Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(3):217-222. doi:10.7150/ijms.22402 This issue Cite
Research Paper
Association of Genetic Variants of Small Non-Coding RNAs with Survival in Colorectal Cancer
1. Department of Pharmacy, Zhongxing Branch, Taipei City Hospital, Taipei, Taiwan
2. Department of Pharmacy, China Medical University, Taichung, Taiwan
3. Department of Colorectal Surgery, China Medical University Hospital, Taichung, Taiwan
4. Division of Colorectal Surgery, Department of Surgery, Chung Shan Medical University Hospital, Taichung, Taiwan
5. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan
6. Department of Obstetrics and Gynecology, China Medical University Hospital, Taichung, Taiwan
7. Sex Hormone Research Center, China Medical University Hospital, Taichung, Taiwan
8. Department of Nursing, Asia University, Taichung, Taiwan
* Jiunn-Bey Pao and Te-Ling Lu contributed equally to this work.
Received 2017-8-16; Accepted 2017-11-22; Published 2018-1-1
Abstract
Background: Single nucleotide polymorphisms (SNPs) of small non-coding RNAs (sncRNAs) can influence sncRNA function and target gene expression to mediate the risk of certain diseases. The aim of the present study was to evaluate the prognostic relevance of sncRNA SNPs for colorectal cancer, which has not been well characterized to date.
Methods: We comprehensively examined 31 common SNPs of sncRNAs, and assessed the impact of these variants on survival in a cohort of 188 patients with colorectal cancer.
Results: Three SNPs were significantly associated with survival of patients with colorectal cancer after correction for multiple testing, and two of the SNPs (hsa-mir-196a-2 rs11614913 and U85 rs714775) remained significant in multivariate analyses. Additional in silico analysis provided further evidence of this association, since the expression levels of the target genes of the hsa-miR-196a (HOXA7, HOXB8, and AKT1) were significantly correlated with colorectal cancer progression. Furthermore, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses indicated that hsa-miR-196a is associated with well-known oncogenic pathways, including cellular protein modification process, mitotic cell cycle, adherens junction, and extracellular matrix receptor interaction pathways.
Conclusion: Our results suggest that SNPs of sncRNAs could play a critical role in cancer progression, and that hsa-miR-196a might be a valuable biomarker or therapeutic target for colorectal cancer patients.
Keywords: colorectal cancer, survival, small non-coding RNA, single nucleotide polymorphism, biomarker
Introduction
Colorectal cancer is the third most frequent cause of cancer-related deaths in the United States [1]. Surgical resection is the primary treatment for patients with colorectal cancer, but ~30-50% of these patients will relapse and ultimately die after surgery [2]. Given the limited therapeutic options and molecular targets currently approved for the treatment of colorectal cancer, there is an urgent need for the identification of novel therapeutic targets to improve clinical outcomes.
Small non-coding RNAs (sncRNAs) have recently gained attention owing to their involvement in multiple biological processes by directly or indirectly interfering with gene expression, and dysregulation of sncRNAs has been implicated in a variety of disorders, including cancer [3]. sncRNAs represent a large group of RNAs that are generally less than 200 nucleotides in length, including microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), small Cajal body-specific RNAs (scaRNAs), and others. miRNAs bind to mRNA targets and negatively regulate gene expression through the degradation of target transcripts and inhibition of translation [4]. snoRNAs comprise a class of nucleolus-enriched sncRNAs that largely regulate the post-transcriptional maturation of ribosomal RNAs. snoRNAs are classified into C/D box snoRNAs and H/ACA box snoRNAs based on their structure and function, responsible for 2'-O-methylation and guiding the pseudo-uridylation of nucleotides, respectively [5, 6]. In addition, certain snoRNAs can also be processed into miRNAs to influence mRNA expression [7]. Similarly, scaRNAs guide the methylation and pseudo-uridylation of small nuclear RNAs [8, 9]. The expression profiles of these sncRNAs tend to vary among different types of cancers, and are potential or prognostic biomarkers with possible clinical applications. To determine their prognostic or therapeutic value in colorectal cancer, in this study, we comprehensively analyzed 31 common single nucleotide polymorphisms (SNPs) of sncRNAs and their impact on the survival of patients with colorectal cancer.
Patients and Methods
Patient recruitment and data collection
The present study cohort included 188 patients with colorectal cancer who underwent surgical resection at China Medical University Hospital, Taiwan between 2001 and 2007. Disease characteristics and follow-up data of these patients were obtained from their medical records. Overall survival was defined as the duration from diagnosis to death from any cause or until the last follow-up. All participants provided written informed consent before surgery. This study was approved by the Institutional Review Board of China Medical University Hospital, and was performed according to the approved guidelines.
SNP selection and genotyping
We identified SNPs of sncRNAs by intersection sno/miRNA table [10] with HapMap SNPs CHB (Han Chinese in Beijing, China) table from the UCSC table browser (NCBI36/hg18) [11]. SNPs with a minor allele frequency less than 0.05 and those in linkage disequilibrium in the HapMap CHB population were excluded. Thirty-eight sncRNA SNPs were initially selected for analysis. Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) and stored at -80°C until analysis. Genotyping was performed at the National Center for Genome Medicine, Taiwan, using the Agena Bioscience iPLEX matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry technology, as described previously [12]. Any SNP that failed the genotyping assay design, did not conform to Hardy-Weinberg equilibrium (P < 0.05), was below a genotyping call rate of 0.8, or had a minor allele frequency less than 0.03 was removed (N = 7). Thus, a total of 31 SNPs of sncRNAs, including 10 in C/D box snoRNAs, 9 in H/ACA box snoRNAs, 7 in miRNAs, and 5 in scaRNAs, were included for further statistical analyses.
Statistical analysis
Patient clinicopathological characteristics are summarized as the number and percentage of patients or the median and interquartile range of values. The hazard ratio (HR) and 95% confidence interval were estimated by Cox regression analysis to assess the association of clinicopathological characteristics with overall survival. The difference in overall survival according to the SNP genotypes was compared using Kaplan-Meier analysis and log-rank tests. We also performed multivariate Cox regression to determine the interdependency of genetic interactions and known prognostic factors such as age, gender, carcinoembryonic antigen (CEA) levels, tumor differentiation, stage, lymphovascular invasion, perineural invasion, and lymph node involvement [13-16]. The Statistical Package for the Social Sciences software version 22.0.0 (IBM, Armonk, NY, USA) was used for statistical analyses. A two-sided P value of <0.05 was considered nominally significant. As we were testing 31 SNPs, the q value was applied to control the false discovery rate [17] and the association was deemed significant at the q < 0.20 level.
Bioinformatics analysis
The publicly available SurvExpress database [18] was utilized to analyze the expression levels of the hsa-miR-196a target genes HOXA7, HOXB8, and AKT1 on the survival of colorectal cancer patients. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathway enrichment analyses were performed by DIANA-miRPath v3.0 [19]. This software calculates enrichment based on experimentally supported miRNA-gene interactions derived from DIANA-TarBase v7.0 [20].
Results
The clinical and pathologic characteristics of patients with colorectal cancer and the association with overall survival are shown in Table S1. At the median follow-up duration of 60.1 months, 61 patients had died and the estimated 5-year overall survival rate was 69.3%. Univariate analysis showed that CEA levels, tumor stage, lymphovascular invasion, perineural invasion, and lymph node involvement were respectively significantly associated with overall survival (P ≤ 0.006).
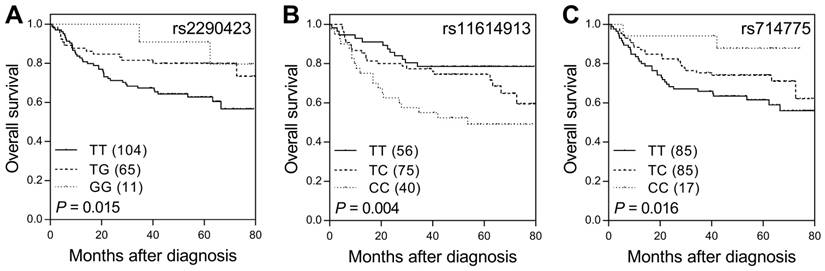
Of the 31 sncRNA SNPs analyzed in the study, three (ACA23 rs2290423, hsa-mir-196a-2 rs11614913, and U85 rs714775) showed significant effects on survival (nominal P ≤ 0.016, q < 0.20 in all cases; Table 1 and Figure 1). In the multivariate analysis adjusted for age, gender, CEA levels, tumor differentiation, stage, lymphovascular invasion, perineural invasion, and lymph node involvement, hsa-mir-196a-2 rs11614913 and U85 rs714775 remained significantly associated with survival (P ≤ 0.012, Table 2). The HRs for death were 1.84 and 0.50 for genotypes at the rs11614913 and rs714775 locus, respectively.
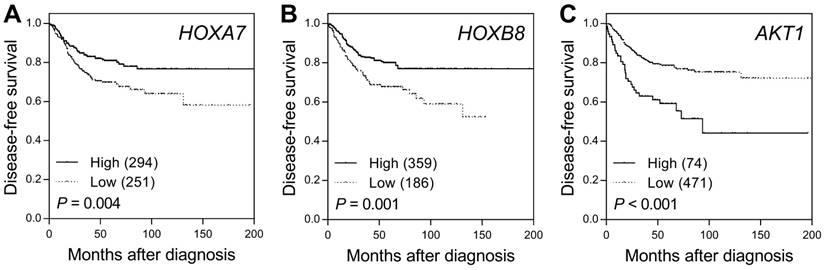
SNPs of miRNAs might affect miRNA binding, the expression of its target genes, and subsequent disease progression. Previous studies have shown that hsa-miR-196a, the mature miRNA of hsa-mir-196a-2, restricts the expression of HOXA7 and HOXB8, leading to activation of the AKT1 survival pathway in colorectal cancer. Therefore, we further evaluated the prognostic role of the expression levels of the hsa-miR-196a target genes HOXA7, HOXB8, and AKT1 on the survival of colorectal cancer patients using the publicly available SurvExpress Colon-Metabase dataset. The risk score for each patient was derived from each gene expression level and their Cox regression coefficients, and was used to divide patients into low- or high-risk groups using an optimization algorithm for the minimum P value. Low expression levels of HOXA7 and HOXB8 and high AKT1 expression levels were significantly associated with poor survival in colorectal cancer patients (P ≤ 0.004, Figure 2).
Association of sncRNA SNPs with overall survival in colorectal cancer patients
| Type | Name | SNP | Chromosome | Position | P | q |
|---|---|---|---|---|---|---|
| C/D box | U94 | rs3770069 | 2 | 86216585 | 0.824 | 0.818 |
| C/D box | HBII-429 | rs9389034 | 6 | 133179703 | 0.714 | 0.818 |
| C/D box | U13 | rs2732289 | 8 | 33490604 | 0.973 | 0.852 |
| C/D box | U14B | rs1541533 | 11 | 17052787 | 0.639 | 0.818 |
| C/D box | U25 | rs1059292 | 11 | 62379627 | 0.650 | 0.818 |
| C/D box | 14q(II-18) | rs12893444 | 14 | 100511971 | 0.451 | 0.818 |
| C/D box | U49B | rs11871958 | 17 | 16283558 | 0.403 | 0.818 |
| C/D box | HBII-135 | rs4622550 | 17 | 16285326 | 0.369 | 0.818 |
| C/D box | U105 | rs2305789 | 19 | 10079332 | 0.418 | 0.818 |
| C/D box | HBI-43 | rs753213 | 20 | 17891492 | 0.109 | 0.493 |
| H/ACA box | U98b | rs792442 | 1 | 210592911 | 0.210 | 0.712 |
| H/ACA box | U19-2 | rs17075508 | 5 | 172380368 | 0.708 | 0.818 |
| H/ACA box | ACA20 | rs2295901 | 6 | 160121285 | 0.087 | 0.472 |
| H/ACA box | ACA29 | rs3818299 | 6 | 160126621 | 0.201 | 0.712 |
| H/ACA box | ACA29 | rs3818298 | 6 | 160126706 | 0.448 | 0.818 |
| H/ACA box | ACA17 | rs2275159 | 9 | 138741147 | 0.399 | 0.818 |
| H/ACA box | ACA23 | rs2290423 | 11 | 9406953 | 0.015 | 0.145 |
| H/ACA box | ACA32 | rs1944108 | 11 | 93103823 | 0.874 | 0.818 |
| H/ACA box | U71d | rs788347 | 20 | 36495997 | 0.646 | 0.818 |
| miRNA | hsa-mir-943 | rs1077020 | 4 | 1957991 | 0.648 | 0.818 |
| miRNA | hsa-mir-604 | rs2368392 | 10 | 29874009 | 0.591 | 0.818 |
| miRNA | hsa-mir-608 | rs4919510 | 10 | 102724768 | 0.862 | 0.818 |
| miRNA | hsa-mir-196a-2 | rs11614913 | 12 | 52671866 | 0.004 | 0.109 |
| miRNA | hsa-mir-492 | rs2289030 | 12 | 93752417 | 0.048 | 0.326 |
| miRNA | hsa-mir-423 | rs6505162 | 17 | 25468309 | 0.783 | 0.818 |
| miRNA | hsa-mir-923 | rs4796042 | 17 | 30502298 | 0.945 | 0.852 |
| scaRNA | U87 | rs3792109 | 2 | 233849156 | 0.666 | 0.818 |
| scaRNA | U92 | rs13302453 | 9 | 19053765 | 0.729 | 0.818 |
| scaRNA | U85 | rs714775 | 12 | 6489663 | 0.016 | 0.145 |
| scaRNA | U93 | rs3825546 | 14 | 95069573 | 0.802 | 0.818 |
| scaRNA | ACA47 | rs2678770 | 17 | 72597029 | 0.802 | 0.818 |
Abbreviations: SNP, single nucleotide polymorphism; sncRNA, small non-coding RNA; miRNA, microRNA; scaRNA, small Cajal body-specific RNA.
q < 0.20 are in boldface.
SNPs of sncRNA correlate with overall survival in colorectal cancer patients. Kaplan-Meier analysis of overall survival according to genotypes at (A) ACA23 rs2290423, (B) hsa-mir-196a-2 rs11614913, and (C) U85 rs714775. Numbers in parentheses indicate the number of patients.
Univariate and multivariate analyses of sncRNA SNPs with overall survival
| SNP Genotype | N (%) | N of deaths | 5-year survival rate, % | P | Univariate analysis | Multivariate analysis* | ||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||||
| rs2290423 | ||||||||
| TT | 104 (57.8) | 40 | 62.7 | 0.015 | 1.00 | 1.00 | ||
| TG | 65 (36.1) | 14 | 79.9 | 0.51 (0.28-0.95) | 0.032 | 0.67 (0.32-1.40) | 0.282 | |
| GG | 11 (6.1) | 2 | 90.9 | 0.35 (0.09-1.47) | 0.152 | 0.47 (0.11-2.07) | 0.319 | |
| Trend | 0.54 (0.33-0.90) | 0.017 | 0.68 (0.39-1.18) | 0.166 | ||||
| rs11614913 | ||||||||
| TT | 56 (32.7) | 12 | 78.6 | 0.004 | 1.00 | 1.00 | ||
| TC | 75 (43.9) | 23 | 74.5 | 1.47 (0.73-2.96) | 0.280 | 1.58 (0.63-3.97) | 0.327 | |
| CC | 40 (23.4) | 20 | 49.1 | 2.79 (1.36-5.70) | 0.005 | 3.26 (1.27-8.38) | 0.014 | |
| Trend | 1.69 (0.18-2.44) | 0.004 | 1.84 (1.15-2.96) | 0.012 | ||||
| rs714775 | ||||||||
| TT | 85 (45.5) | 34 | 61.5 | 0.016 | 1.00 | 1.00 | ||
| TC | 85 (45.5) | 24 | 74.1 | 0.66 (0.39-1.11) | 0.115 | 0.61 (0.33-1.14) | 0.122 | |
| CC | 17 (9.1) | 2 | 87.8 | 0.25 (0.06-1.04) | 0.056 | 0.12 (0.02-0.91) | 0.040 | |
| Trend | 0.59 (0.39-0.91) | 0.017 | 0.50 (0.30-0.84) | 0.009 | ||||
Abbreviations: SNP, single nucleotide polymorphism; sncRNA, small non-coding RNA; CEA, carcinoembryonic antigen; HR, hazard ratio; CI, confidence interval.
*Adjusted by age, gender, CEA levels, tumor differentiation, stage, lymphovascular invasion, perineural invasion, and lymph node involvement.
P < 0.05 are in boldface.
Prognostic value of the hsa-miR-196a target genes HOXA7, HOXB8, and AKT1 in colorectal cancer. Kaplan-Meier analysis of disease-free survival based on the risk score generated from (A) HOXA7, (B) HOXB8, and (C) AKT1 expression levels obtained from the SurvExpress Colon-Metabase dataset. Numbers in parentheses indicate the number of patients.
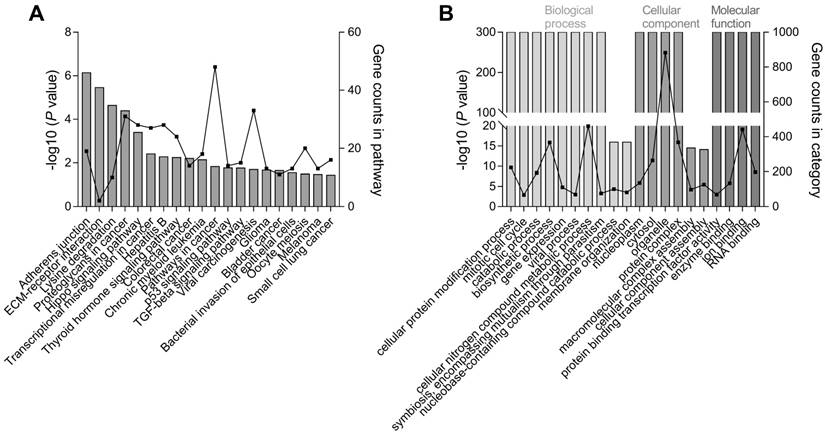
In addition to the known hsa-miR-196a target genes, DIANA-miRPath analysis further revealed the pathways with significant enrichment of target mRNAs of hsa-miR-196a-5p and hsa-miR-196a-3p, two mature miRNAs of hsa-mir-196a-2. We annotated the 20 top-ranked KEGG and GO pathways through Fisher's exact test and false-discovery rate correction (Figure 3). Adherens junction, extracellular matrix receptor, proteoglycans in cancer, Hippo signaling, transcriptional misregulation in cancer, colorectal cancer, pathways in cancer, and p53 signaling pathways were the top enriched KEGG pathways.
The top biological processes in GO enrichment analysis were associated with cellular protein modification, mitotic cell cycle, and gene expression, and the top molecular functions were protein binding transcription factor activity and enzyme binding in cellular components of the nucleoplasm and cytosol. Importantly, these KEGG/GO pathways are known to be critical for controlling the development of cancers.
Discussion
We conducted a comprehensive study to identify genetic variants of sncRNAs associated with the prognosis of patients with surgically resected colorectal cancer. The SNPs hsa-mir-196a-2 rs11614913 and U85 rs714775 were significantly associated with survival, even after adjusting for other clinical predictors and multiple testing. Further functional annotation of the predicted downstream targets demonstrated that these SNPs are associated with significant dysregulation in protein modification, cell cycle, and cell-cell interactions, which are closely related to the biological characteristics of cancer cells. Our work therefore contributes to gaining a better understanding of the role of sncRNAs in cancer, and highlights their potential as valuable molecular biomarkers in colorectal cancer.
The SNP rs11614913 is located in the 3′ passenger (3p) strand of the mature sequence of hsa-mir-196a-2. Patients with lung cancer carrying a variant homozygous CC genotype of rs11614913 were reported to have relatively poor survival, and the mechanism was determined to be elevated expression of mature hsa-miR-196a with weakened target binding of hsa-miR-196a-3p [21]. These data are consistent with our findings of a trend toward poorer survival in patients with colorectal cancer carrying an increased number of the C risk allele at rs11614913 (Table 2 and Figure 1). In addition, this functional SNP has been associated with susceptibility to a wide variety of cancers, including lung [22], breast [23], gastric [24], head and neck [25], liver [26], and colorectal cancer [27], suggesting its important general role in carcinogenesis. The miRNA hsa-miR-196a is encoded at three locations in the mammalian homeobox (HOX) A, B, and C clusters, and the hsa-miR-196a-binding sites show evolutionarily conserved complementarity to mRNAs of several HOX genes [28]. hsa-miR-196a-directed cleavage of HOXB8 mRNA was observed in mouse embryos [28], and additional experiments revealed the down-regulated expression of HOXA7, HOXB8, HOXC8, and HOXD8 in colorectal cancer cells [29]. HOX genes are known critical regulators of carcinogenesis [30], and our in silico analysis supports that altered HOXA7, HOXB8, and AKT1 expression levels are correlated with colorectal cancer patient prognosis (Figure 2). Up-regulation of hsa-miR-196a expression has also been reported to be associated with a more aggressive disease course and unfavorable treatment outcomes in colorectal cancer [31]. However, the role of U85 in cancer is currently unknown, thus warranting further study.
The significant association of the SNPs hsa-mir-196a-2 rs11614913 and U85 rs714775 with survival outcomes persisted after controlling for known prognostic factors and multiple testing, which should reduce the possibility of false-positive findings in this study. In addition, functional annotation supported the detected relationships between these sncRNA SNPs and the progression of colorectal cancer. However, these results should be interpreted with caution owing to several limitations of the present study, such as the relatively small sample size and retrospective nature, as well as the limitation of the sample including only cases from the Taiwanese population. Therefore, further studies are required to validate our results.
Nevertheless, the results of this study demonstrate that genetic variants of sncRNAs influence the survival of patients with colorectal cancer. Further studies are required to confirm the effects of these SNPs in larger and diverse populations and to understand the biological functions of sncRNAs in the progression of colorectal cancer.
Pathway enrichment analysis of hsa-mir-196a-2 target genes. The top 20 significantly enriched (A) KEGG and (B) GO pathways of target genes for two mature miRNAs, hsa-miR-196a-5p and hsa-miR-196a-3p, originating from hsa-mir-196a-2. The left y-axis represents the significance according to the P value (log-scaled) and the right y-axis presents the number of genes in a particular pathway; the x-axis indicates the KEGG/GO pathways.
Abbreviations
CEA, carcinoembryonic antigen; GO, Gene Ontology; HOX, homeobox; HR, hazard ratio; IQR, interquartile range; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA; scaRNA, small Cajal body-specific RNA; sncRNA, small non-coding RNA; snoRNA, small nucleolar RNA; SNP, single nucleotide polymorphism.
Supplementary Material
Table S1.
Acknowledgements
This work was supported by the Ministry of Science and Technology of Taiwan (grant number: 106-2314-B-039-018), and the China Medical University (grant number: CMU106-S-24). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the National Center for Genome Medicine, Ministry of Science and Technology of Taiwan, for technical support.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
2. Schmoll HJ, Van Cutsem E, Stein A. et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23:2479-516
3. Taft RJ, Pang KC, Mercer TR. et al. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126-39
4. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-5
5. Cavaille J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732-5
6. Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799-809
7. Ender C, Krek A, Friedlander MR. et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519-28
8. Darzacq X, Jady BE, Verheggen C. et al. Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746-56
9. Richard P, Darzacq X, Bertrand E. et al. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283-93
10. Griffiths-Jones S, Saini HK, van Dongen S. et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154-8
11. Karolchik D, Hinrichs AS, Furey TS. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493-6
12. Huang SP, Huang LC, Ting WC. et al. Prognostic significance of prostate cancer susceptibility variants on prostate-specific antigen recurrence after radical prostatectomy. Cancer Epidemiol Biomarkers Prev. 2009;18:3068-74
13. Ting WC, Chen LM, Huang LC. et al. Impact of interleukin-10 gene polymorphisms on survival in patients with colorectal cancer. J Korean Med Sci. 2013;28:1302-6
14. Ting WC, Chen LM, Pao JB. et al. Genetic polymorphisms of matrix metalloproteinases and clinical outcomes in colorectal cancer patients. Int J Med Sci. 2013;10:1022-7
15. Ting WC, Chen LM, Pao JB. et al. Common genetic variants in Wnt signaling pathway genes as potential prognostic biomarkers for colorectal cancer. PLoS One. 2013;8:e56196
16. Yang YP, Ting WC, Chen LM. et al. Polymorphisms in MicroRNA Binding Sites Predict Colorectal Cancer Survival. Int J Med Sci. 2017;14:53-7
17. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440-5
18. Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E. et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250
19. Vlachos IS, Zagganas K, Paraskevopoulou MD. et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460-6
20. Vlachos IS, Paraskevopoulou MD, Karagkouni D. et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153-9
21. Hu Z, Chen J, Tian T. et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600-8
22. Tian T, Shu Y, Chen J. et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183-7
23. Hu Z, Liang J, Wang Z. et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79-84
24. Peng S, Kuang Z, Sheng C. et al. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2010;55:2288-93
25. Christensen BC, Avissar-Whiting M, Ouellet LG. et al. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res. 2010;16:3713-20
26. Qi P, Dou TH, Geng L. et al. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol. 2010;71:621-6
27. Zhan JF, Chen LH, Chen ZX. et al. A functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese population. Arch Med Res. 2011;42:144-8
28. Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594-6
29. Schimanski CC, Frerichs K, Rahman F. et al. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089-96
30. Blatt C. The betrayal of homeo box genes in normal development: the link to cancer. Cancer Cells. 1990;2:186-9
31. Ge J, Chen Z, Li R. et al. Upregulation of microRNA-196a and microRNA-196b cooperatively correlate with aggressive progression and unfavorable prognosis in patients with colorectal cancer. Cancer Cell Int. 2014;14:128
Author contact
![]() Corresponding author: Bo-Ying Bao, Department of Pharmacy, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22053366 ext. 5126; Fax: +886-4-22031075; E-mail: baocmu.edu.tw
Corresponding author: Bo-Ying Bao, Department of Pharmacy, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22053366 ext. 5126; Fax: +886-4-22031075; E-mail: baocmu.edu.tw

 Global reach, higher impact
Global reach, higher impact