell adhesion assay was also used to confirm this finding. Consistently, knockdown of YB1 inhibited the 786-0 cells adhesion (Figure 3B).
YB1 regulated ITGB8 to affect the RCC cell adhesion
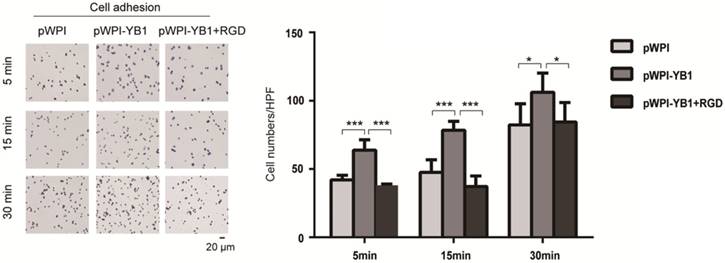
The ITGB8 is a cell surface receptor for the latent domain of the TGF-β [17]. Through non-covalent combination with latency-associated peptide (LAP), TGF-β is maintained in a latent form that must be activated to function [18]. ITGB8 has been shown to interact with Arg-Gly-Asp (RGD) sequence of LAP and then recruitment of a metalloprotease, which causes leavage of LAP and release of the active (mature) TGF-β peptide [19].
In order to investigate the role of ITGB8 in YB1 regulated RCC cell adhesion, we treated 786-0-YB1 cells with RGD peptide to mimic the disruption of ITGB8 and LAP. The results showed that the YB1 induced cancer cell adhesion ability was inhibited when RGD peptide was added (Figure 4). Thus, these results indicated that YB1 regulated RCC cell adhesion by modulating ITGB8.
The effect of YB1 on ITGB8 downstream signaling molecule TGF-β
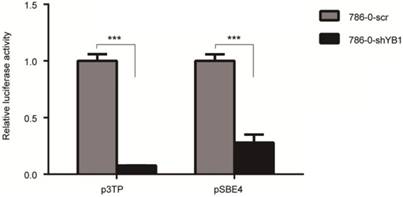
To examine the effect of YB1 on ITGB8 downstream signaling molecule TGF-β, we performed the luciferase assay to determine the potential impact of YB1 on TGF-β-responsive promoter, p3TP activation. As shown in Figure 5, YB1 knockdown could decrease the TGF-β transactivation. Then, the pSBE4 reporter plasmid was used to further explore the involvement of Smads in TGF-β responsiveness. Consistently, the results showed that pSBE4 transcriptional activity were restrained in 786-0-shYB1 cells. In conclusion, TGF-β activity was regulated by YB1.
YB1 regulated renal cell adhesion. A. Knockdown of YB1 decreased cell adhesion ability of 786-0 cells. Left: Representative microscopic photographs of the cells. Right: Quantization of cell migration. B. Another adhesion assay was measured by determined cell counts that adhered to BD-coated 96-well plates after washing with PBS by CCK8 assay. Bars indicate SD. (* indicate p < 0.05, ** indicate p < 0.01, *** indicate p < 0.001, NS: no significant difference,)
YB1 regulated ITGB8 to affect the RCC cell adhesion. Increased adhesion capability of 786-0-YB1 cells was rescued by adding RGD. The RGD peptide (Dalian Meilun Biotechnology Co. LTD) was dissolved in ddH2O at 10 mg/ml. 786-0-YB1 cells were incubated with 0.1 mg/ml RGD for 24 h before harvested. (* indicate p < 0.05, *** indicate p < 0.001)
YB1 knockdown decreased the TGF-β activation. 786-0-shYB1 and control cells were transfected with Renilla and p3TP-luci/pSBE4-luci reporter plasmid. Luciferase activities were measured. (*** indicate p < 0.001)
Discussion
YB1, a 36 kDa multifunctional protein, plays a fundamental role in tumorigenesis, progression, and drug resistance [4, 7-14, 20-22]. As the main component of messenger ribonucleoproteins (mRNPs), cytosolic YB1 can regulate mRNA translation and stability [23]. More recently, studies show that YB1 plays diverse pro-oncogenic roles in tumors. For example, YB1 was involved in cell migration, invasion, and promoted epithelial-to-mesenchymal transition (EMT) in human breast cancer cell lines [11]. Furthermore, in human bladder cancer, YB1 was able to regulate the expression of c-Myc and HIF1α to enhance glycolysis, and therefore promoted tumor growth and inhibit apoptosis [24]. YB1 promoted lung cancer stem-like properties and metastasis both in vitro and in vivo [25]. Our previous study [5, 6] showed that expression of YB1 in renal cell carcinoma tissues were higher compared with para-carcinoma tissues and nuclear expression of YB1 was correlated with RCC stage, Fuhrman tumor grade and metastasis. These findings promote us to further test the biological function of YB1 in RCC.
Microarray analysis in RCC 786-0 cells identified a number of genes regulated by YB1, which could help to discover new signaling pathways or molecular mechanisms of tumorigenesis in RCC. At the same time, subsequent pathway analysis found that the most of down-regulated genes were enriched in CAMs, axon guidance, ECM-receptor interaction, chemical carcinogenesis, sphingolipid metabolism, and tryptophan metabolism. Cancer cell adhesion is considered to be the common tumor cellular development processes that including migration of tumor cells within the primary site, degradation of the ECM, invasion into the surrounding blood vessels and lymphatics, adherence to vascular endothelial cells at secondary sites [26]. So cancer cell adhesion is of importance in tumor metastasis. Both the clinic data in our previous study and the pathway analysis in microarray supported that YB1 might participate in RCC metastasis. Focused on the CAMs pathway, we uncovered a significantly down-regulated gene ITGB8 after YB1 knockdown. In addition, ITGB8 was also involved in ECM-receptor interaction pathway. As a CAMs component, ITGB8 is most abundantly expressed in the kidney, brain, and placenta [27]. Kidney ITGB8 was located in mesangial cells, which maintained glomerular capillary morphology and repaired after mesangial injury [28]. Sujata Lakhe-Reddy [29] also found that ITGB8 regulated adhesion to the mesangial matrix and glomerular capillary architecture. In glioblastoma cells, complex formation of ITGB8 association with Rho-GDP dissociation inhibitor 1 (RhoGDI1) was critical for regulating Rho GTPase activation and promoting GBM cell invasion [30]. In the present study, knockdown of YB1 inhibited RCC cell adhesion. Consistently, our previous studies showed that YB1 postively regulated RCC tumor metastasis using xenograft model in vivo [4]. Taken together, YB1 is an important molecule in modulating RCC cell adhesion, which may be mediated by ITGB8.
TGF-β pathway is known to engage in a number of biological processes, including cell proliferation, differentiation, migration, and epithelial mesenchymal transition (EMT) in various cancer cells [31]. In RCC cells, TGF-β signaling inhibition attenuated the invasive capacity [32]. Importantly, ITGB8 have been shown to regulate the TGF-β signaling by directly regulate the liberation of active TGF-β from LAP complex. In our current study, we showed that ITGB8 was the downstream target of YB1. Bin Ha [33] found that YB1 was involved in the TGF-β-induced EMT and cell migration. This finding promoted us to investigate the role of YB1 in regulating TGF-β signaling pathway. Of note, our results demonstrated that YB1 could regulate TGF-β activity, supporting that YB1 functions through ITGB8/TGF-β signaling and eventually contributes to RCC cell adhesion.
In a word, the results of this study reveal YB1/ITGB8/TGF-β pathway may act as a novel role in regulating RCC cell adhesion. Indeed, targeting the molecules of YB1/ITGB8/TGF-β axis provides a novel and potential treatment strategy for RCC.
Abbreviations
YB1: Y-box binding protein 1
RCC: renal cell carcinoma
TGF-β: transforming growth factor-β
CAMs: cell adhesion molecules
EMT: epithelial-to-mesenchymal transition
ITGB8: integrin beta 8
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant numbers 81772945, 81402094 and 21575103], the Scientific Research Foundation for the Returned Overseas Chinese scholars, Burea of personnel of China, Tianjin [grant number 2016015], Tianjin Natural Science Foundation [grant number 18JCYBJC26700], National Training Program of innovation and Entrepreneurship for undergraduates [grant number 201710062013] and Youth Innovation Foundation of Inner Mongolia Medical University [grant number YKD2017QNCX034].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sircar K, Rao P, Jonasch E, Monzon FA, Tamboli P. Contemporary approach to diagnosis and classification of renal cell carcinoma with mixed histologic features. Chin J Cancer. 2013;32(6):303-311
2. Haddad H, Rini BI. Current Treatment Considerations in Metastatic Renal Cell Carcinoma. Curr Treat Options Oncol. 2012;13(2):212-229
3. De Meerleer G, Khoo V, Escudier B, Joniau S, Bossi A, Ost P, Briganti A, Fonteyne V, Van Vulpen M, Lumen N, Spahn M, Mareel M. Radiotherapy for renal-cell carcinoma. Lancet Oncol. 2014;15(4):e170-e177
4. Yue D, Wang Y, Sun Y, Niu Y, Chang C. C1QBP Regulates YBX1 to Suppress the Androgen Receptor (AR)-Enhanced RCC Cell Invasion. Neoplasia. 2017;19(2):135-144
5. Wang Y, Chen Y, Geng H, Qi C, Liu Y, Yue D. Overexpression of YB1 and EZH2 are associated with cancer metastasis and poor prognosis in renal cell carcinomas. Tumour Biol. 2015;36(9):7159-7166
6. Wang Y, Yue D, Xiao M, Qi C, Chen Y, Sun D, Zhang N, Chen R. C1QBP negatively regulates the activation of oncoprotein YBX1 in the renal cell carcinoma as revealed by interactomics analysis. J Proteome Res. 2015;14(2):804-813
7. Dolfini D, Mantovani R. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 2013;20(5):676-685
8. Prabhu L, Mundade R, Wang B, Wei H, Hartley AV. Critical role of phosphorylation of serine 165 of YB1 on the activation of NF-κB in colon cancer. Oncotarget. 2015;6(30):29396-29412
9. Giménez-Bonafé P, Fedoruk MN, Whitmore TG, Akbari M, Ralph JL, Ettinger S, Gleave ME, Nelson CC. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate. 2004;59(3):337-349
10. Cybulski M, Jarosz B, Nowakowski A, Jeleniewicz W, Kutarska E, Bednarek W, Stepulak A. Cyclin A correlates with YB1, progression and resistance to chemotherapy in human epithelial ovarian cancer. Anticancer Res. 2015;35(3):1715-1721
11. Castellana B, Aasen T, Moreno-Bueno G, Dunn SE, Ramón y Cajal S. Interplay between YB-1 and IL-6 promotes the metastatic phenotype in breast cancer cells. Oncotarget. 2015;6(35):38239-38256
12. Martin M, Hua L, Wang B, Wei H, Prabhu L, Hartley AV, Jiang G, Liu Y, Lu T. Novel Serine 176 Phosphorylation of YBX1 Activates NF-κB in Colon Cancer. J Biol Chem. 2017;292(8):3433-3444
13. Stope MB, Weiss M, Koensgen D, Popp SL, Joffroy C, Mustea A, Buck MB, Knabbe C. Y-box Binding Protein-1 Enhances Oncogenic Transforming Growth Factor β Signaling in Breast Cancer Cells via Triggering Phospho-Activation of Smad2. Anticancer Res. 2017;37(12):6745-6748
14. Shiota M, Fujimoto N, Imada K, Yokomizo A, Itsumi M, Takeuchi A, Kuruma H, Inokuchi J, Tatsugami K, Uchiumi T, Oda Y, Naito S. Potential Role for YB-1 in castration-resistant prostate cancer and resistance to Enzalutamide through the androgen receptor V7. J Natl Cancer Inst. 2016:108 (7). doi: 10.1093/jnci/djw005
15. Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32(3-4):643-671
16. Lee JG, Ahn JH, Jin Kim T, Ho Lee J, Choi JH. Mutant p53 promotes ovarian cancer cell adhesion to mesothelial cells via integrin β4 and Akt signals. Sci Rep. 2015;5:12642
17. Cambier S, Gline S, Mu D, Collins R, Araya J, Dolganov G, Einheber S, Boudreau N, Nishimura SL. Integrin alpha(v)beta8-mediated activation of transforming growth factor-beta by perivascular astrocytes: an angiogenic control switch. Am J Pathol. 2005;166(6):1883-1894
18. Kumar V, Maurya VK, Joshi A, Meeran SM, Jha RK. Integrin beta 8 (ITGB8) regulates embryo implantation potentially via controlling the activity of TGF-β1 in mice. Biol Reprod. 2015;92(4):109
19. Jennifer A. Markovics, Jun Araya, Stephanie Cambier, David Jablons, Arthur Hill, Paul J. Wolters, Stephen L. Transcription of the Transforming Growth Factor β Activating Integrin β8 Subunit Is Regulated by SP3, AP-1, and the p38 Pathway Nishimura.J Biol Chem. 2010;285(32):24695-24706
20. El-Naggar AM, Veinotte CJ, Cheng H, Grunewald TG, Negri GL, Somasekharan SP, Corkery DP, Tirode F, Mathers J, Khan D, Kyle AH, Baker JH, LePard NE, McKinney S, Hajee S, Bosiljcic M, Leprivier G, Tognon CE, Minchinton AI, Bennewith KL, Delattre O, Wang Y, Dellaire G, Berman JN, Sorensen PH. Translational Activation of HIF1α by YB-1 Promotes Sarcoma Metastasis. Cancer Cell. 2015;27(5):682-697
21. Ha B, Lee EB, Cui J, Kim Y, Jang HH. YB-1 overexpression promotes a TGF-β1-induced epithelial-mesenchymal transition via Akt activation. Biochem Biophys Res Commun. 2015;458(2):347-351
22. Zhao S, Guo W, Li J, Yu W, Guo T, Deng W, Gu C. High expression of Y-box-binding protein 1 correlates with poor prognosis and early recurrence in patients with small invasive lung adenocarcinoma. Onco Targets Ther. 2016;9:2683-2692
23. Nekrasov MP, Ivshina MP, Chernov KG, Kovrigina EA, Evdokimova VM, Thomas AA, Hershey JW, Ovchinnikov LP. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J Biol Chem. 2003;278(16):13936-13943
24. Xu L, Li H, Wu L, Huang S. YBX1 promotes tumor growth by elevating glycolysis in human bladder cancer. Oncotarget. 2017;8(39):65946-65956
25. Guo T, Kong J, Liu Y, Li Z, Xia J, Zhang Y, Zhao S, Li F, Li J, Gu C. Transcriptional activation of NANOG by YBX1 promotes lung cancer stem-like properties and metastasis. Biochem Biophys Res Commun. 2017;487(1):153-159
26. Xin M, Dong XW, Guo XL. Role of the interaction between galectin-3 and cell adhesion molecules in cancer metastasis. Biomed Pharmacother. 2015;69:179-185
27. Moyle M, Napier MA, McLean JW. Cloning and expression of a divergent integrin subunit beta 8. J Biol Chem. 1991;266(29):19650-19658
28. Khan S, Lakhe-Reddy S, McCarty JH, Sorenson CM, Sheibani N, Reichardt LF, Kim JH, Wang B, Sedor JR, Schelling JR. Mesangial cell integrin alpha(v)beta(8) provides glomerular endothelial cell cytoprotection by sequestering TGF-β and regulating PECAM-1. Am J Pathol. 2011;178(2):609-620
29. Lakhe-Reddy S, Li V, Arnold TD, Khan S, Schelling JR. Mesangial cell alpha(v)beta(8)-integrin regulates glomerular capillary integrity and repair. Am J Physiol Renal Physiol. 2014;306(12):F1400-F1409
30. Reyes SB, Narayanan AS, Lee HS, Tchaicha JH, Aldape KD, Lang FF, Tolias KF, McCarty JH. alpha(v)beta(8) integrin interacts with RhoGDI1 to regulate Rac1 and Cdc42 activation and drive glioblastoma cell invasion. Mol Biol Cell. 2013;24(4):474-482
31. Li J, Qu Y, Li X, Li D, Zhao F, Mao M, Ferriero D, Mu D. The role of integrin alpha(v)beta(8) in neonatal hypoxic-ischemic brain injury. Neurotox Res. 2010;17(4):406-417
32. Boström AK, Lindgren D, Johansson ME, Axelson H. Effects of TGF-β signaling in clear cell renal cell carcinoma cells. Biochem Biophys Res Commun. 2013;435(1):126-133
33. Ha B, Lee EB, Cui J, Kim Y, Jang HH. YB-1 overexpression promotes a TGF-β1-induced epithelial-mesenchymal transition via Akt activation. Biochem Biophys Res Commun. 2015;458(2):347-351
Author contact
![]() Corresponding author: Dan Yue, Ph.D. Department of Microbiology, School of Medical Laboratory, Tianjin Medical University, Tianjin 300070, China. Email: yuedanedu.cn
Corresponding author: Dan Yue, Ph.D. Department of Microbiology, School of Medical Laboratory, Tianjin Medical University, Tianjin 300070, China. Email: yuedanedu.cn

 Global reach, higher impact
Global reach, higher impact