Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(7):932-937. doi:10.7150/ijms.5708 This issue Cite
Research Paper
Influence of CYP2D6 Polymorphisms on Serum Levels of Tamoxifen Metabolites in Spanish Women with Breast Cancer
1. Clinical Biochemistry Services, San Cecilio University Hospital. Avd/ Doctor Olóriz s/n 18012, Granada, Spain.
2. Department of Biochemistry and Molecular Biology III and Immunology, Faculty of Medicine Avd/Madrid s/n 18071, Granada, Spain.
3. Department of Oncology, San Cecilio University Hospital. Avd/Doctor Olóriz s/n 18012, Granada, Spain.
4. Aula de Estudios Avanzados. Fundación Tejerina. Madrid, Spain.
5. Department of Oncology, Hospital Provincial de Castellón. Avd/ Dr. Clará, nº 19, 12002 Castellón. Spain.
6. Molecular Biopathology Laboratory. Hospital Provincial de Castellón. Avd/ Dr. Clará, nº 19 12002. Castellón, Spain.
7. Department of Biochemistry and Molecular Biology II, Institute of Nutrition and Food Technology, Center for Biomedical Research, University of Granada, Granada. Avd/Conocimiento s/n 18100 Armilla, Granada, Spain.
Received 2012-12-14; Accepted 2013-5-8; Published 2013-5-27
Abstract
Background Estrogen receptor-positive breast cancer tumors depend on estrogen signaling for their growth and replication and can be treated by anti-estrogen therapy with tamoxifen. Polymorphisms of the CYP2D6 and CYP2C19 genes are associated with an impaired response to tamoxifen. The study objective was to investigate the impact of genetic polymorphisms in CYP2D6 and CYP2C19 on the pharmacokinetics of tamoxifen and its metabolites in Spanish women with estrogen receptor-positive breast cancer who were candidates for tamoxifen therapy.
Methods: We studied 90 women with estrogen receptor-positive breast cancer, using the AmpliChip CYP450 test to determine CYP2D6 and CYP2C19 gene variants. Plasma levels of tamoxifen and its metabolites were quantified by high-performance liquid chromatography.
Results The CYP2D6 phenotype was extensive metabolizer in 80%, intermediate metabolizer in 12.2%, ultra-rapid metabolizer in 2.2%, and poor metabolizer in 5.6% of patients, and the allele frequency was 35.0% for allele *1, 21.0% for *2, and 18.9% for *4. All poor metabolizers in this series were *4/*4, and their endoxifen and 4-hydroxy tamoxifen levels were 25% lower than those of extensive metabolizers. CYP2C19*2 allele, which has been related to breast cancer outcomes, was detected in 15.6% of the studied alleles.
Conclusion CYP2D6*4/*4 genotype was inversely associated with 4-hydroxy tamoxifen and endoxifen levels. According to these results, CYP2D6 and CYP2C19 genotyping appears advisable before the prescription of tamoxifen therapy.
Keywords: CYP2D6, CYP2C19, genetic diagnosis, estrogen-positive breast cancer, endoxifen, tamoxifen
BACKGROUND
Breast cancer is the most common cancer worldwide and the second leading cause of death by cancer. Estrogen and progesterone (steroid hormones) have been implicated in the pathogenesis of breast cancer [1]. Estrogen receptor-positive (ER+) tumors depend on estrogen signaling for their growth and replication and can be treated by anti-estrogen therapy with tamoxifen or an aromatase inhibitor [2].
The use of pharmacogenomic markers is important in oncology because of the frequently narrow therapeutic index of available drugs, the critical need for a favorable drug response, and the potentially life-threatening consequences of drug toxicity [3]. In this respect, genetic polymorphisms of the CYP2D6 gene have been reported as the main cause of variation in the metabolism of tamoxifen, which has been the standard treatment for ER+ breast cancer for more than three decades [4-6]. N-desmethyl tamoxifen, the main metabolite found in the serum of treated patients, undergoes secondary metabolism to 4-hydroxy-N-desmethyl tamoxifen (endoxifen). CYP2D6 is the enzyme responsible for this conversion and also converts tamoxifen to 4-hydroxy tamoxifen (4-OH tamoxifen), which undergoes secondary metabolism to endoxifen [5]. The CYP2D6 gene is highly polymorphic and has more than 100 different allele forms (http://www.cypalleles.ki.se/cyp2d6.htm), which can be classified as a function of the intensity of their enzyme activity as extensive metabolizer (EM), intermediate metabolizer (IM), poor metabolizer (PM), or ultra-rapid metabolizer (UM) phenotypes [7, 8]. CYP2C19 gene polymorphisms affect the metabolism of numerous drugs, including anti-depressants, and play an important role in the bioactivation of cyclophosphamide [9].
The enzyme CYP2C19 also participates in the conversion of tamoxifen to active metabolites, and the CYP2C19 gene has more than 35 allele variants (http://www.cypalleles.ki.se/cyo2c19.htm) [7, 8] A considerable variability has been observed among ethnic/racial groups in the frequency of CYP2D6 variants among the general population, with a decrease in the frequency of functional CYP2D6 alleles from 71% in Europeans to 50% in Asians [10, 11]. A high frequency of null and reduced-function variants has been reported worldwide, and ~7% of western Eurasian populations carry defective CYP2D6 alleles that are PM phenotypes encoding for inactive enzyme molecules [12]. CYP2D6*2*3*4*5*6*10 and *41 alleles are more frequent in Caucasians, CYP2D6*2 and *17 alleles in Africans, and CYP2D6*10 allele in Asians [11]. With regard to CYP2C19, two non-functional alleles (*2 and *3) account for 87% and 98% of the PM phenotype in Caucasian and Oriental populations, respectively [7].
In breast cancer patients, frequencies of 3.5%, 21.7%, 1.5%, 1.0%, 1.5% and 6.6% have been reported for CYP2D6 gene alleles *3, *4, *5, *6, *10 and *41, respectively [13]. Frequencies of 60.2% EM, 29.5% IM, and 8.3% PM were recently described in a large cohort of breast cancer patients, although the DNA was isolated from tumor samples, in which frequent losses of heterozygosity have been reported for breast cancer cells [14, 15]. Loss of CYP2D6 function was found to reduce the clinical efficacy of tamoxifen treatment [11], although two more recent articles have stimulated debate about the association of the CYP2D6 genotype with the pharmacodynamics of tamoxifen [14, 16]. Moreover, the clinical validity of CYP2D6 genotyping is not well established in breast cancer patients who are candidates for tamoxifen treatment [17].
Hence, the ability to predict the prevalence of EM, IM, PM, and UM phenotypes in a given population would support the design of a practical genotyping protocol for routine clinical use. The main objective of this pilot study was to determine the allele frequencies of the most relevant variants of CPY2D6 and CPY2C19 genes and their association with tamoxifen metabolite levels as a function of CYP2D6 polymorphisms in patients with ER+ breast cancer from Granada (Spain).
METHODS
Patients
Based on the main outcome, i.e., the prevalence of the PM phenotype in the Spanish population [18], and assuming a type 1 error of α=0.05 and a precision of 5%, the minimum sample size was estimated to be 72 patients. The study included 90 women from San Cecilio University Hospital (Granada, Spain) with a histological diagnosis of ER+ non-invasive breast cancer and a Karnofsky index ≥ 80. All women received adjuvant tamoxifen (20 mg/day) during the study and for at least 4 months before the blood sampling for tamoxifen and metabolite determinations. None of the women in this study were receiving drugs that inhibit CYP2D6 function. All patients were non-metastatic at recruitment. The age range of the patients was 29-72 years, and 93% were premenopausal; the six postmenopausal women in the study received tamoxifen due to adverse effects from the use of aromatase inhibitor, in accordance with the hospital protocol. The study complied with the guidelines laid down in the Declaration of Helsinki, and all procedures involving human subjects were approved by the Clinical Research Ethics Committee of San Cecilio University Hospital. Informed consent was obtained from all participants.
DNA isolation and AmpliChip CYP450 test
Whole blood samples were used for the preparation of human genomic DNA, using a QIAcube automatic extractor (Qiagen Iberia, S.L. Madrid, Spain) according to the manufacturer's instructions. Concentrations of samples were determined spectrophotometrically with a Jenway-Genova UV spectrophotometer (Jenway, Staffordshire, UK).
CYP2D6 and CYP2C19 genotyping was performed by using the AmpliChip CYP450 test according to the manufacturer`s instructions, screening for 29 variants of the CPY2D6 gene and 2 variants of CPY2C19 (Roche AmpliChip® CYP450 test product, Roche Diagnostics, Spain), which has been approved as a diagnostic instrument by the Food and Drug Administration. The data obtained were analyzed by using Molecular Diagnostic (AMDS) v1.0 (Affymetrix) and AmpliChip CYP450 v2.0 software packages (Roche Diagnostics, Spain). The CYP2D6 and CYP2C19 phenotypes were defined according to the AmpliChip algorithm (Roche AmpliChip® CYP450 test product, Roche Diagnostics, Spain), based on published studies (http://www.cypalleles.ki.se/).
Tamoxifen, N-desmethyl tamoxifen, and endoxifen determinations
Blood samples were centrifuged at 3000 rpm within 1 h of collection, and the plasma was stored at -80ºC until analysis. Concentrations of tamoxifen and its main metabolites were tested by high-performance liquid chromatography in an Agilent 1200 system (Agilent technologies) associated with tandem mass spectrometry at the Tejerina Foundation (Madrid, Spain).
Statistical analysis
The Mann-Whitney U test was used to compare differences in the levels of tamoxifen, 4-OH tamoxifen, N-desmethyl tamoxifen, and endoxifen. SPSS v.20 (IBM, Chicago, IL, USA) was used for the statistical analysis.
RESULTS
The phenotype of CYP2D6 variants in our study population was EM in 80%, IM in 12.2%, UM in 2.2%, and PM in 5.6% of cases. There were 17 different genotypes and 9 different alleles for EM, 8 genotypes and 5 alleles for IM, and 2 genotypes and 3 alleles for UM phenotypes. A single genotype (*4/*4) was found for the PM phenotype. Table 1 lists the CYP2D6 genotype frequencies found. CYP2D6 allele frequencies in the study population were 35.0% for *1 allele, 21% for *2 allele, and 18.9%, 6.7%, and 6.1% for alleles *4, *35, and *41, respectively. The frequencies of alleles *9, *10, *1XN, *17, *5 and *2XN were 5.0%, 3.9%, 1.1%, 1.1%, and 0.6%, respectively.
CYP2C19 phenotype and genotype frequencies were 71.1% and 26.7% for the *1/*1 and *1/*2 CYP2C19 genotype (both EM), respectively, and 2.2% for the *2/*2 genotype (PM). CYP2C19 allele frequencies were 84.4% for allele *1 and 15.6% for allele *2.
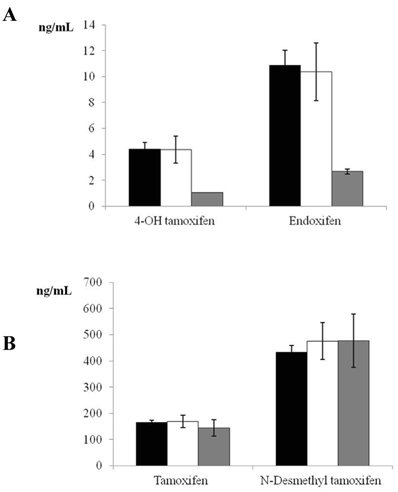
Given that only 5 women were PMs, a sample size of 18 individuals was estimated for the control group, based on a power of 85% and alpha error of 0.05 to detect a difference of one standard deviation between the means of two groups (EM and PM). We therefore randomly selected 20 women for the control group (EM phenotype). Figure 1 depicts tamoxifen, endoxifen, 4-OH tamoxifen, and N-desmethyl tamoxifen levels in EM, IM, and PM CYP2D6 phenotypes. Significant differences were found in the comparison of 4-OH tamoxifen and endoxifen levels between EM and PM phenotypes (P<0.001).
CYP2D6 phenotype and genotype frequencies in 90 women with estrogen receptor positive breast cancer
| CYP2D6 phenotype | CYP2D6 genotype | Frequency (%) | |
|---|---|---|---|
| Extensive metabolizers | *1/*1 | 15.6 | |
| *1/*2 | 14.5 | ||
| *1/*4 | 7.8 | ||
| *1/*35 | 6.7 | ||
| *2/*2 | 6.7 | ||
| *2/*4 | 6.7 | ||
| *4/*35 | 4.5 | ||
| *1/*9 | 3.3 | ||
| *1/*41 | 3.3 | ||
| *2/*10 | 2.2 | ||
| *1/*10 | 2.2 | ||
| *2/*5 | 1.1 | ||
| *2/*41 | 1.1 | ||
| *2XN/*4 | 1.1 | ||
| *2/*9 | 1.1 | ||
| *2/*35 | 1.1 | ||
| *10/*35 | 1.1 | ||
| Intermediate metabolizers | *9/*41 | 2.2 | |
| *4/*10 | 2.2 | ||
| *9/*9 | 1.1 | ||
| *4/*17 | 1.1 | ||
| *4/*41 | 2.2 | ||
| *4/*9 | 1.1 | ||
| *17/*41 | 1.1 | ||
| *41/*41 | 1.1 | ||
| Ultra-rapid metabolizers | *1/*1XN | 1.1 | |
| *1XN/*2 | 1.1 | ||
| Poor metabolizers | *4/*4 | 5.6 | |
DISCUSSION
Genetic polymorphisms of CYP2D6 and CYP2C19 are clinically relevant in drug therapy. The CYP2D6 gene is considered responsible for the metabolism of around a quarter of the drugs available on the market, and its variants affect the metabolism of around half of these [19, 20]. Tamoxifen, which is widely used to treat ER+ breast cancer, is metabolized by the P450 cytochrome enzyme.
Variation in the CYP2D6 gene is high among different populations and among individuals in the same population [11]. Frequencies of 31% for the wild-type allele and 40.47% for allele *2 were reported in a Spanish general population [21]. Caucasian populations have shown frequencies of 33-37% for allele *1 and 22-33% for allele *2 [11, 22, 23]. The present results in breast cancer ER+ positive patients are therefore in broad agreement with findings in general populations, although there was a tendency for the frequency of allele *2 (21.0%) to be lower in our series than in the Spanish population [21] but close to the 16.5% found by Ramon et al in Spanish breast cancer patients [24].
The prevalence of the PM phenotype was reported to be 7-10% in Caucasian populations, and allele *4 was the most frequent null allele [4]. In the present series of women with ER+ breast cancer, 5.6% harbored allele CYP2D6*4 in homozygosis, similar to findings by Rebsamen et al in Caucasian women (5.5%) [25] and by Fernández-Santander et al in a cohort from southern Spain (3.9%) [10]. Breast cancer patients in Spain were found to have a frequency of 5.5% for the *4/*4 genotype [24]. Furthermore, the CYP2D*4 genotyping frequencies were in Hardy-Weinberg equilibrium. The frequency of this allele was 17.7%, comparable to frequencies observed in the Spanish general population (13.8-15.9%) [10, 21] and in other Caucasian populations (19%) [21, 22]. Interestingly, all PMs in our study were *4/*4. The frequency of non-functional allele CYP2D6*5 was only 0.6%, below the range of 2-7% described in Spanish and Caucasian populations, although closer to the percentages found in Sardinian (1.04%), central Italian (0.0%), and Syrian (0.98%) populations [10, 11, 21, 26]. It is well known that CYP2D6 diversity is far greater within than between populations and groups [27]. The molecular heterogeneity in Spain is higher than in other populations, as has been shown by our group for cystic fibrosis and for glucose 6- phosphate dehydrogenase [28, 29], which may explain the low frequency observed for allele *5.
It has been reported that tamoxifen-treated Caucasian patients who are heterozygous or homozygous for the CYP2D6*4 allele have a significantly increased risk of breast cancer recurrence, a shorter relapse-free period, and a lower event-free survival rate in comparison to carriers of functional alleles [30, 31]. In this context, serum levels of 4-OH tamoxifen and endoxifen, both dependent on CYP2D6 activity, were four-fold lower in the PMs than in the EMs in the present study, whereas no differences were found in the levels of tamoxifen or N-desmethyl tamoxifen, which are independent of CYP2D6 activity (Figure 1). Similar results were reported for Asian patients with CYP2D6*10/10 [32].
Plasma tamoxifen, endoxifen, 4-hydroxy (4-OH) tamoxifen, and N-desmethyl tamoxifen levels in women with estrogen receptor-positive breast cancer receiving adjuvant tamoxifen (20 mg/day) for 4 months. Data are expressed as means ± standard error of the mean. Panel A shows the significant difference (P <0.001) in 4-OH tamoxifen and endoxifen between extensive and poor CYP2D6 metabolizers (black bars = extensive metabolizers, white bars= intermediate metabolizers, grey bars = poor metabolizers). Panel B depicts tamoxifen and N-desmethyl tamoxifen levels in extensive, intermediate, and poor CYP2D6 metabolizers (non-significant difference) (black bars = extensive metabolizers, white bars= intermediate metabolizers, grey bars = poor metabolizers).
Some researchers found no association between CYP2C19 and tamoxifen efficacy [32, 33] , whereas others demonstrated that CYP2C19*2 in heterozygosis or homozygosis is relevant for tamoxifen therapy in advanced cancer disease and can serve as a predictive factor for survival in breast cancer patients treated with this drug [34, 35]. The frequency of CYP2C19 polymorphisms in our population was 84.4% for allele *1 and 15.6% for allele *2. A frequency of 16.5% was recently reported for allele *2 in a Hungarian general population, and a similar frequency of this CYP2C19 variant has been observed in various ethnic groups [7]. All of the CYP2D6 PMs in the present study were EMs for CYP2C19 (*1/*1 and *1/*2), which may indicate that CYP2C19 has no influence on the plasma endoxifen levels.
Our findings on CYP2D6 and CYP2C19 polymorphisms in a group of Spanish women with ER+ breast cancer are broadly similar to reports in general Caucasian populations. Without previous genotyping, incorrect treatment would likely have been ordered for the appreciable number (5.6%) of our patients who had PM phenotype (CYP2D6*4 allele in homozygosis in all cases) and were found to have lower endoxifen and 4-OH tamoxifen levels.
Our findings suggest that genotyping studies may be desirable before tamoxifen is prescribed. However, our study has some limitations. We measured tamoxifen and its main metabolites, but it was recently shown that N-desmethyl tamoxifen is converted into Z-endoxifen by the CYP2D6 enzyme and into Z'-endoxifen by an unknown hepatic enzyme, and both have different levels of anti-estrogenic activity [36]. It would be of interest to explore the relationship between the CYP2D6 genotype and the levels of these isomers. In addition, although the sample size was adequate to yield significant differences, the results of this pilot study need to be confirmed in larger samples. Further studies are in progress by our group to develop guidelines on drug dosage as a function of CYP2D6 and CYP2C19 genotypes.
Acknowledgements
The authors are grateful to Dr. Josep Solé from Roche Diagnostics S.L (Spain) for his collaboration and comments on this paper and to Richard Davies for assistance with the English version. CGLL has a postdoctoral fellowship from the Plan Propio of the University of Granada.
Abbreviations
ER+: estrogen-receptor positive; EM: extensive metabolizer; IM: intermediate metabolizer; UM: ultra-rapid metabolizer; PM: poor metabolizer; 4-OH tamoxifen: 4-hydroxy tamoxifen.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Russell RC. Bailey and Love´s short practice of surgery. Chapter on breast cancer; 23rd ed. London: Arnold. 2000
2. Del Re M, Michelucci A, Simi P. et al. Pharmacogenetics of anti-estrogen treatment of breast cancer. Cancer. Treat. Rev. 2012;38:442-450
3. O'Donnell PH, Ratain MJ. Germline pharmacogenomics in oncology: decoding the patient for targeting therapy. Mol. Oncol. 2012;6:251-259
4. Teh LK, Bertilsson L. Pharmacogenomics of CYP2D6: Molecular Genetics, interethnic differences and clinical importance. Drug. Metab. Pharmacokinet. 2012;27:55-67
5. Abraham JE, Maranian MJ, Driver KE. et al. CYP2D6 gene variants: association with breast cancer specific survival in a cohort of breast cancer patients from the United Kingdom treated with adjuvant tamoxifen. Breast. Cancer. Res. 2010;12:R64
6. Kelly CM, Pritchard KI. CYP2D6 genotype as a marker for benefit of adjuvant tamoxifen in postmenopausal women: lessons learned. J. Natl. Cancer. Inst. 2012;104:427-428
7. Rideg O, Háber A, Botz L A. et al. Pilot study for the characterization of pharmacogenetically relevant CYP2D6, CYP2C19 and ABCB1 gene polymorphisms in the Hungarian population. Cell. Biochem. Funct. 2011;29:562-568
8. Singh MS, Francis PA, Michael M. Tamoxifen, cytochrome P450 genes and breast cancer clinical outcomes. Breast. 2011;20:111-118
9. Ingelman-Sundberg M, Sim SC Gomez A. et al. Influence of cytochrome P450 polymorphisms on drug therapies: Pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacolo. Ther. 2007;116:496-526
10. Fernandez-Santander A, Luna F, Santiago C. et al. CYP2D6 polymorphism screening in a selected population of Spain (La Alpujarra): no effect of geographical isolation. Ann. Hum. Biol. 2010;37:267-273
11. Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229-243
12. De Leon J, Armstrong SC, Cozza KL. Clinical guidelines for psychiatrists for the use of pharmacogenetic testing for CYP450 2D6 and CYP450 2C19. Psychosomatics. 2006;47:75-85
13. Lammers LA, Mathijssen RHJ, Gelder T van. et al. Impact of CYP2D6-predicted phenotype on tamoxifen treatment outcome in patients with metastatic breast cancer. Br J Cancer. 2010;103:765-771
14. Rae JM, Drury S, Hayes DF. et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst. 2012;104:452-460
15. Nakamura Y, Ratain MKJ, Cox NJ. et al. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial. J Natl Cancer Inst. 2012;104:1264
16. Regan MM, Leyland-Jones B, Bouzyk M. et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: The breast International Group 1-98 trial. J Natl Cancer Inst. 2012;104:441-451
17. Lyon EG, Gastier FJ, Palomaki GE. et al. Laboratory testing of CYP2D6 alleles in relation to tamoxifen theraphy. Genet Med. 2012;14:990-1000
18. González I, Perez Picañol B, Alvarez M. et al. Study of debrisoquine hydroxylation polymorphism (CYP2D6) in the Cuban population compared to Spaniards. Med Clin (Barc). 2007;128:772-774
19. Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu. Rev. Med. 2006;57:119-137
20. Bernard S, Neville KA, Nguyen AT. et al. Interethnic differences in genetic polymorphism of CYP2D6 in the U.S. population: clinical implications. Oncologist. 2006;11:126-135
21. Menoyo A, Del Rio E, Baiget M. Characterization of variant alleles of cytochrome CYP2D6 in a Spanish population. Cell. Biochem. Funct. 2006;24:381-385
22. Sachse C, Brockmöller J, Bauer S. et al. Cytochrome P450 2D6 variants in a Caucasian population: Allele frequencies and phenotypic consequences. Am. J. Hum. Genet. 1997;60:284-295
23. Marez D, Legrand M, Sabbagh N. et al. Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics. 1997;7:193-202
24. Ramón y Cajal T, Altés A, Paré L. et al. Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res Treat. 2010;119:33-38
25. Rebsamen MC, Desmeules J, Daali Y. et al. The AmpliChip CYP450 test: cytochrome P450 2D6 genotype assessment and phenotype prediction. Pharmacogenomics J. 2009;9:34-41
26. Fuselli S, Dupanloup I, Frigato E. Molecular diversity at CYP2D6 locus in the Mediterranean region. Eur J Hum Genet. 2004;12:916-924
27. Sistonen J, Sajantila A, Lao O. et al. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenetics and Genomics. 2007;17:93-101
28. Farez-Vidal ME, Gomez-Llorente C, Blanco S. et al. Multimutational analysis of eleven cystic fibrosis mutations common in the Mediterranean areas. Clin. Chem. 2004;50:2155-2157
29. Farez-Vidal ME, Gandia-Pla S, Blanco S. et al. Multi-mutational analysis of fifteen common mutations of the glucose 6-phosphate dehydrogenase gene in the Mediterranean population. Clin. Chim. Acta. 2008;395:94-98
30. Schorth W, Antoniadou L, Fritz P. et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J. Clin. Oncol. 2007;25:5187-5193
31. Schroth W, Goetz MP, Hamann U. et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429-1436
32. Lim JS, Chen XA, Singh O. et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br. J. Clin. Pharmacol. 2011;71:737-750
33. Serrano D, Lazzeroni M, Zambon CF. et al. Efficacy of tamoxifen based on cytochrome P450 2D6, CYP2C19 and SULT1A1 genotype in the Italian tamoxifen prevention trial. Pharmacogenomics. J. 2011;11:100-107
34. Ruiter R, Bijl MJ, van Schaik RH. et al. CYP2C19*2 polymorphism is associated with increased survival in breast cancer patients using tamoxifen. Pharmacogenomics. 2010;11:1367-1375
35. Van Schaik RH, Kok M, Sweep FC. et al. The CYP2C19*2 genotyped predicts tamoxifen treatment outcome in advanced breast cancer patients. Pharmacogenomics. 2011;12:1137-1146
36. Barginear MF, Jaremko M, Peter I. et al. Increasing tamoxifen dose in breast cancer patients based on CYP2D6 genotypes and endoxifen levels: effect on active metabolite isomers and the antiestrogenic activity score. Clin Pharmacol Ther. 2011;90:605-611
Author contact
![]() Corresponding author: Dr. Carolina Gomez-Llorente, Department of Biochemistry and Molecular Biology II, Institute of Nutrition and Food Technology, Center of Biomedical Research, University of Granada, Avd/Conocimiento s/n, 18100 Armilla, Granada, Spain. Tel +34 958241000 ext.20322 Mobile: +34 686458391 Fax number: +34 958243391 E-mail: gomezlles.
Corresponding author: Dr. Carolina Gomez-Llorente, Department of Biochemistry and Molecular Biology II, Institute of Nutrition and Food Technology, Center of Biomedical Research, University of Granada, Avd/Conocimiento s/n, 18100 Armilla, Granada, Spain. Tel +34 958241000 ext.20322 Mobile: +34 686458391 Fax number: +34 958243391 E-mail: gomezlles.

 Global reach, higher impact
Global reach, higher impact