Impact Factor
ISSN: 1449-1907
Int J Med Sci 2006; 3(4):135-140. doi:10.7150/ijms.3.135 This issue Cite
Research Paper
Eradication rate of Helicobacter pylori according to genotypes of CYP2C19, IL-1B, and TNF-A
Department of Preventive Medicine / Biostatistics and Medical Decision Making, Nagoya University Graduate School of Medicine, Nagoya 466-8550, Japan
Received 2006-8-1; Accepted 2006-9-10; Published 2006-9-10
Abstract
Background: Lansoprazole, amoxicillin, and clarithromycin are commonly used drugs for eradication of Helicobacter pylori (H. pylori). A few studies reported that the eradication rate was influenced by the functional polymorphism of CYP2C19, whose product metabolizes proton pomp inhibitors including lansoprazole.
Methods: This study examined the eradication rate among 67 participants in the polymorphism study who visited Daiko Medical Center, Nagoya University from July 2004 to October 2005. The participants aged 20 to 69 years were classified into three group according to CYP2C19 genotype; rapid metabolizers (RM) with *1*1 genotype, intermediate metabolizers (IM) with *1*2 or *1*3 genotype, and poor metabolizers (PM) with *2*2, *2*3, or *3*3 genotype. For the genotype classification, G681A (681G for *1 and 681A for *2) and G636A (636G for *1 and 636A for *3) were genotyped by PCR with confronting two-pair primers (PCR-CTPP). They were also genotyped for IL-1B T-31C and TNF-A T-1031C by a duplex PCR-CTPP.
Results: The eradication rate was 70.0% for RM, 93.9% for IM, and 85.7% for PM. The difference in the rate between RM and IM+PM was statistically significant (p=0.025). The eradication rate was highest for those with IL-1B -31CC; the p value was marginal among the whole subjects (χ2=3.78, p=0.05) and not significant among the RM group (χ2=1.60, p=0.21). The genotypes of TNF-A T-1031C had no associations with the eradication rate. But among the RM group, the odd ratio (OR) of the TNF-A CT for the eradication rate relative to TT was marginally reduced (OR=0.05, 95% confidence interval, 0.002-1.19).
Conclusions: The present study confirmed the low eradication rate for RM. The reproduced finding provides evidence that the CYP2C19 genotype is useful to predict the success of the treatment. For the RM group, alternative regimens expected to be with a higher eradication rate will be recommended, especially to those with the TNF-A -1031C allele.
Keywords: Helicobacter pylori, Eradication, CYP2C19, IL-1B, TNF-A
1. Introduction
Helicobacter pylori (H. pylori) infection is an established cause of digestive diseases including ulcer, atrophic gastritis, stomach cancer, and mucosa-associated lymphoid tissue lymphoma [1-3]. Recent studies demonstrated that H. pylori eradication caused increase in platelets of patients with idiopathic thrombocytopenic purpura (ITP) [4,5]. Another study reported that the eradication may be useful to treatment of chronic urticaria, an autoimmune disease [6]. Arteriosclerosis is also presumed to be H. pylori –associated disease, though there is still controversy [7]. A study reported that infertility was associated with the seropositivity among men and women. In addition, they showed biological evidence that follicular fluid from infected women contained anti-H. pylori antibody, which reacted with spermatozoa [8].
In Japan, health insurance covers H. pylori tests and eradication treatment for digestive ulcers, but not for the other diseases. Since are convinced the effect of the eradication on ITP treatment and the possibility of stomach cancer prevention at least among those who are successfully treated for early stomach cancer [9], there are increasing demands for H. pylori eradication treatment in Japan. In an international workshop held in Sweden, “the majority of the scientific task force favored a search-and-treat strategy in first-degree relatives of gastric cancer patients and an overwhelming majority felt that a more general screen-and-treat strategy should be focused in the first instance on a population with a high incidence of H. pylori-associated diseases” [10], which may include Japan, a high stomach cancer incidence country. The eradication was reported to be effective to reduce the medical care costs in the United Kingdom [11], and in Japan [12]. These expert recommendations/opinions may partly justify the H. pylori eradication for the more generalized populations in Japan.
The treatments against H. pylori infection approved in Japanese health insurance are 7-day triple therapies of lansoprazole (60mg) + amoxicillin (1,500mg) + clarithromycin (400mg) and omeprazole (40mg) + amoxicillin (1,500mg) + clarithromycin (800mg). The reported eradication rate for the former treatment was 87.5% in 96 patients with gastric ulcer and 91.1% in 90 patients with duodenal ulcer [13]. The rate for the latter treatment was reported to be 78.8% (n=113) by Kuwayama et al at the 7th annual scientific meeting of the Japanese Society of Helicobacter Research 2001. Since lansoprazole and omeprazole are metabolized by CYP2C19 in the liver, the functional genotypes of CYP2C19 could influence the eradication rate, as well as clarithromycin resistance of H. pylori. Concerning the lansoprazole-based triple therapy, there are two studies reporting the significant difference in the rate among those with different CYP2C19 genotypes [14] or with the different genotype combinations of CYP2C19 and IL-1B [15], while other studies showed no significant association [16,17]. It was reported that those with the -511TT genotype of IL-1B encoding interleukin 1β, a potent gastric acid inhibiting proinflammatory cytokine, had a higher eradication rate relative to those with the -511CC genotype [18].
This study aimed to examine the associations of the eradication rate with the functional polymorphisms of CYP2C19 G681A (*2) and G636A (*3), IL-1B T-31C tightly linked with C-511T [19], and TNF-A T-1031C. Since TNF-α encoded by TNF-A, is also a proinflammatory cytokine inhibiting gastric acid secretion [20], we hypothesized that the functional polymorphism of TNF-A may influence the eradication rate. The TNF-A gene encoding TNF-α is known to have five polymorphism in the promoter region; G-238A, G-308A, C-857T, C-863A, and T-1031C. Among Japanese, the -238A and -308A alleles are rare and C-863A is tightly linked with T-1031C. With respect to H. pylori seroposotivity, we have reported that the decreased OR of TNF-A -1031CC for H. pylori seropositivity relative to TT was significant, but the association with C-857T was not significant [21].
This study was approved by the Ethics Committee of Nagoya University Graduate School of Medicine (approval number 155) on June 16, 2004.
2. Subjects and methods
Study subjects
Subjects were sampled from those visited Daiko Medical Center, Nagoya University, Nagoya, Japan to seek H. pylori tests and eradication between July 2004 and October 2005. The visitors aged 20 to 69 years were asked to participate in the polymorphism study. Those who agreed with a written informed consent form to provide a 7ml of blood sample and to answer a questionnaire form on lifestyle including smoking habit, were enrolled in the present study. Any genotypes were not disclosed to the participants. The study protocol was approved by the ethics committee of the Nagoya University Graduate School of Medicine.
Treatment for H. pylori infection
Lansoprazole (30mg), amoxicillin (750mg), and clarithromycin (200mg) twice a day for 7 days (LAC regimen) were prescribed for those found to be infected with H. pylori by a 13C-urea breath test or serum anti-H. pylori antibody. More than one month after the medication, a 13C-urea breath test was conducted to examine the success/failure of the eradication treatment.
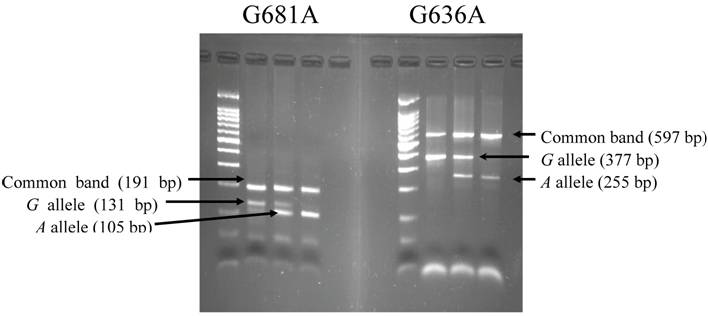
The agarose gel electrophoresis for polymorphism at G681A and G636A of CYP2C19 by PCR with confronting.
Genotyping
DNA was extracted from the buffy coat by a BioRobot® EZ1 (QIAGEN Group, Tokyo) for genotyping CYP2C19 G681A (*2) and G636A (*3), IL-1B T-31C, and TNF-A T-1031C. Genotypes were determined separately for the CYP2C19 polymorphisms by polymerase chain reaction with confronting two-pair primers (PCR-CTPP) [22]. The primers for CYP2C19 G681A and G636A were *2F1: 5' AGA GCT TGG CAT ATT GTA TCT, *2R1: 5' TAA GTA ATT TGT TAT GGG TTC CC, *2F2: 5' CCA CTA TCA TTG ATT ATT TCC CA, *2R2: 5' TCG ATT CTT GGT GTT CTT TTA C, and *3F1: 5' AAC CAG CTA GGC TGT AAT TGT, *3R1: 5' CTT GGC CTT ACC TGG ATC, *3F2: 5' ATT GTA AGC ACC CCC TGA, *3R2: 5' CAC TGA TCA GGG AGC TAA TG, respectively. The underlined are the bases of the single nucleotide polymorphism. Genomic DNA (30ng to 100ng) was used per 25μl of reaction with 0.18mM dNTPs, 12.5 pmol of each primer, 0.5 units of "AmpliTaq Gold", and 2.5μl GeneAmp 10×PCR Buffer including 15mM MgCl2 (Perkin-Elmer Corp., Foster City, CA). Amplification conditions were 10 minutes of initial denaturation at 95°C, followed by 30 cycles of 1 minute at 95°C, 1 minute at 59°C for G681A and 58°C for G636A, and 1 minute at 72°C, then a 5 minutes final extension at 72°C. The amplified DNA was visualized on a 2% agarose gel with ethidium bromide staining. A representative gel is shown in Fig. 1. The alleles were distinguished as follows: a 131-bp band for the 681G allele and a 105-bp band for 681A allele, with a 191-bp their common band, and a 377-bp band for 636G allele and a 255-bp band for 636A allele, with a 597-bp their common band. IL-1B T-31C and TNF-A T-1031C were genotyped with a duplex PCR-CTPP described previously [23].
Statistical analysis
Statistical analysis was performed using STATA Ver. 8 (College Station, TX) statistical software. The Hardy-Weinberg equilibrium, which indicates an absence of discrepancy between genotype and allele frequencies, was checked for the study subjects using a χ2 test. The odds ratios (ORs) and 95% confidence intervals (95% CIs) were adjusted for sex (dichotomous) and age (a continuous variable) using an unconditional logistic regression model.
3. Results
Table 1 describes the backgrounds of the whole visitors and participants.
From July 2004 to October 2005, 210 persons visited the center. Among them, 142 were found to be infected, 165 participated in the polymorphism study and LAC were prescribed for 93 participants with H. pylori infection (in fact, 97 were prescribed eradication drug but 4 subjected who failed the eradication by LAC at other clinics were excluded because we dosed them metronidazole instead of clarithromycin). The distribution of the IL-1B T-31C, CYP2C19 G681A and G636A genes were in Hardy-Weinberg equilibrium (χ2=0.83 and p=0.36, χ2=0.72 and p=0.40, χ2=0.54 and p=0.54, respectively), but the distribution of the TNF-A T-1031C gene was not (χ2=4.05, p=0.04). The difference of the distributions between the expected and observed from the TNF-A T-1031C gene was not substantial; 128.3 for 131, 34.4 for 29, and 2.3 for 5. The participant subjects were classified into three groups according to CYP2C19 genotype; rapid metabolizers (RM) with *1*1 genotype, intermediate metabolizers (IM) with *1*2 or *1*3 genotype, and poor metabolizers (PM) with *2*2, *2*3, or *3*3 genotype; *1*1, with the 681GG and 636GG; *1*2, with the 681GA and 636GG; *1*3 with the 681GG and 636GA; *2*2, with the 681AA and 636GG; *2*3, with the 681GA and 636GA; *3*3, with the 681GG and 636AA.
Characteristics of the visitors and the participants in the polymorphism study
| Characteristics | Visitors (n=210) n (%) | Participants (n=165) n (%) | |
|---|---|---|---|
| Age | < 30 years | 23 (11.0) | 15 (9.1) |
| 31-49 years | 52 (24.8) | 42 (25.5) | |
| 50-69 years | 126 (60.0) | 106 (64.2) | |
| 70 < years | 9 (4.3) | 2 (1.2) | |
| Sex | Males | 91 (43.3) | 74 (55.2) |
| Females | 119 (56.7) | 91 (44.9) | |
| Smokinga | Never | 160 (76.6) | 127 (77.0) |
| Former | 25 (12.0) | 19 (11.5) | |
| Current | 24 (11.5) | 19 (11.5) | |
| H. pylorib | Uninfected | 65 (31.6) | 55 (33.3) |
| Infecteted | 142 (68.6) | 110 (66.8) | |
| CYP2C19 | RM | - | 51 (30.9) |
| IM | - | 81 (49.1) | |
| PM | - | 33 (20.0) | |
| IL-1B C-31Tc | TT | - | 48 (29.3) |
| CT | - | 76 (46.3) | |
| CC | - | 40 (24.4) | |
| TNF-A C-1031T | TT | - | 131 (79.4) |
| CT | - | 29 (17.6) | |
| CC | - | 5 (3.0) |
a The information on the status of smoking was not available for one subject. b Three subjects among visitors did not take the examination on H. pylori infection. c One subject was not genotyped
The rate and odds ratios (ORs) of the eradication among the participants in the polymorphism study who were prescribed LAC and examined for the success/failure of the eradication.
| Factors | Treateda n | Eradicated n (%) | ORb 95% CI | |
|---|---|---|---|---|
| Total | 67 | 57 (85.1) | ||
| Agec | < 30 years | 1 | 0 (0.0) | |
| 31-49 years | 14 | 10 (71.4) | ||
| 50-69 years | 52 | 47 (90.4) | ||
| 70 < years | 0 | - | ||
| Sex | Females | 33 | 29 (87.9) | 1 (Reference) |
| Males | 34 | 28 (82.4) | 0.69 (0.17-2.82) | |
| CYP2C19 | RM | 20 | 14 (70.0) | 1 (Reference) |
| IM | 33 | 31 (93.9) | 6.69 (1.12-39.9) | |
| PM | 14 | 12 (85.7) | 4.35 (0.55-34.3) | |
| IM+PM | 47 | 43 (91.5) | 5.72 (1.24-26.3) | |
| IL-1B | -31TT | 17 | 13 (76.5) | 1 (Reference) |
| -31CT | 36 | 30 (83.3) | 1.36 (0.31-5.96) | |
| -31CC | 14 | 14 (100) | ∞e | |
| IL-1Bd | -31TT | 5 | 3 (60.0) | 1 (Reference) |
| -31CT | 12 | 8 (66.7) | 0.45 (0.03-7.99) | |
| -31CC | 3 | 3 (100) | ∞f | |
| TNF-A | -1031TT | 51 | 45 (85.1) | 1 (Reference) |
| -1031CT | 14 | 10 (71.4) | 0.33 (0.07-1.51) | |
| -1031CC | 2 | 2 (100) | ∞g | |
| TNF-Ad | -1031TT | 16 | 13 (81.3) | 1 (Reference) |
| -1031CT | 4 | 1 (25.0) | 0.05 (0.002-1.19) | |
| -1031CC | 0 | - | - | |
| Smoking | Never | 49 | 43 (87.8) | 1 (Reference) |
| Former | 12 | 9 (75.0) | 0.49 (0.08-3.00) | |
| Current | 6 | 5 (83.3) | 0.82 (0.07-10.0) | |
| Smokingd | Never | 16 | 12 (75.0) | 1 (Reference) |
| Former | 4 | 2 (50.0) | 2.16 (0.12-40.5) | |
| Current | 0 | - | - |
a participants in the polymorphism study who were prescribed LAC and examined for the success/failure of the eradication. b adjusted for sex and age. c p=0.012 for 2 by 3 table using a χ2 test. d Among the participants with the rapid metabolism. e,f,g Compared with reference by a χ2 test, the p values were 0.05, 0.21 and 0.61, respectively
Table 2 shows the eradication rate for 67 participants in the polymorphism study who were prescribed LAC and examined for the success/failure of the eradication. The combination of CYP2C19 IM and PM had a significantly high rate relative to RM. The eradication rate was highest for those with IL-1B -31CC but the p value was marginal (χ2=3.78, p=0.05). There were no differences in the eradication rate among those with different genotypes of TNF-A T-1031C. Among the rapid metabolizers, however, the decreased OR of TNF-A CT relative to TT was marginal (OR=0.05, 95% CI, 0.002-1.19). The OR for current and former smokers at the first visit was 0.82 (95% CI, 0.07-10.0) and 0.49 (95% CI, 0.08-3.00), respectively. During the medication, two out of six current smokers quit smoking, three continued smoking, and one did not answer about smoking habit.
4. Discussion
In the present study, CYP2C19 IM+PM showed a significantly high eradication rate relative to RM. Among the 10 failed, 6 were RM. The IL-1B T-31C polymorphism was associated with the eradication rate. Smoking during non-medication period did not affect the eradication rate. This was the first report on the possible association between the TNF-A polymorphism and the eradication rate among those with RM.
The percentage of RM among those infected with H. pylori (31.2%) was similar to that of Japanese populations; by Furuta et al (32.6% of 141 subjects) [14], by Take et al (32.5% of 249 subjects) [15], by Kawabata et al (41.3% of 80 subjects) [16], and by Sugimoto et al (38.1% of 315 controls infected with H. pylori) [24]. Those percentages are much lower than those in Europe (81.1% of 143 subjects in Italy [25] and 73.3% of 60 subjects in Germany [26]). The allele frequencies for IL-1B -31T (0.536) and TNF-A -1031T (0.871) in this study were also similar to those among Japanese (0.558 of 1,062 chromosomes [27], and 0.835 of 2,742 chromosomes [21], respectively), and lower and higher than that in Italy (0.669 and 0.768 of 1,288 chromosomes, respectively) [28].
To date, four studies examined the association of the eradication rate with the CYP2C19 genotypes. Two studies demonstrated non-significant results contrary to ours; in a randomized controlled study of lansoprazole versus rabeprazole for 187 patients with digestive ulcer by Kawabata et al [16], the eradication rate was 73% for 33 RM, 74% for 35 IM, and 83% for 12 PM in the LAC group, and in the study by Miki et al [17], 83% for 12 RM, 85% for 26 IM, and 78% for 9 PM. Although the corresponding association is controversial, our present study confirmed that the RM group relatively resistant to LAC should be treated with a more effective alternative regimen. Accordingly, the genotyping of CYP2C19 before eradication treatment could be useful to avoid the failure of treatment.
The present study indicated that the polymorphism of IL-1B T-31C tightly linked with C-511T is one of the determinants on the result of H. pylori eradication. Among the RM group, IL-1B -31CC tended to show the highest eradication rate but the p value was not significant (χ2=1.60, p=0.21). Although the association with combinations of CYP2C19 and IL-1B C-511T was reported [15], our data did not reach significance, probably due to the small number of subjects analyzed.
TNF-A T-1031C did not provide useful information to predict the result of the eradication treatment. But among the RM group, TNF-A T-1031C was useful as the indication of the success/failure of the eradication. TNF-A C-863A polymorphism linked tightly to T-1031C affects TNF-α expression; -863 A was associated with a lower serum TNF-α level.29 Previously we have reported that TNF-A -1031TT was associated with persistent H. pylori infection and hypothesized that TNF-A -1031TT produces the high level of TNF-α, resulting in low gastric acid [21]. This study also indicated that those with the RM and TNF-A -1031CT genotype could not suppress gastric acid enough to eradicate H. pylori. Because this result was based on the small numbers, studies of a larger size are needed to confirm our finding.
The problem of clarithromycin-resistant H. pylori is substantial in the eradication, but we could not determine the drug resistance because endoscopy was not conducted in our clinic. This is one of the limitations of this study. It was reported that the eradication rate was quite low among the RM group infected with clarithromycin-resistant H. pylori [14]. Take et al reported that in a mulitivariate analysis, susceptibility to clarithromycin and CYP2C19 were significant factors, with IL-1B C-511T and smoking not significant, but the association of CYP2C19 genotype was marked only among those with IL-1B -511CC [15]. The relation between the TNF-A genotype and the eradication rate according to the status of H. pylori drug resistance remains to be elucidated.
It has been well documented that smokers had a lower success probability of the eradication [30-32]. On the other hand, smokers who quit smoking during the medication reportedly had a higher eradication rate [32]. We strongly recommended to quit smoking during the medication, so two quit smoking at least during the medication and succeeded in H. pylori eradication. Since the smokers during the medication were four at most, eradication rate for smokers could not be estimated with an enough statistical power.
In conclusion, the present study confirmed the low eradication rate among the RM who were treated with LAC. The reproduced finding provides evidence that the CYP2C19 and IL-1B genotype are useful to predict the success of the treatment. For the RM group, alternative regimens expected to be with a higher eradication rate will be recommended especially to those with the TNF-A -1031C allele. Further biological investigation will be required to elucidate the association of these polymorphisms and the eradication rate. Based on the finding, we began in November 2005 to inform the participants of the CYP2C19 genotype to recommend another triple therapy (rabeprazole, metronidazole, and amoxicillin) for those with RM [16,33].
Acknowledgements
The authors are grateful to Ms. Yoko Mitsuda and Ms. Mayumi Kato for their technical assistance. This work was supported in part by a Grant-in-Aid for Cancer Research (17Shitei-6) from the Ministry of Health, Labour and Welfare of Japan.
Conflicts of interest
The authors have declared that no conflict of interest exists.
References
1. Labenz J, Borsch G. Evidence for the essential role of Helicobacter pylori in gastric ulcer disease. Gut. 1994 ;35:19-22
2. IARC. Infection with Helicobacter pylori. IARC monographs on the evaluation of the carcinogenic risks to humans. Lyon, France: International Agency for Research on Cancer. 1994:177-241
3. Wundisch T, Thiede C, Morgner A. et al. Long-term follow-up of gastric MALT lymphoma after Helicobacter pylori eradication. J Clin Oncol. 2005 ;23:8018-24
4. Kohda K, Kuga T, Kogowa K. et al. Effective of Helicobacter pylori eradication on platelet recovery in Japanese patients with chronic idiopathic thrombocytopenic purpura and secondary autoimmune thrombocytopenic purpura. Br J Haematol. 2002 ;118:584-8
5. Suzuki T, Matsushima M, Masui A. et al. Effect of Helicobacter pylori eradication in patients with chronic idiopathic thrombocytopenic purpura - a randomized controlled trial. Am J Gastroenterol. 2002 ;100:1271-3
6. Fukuda S, Shimoyama T, Umegaki N. et al. Effect of Helicobacter pylori eradication in the treatment of Japanese patients with chronic idiopathic urticaria. J Gastroenterol. 2004 ;29:827-30
7. Franceschi F, Leo D, Fini L. et al. Helicobacter pylori infection and ischaemic heart disease: an overview of the general literature. Dig Liver Dis. 2005 ;37:301-8
8. Figura N, Piomboni P, Ponzetto A. et al. Helicobacter pylori infection and infertility. Eur J Gastroenterol Hepatol. 2002 ;14:663-9
9. Uemura N, Mukai T, Okamoto S. et al. Effect of Helicobacter pylori eradication on subsequent development of cancer after endoscopic resection of early gastric cancer. Cancer Epidemiol Biomarkers Prev. 1997 ;6:639-42
10. Malfertheiner P, Sipponen P, Naumann M. et al. H. pylori-Gastric Cancer Task Force. Helicobacter pylori eradication has the potential to prevent gastric cancer: a state-of-the-art critique. Am J Gastroenterol. 2005 ;100:2100-115
11. Ford A.C, Forman D, Bailey A.G. et al. A community screening program for Helicobacter pylori saves money: 10-year follow-up of a randomized controlled trial. Gastroenterology. 2005 ;129:1910-7
12. Sone Y, Kumada T, Kiriyama S. et al. Helicobacter pylori eradication reduces medical care costs: a community-based study. Helicobacter Res. 2002 ;6:299-304
13. Asaka M, Sugiyama T, Kato M. et al. A multicenter, double-blind study on triple therapy with lansoprazole, amoxicillin and clarithromycin for eradication of Helicobacter pylori in Japanese peptic ulcer patients. Helicobacter. 2001 ;6:254-61
14. Furuta T, Sagehashi Y, Shirai N. et al. Influence of CYP2C19 polymorphism and Helicobacter pylori genotype determined from gastric tissue samples on response to triple therapy for H pylori infection. Clin Gastroenterol Hepatol. 2005 ;3:564-73
15. Take S, Mizuno M, Ishiki K. et al. Interleukin-1β genetic polymorphism influences the effect of cytochrome P 2C19 genotype on the cure rate of 1-week triple therapy for Helicobacter pylori infection. Am J Gastroenterol. 2003 ;98:2403-8
16. Kawabata H, Habu Y, Tomioka H. et al. Effect of different proton pump inhibitors, differences in CYP2C19 genotype and antibiotic resistance on the eradication rate of Helicobacter pylori infection by a 1-week regimen of proton pump inhibitor, amoxicillin and clarithromycin. Aliment Pharmacol Ther. 2003 ;17:259-65
17. Miki I, Aoyama N, Sakai T. et al. Impact of clarithromycin resistence and CYP2C19 genetic polymorphism on treatment efficacy of Helicobacter pylori infection with lansoprazole- or rabeprazole-based triple therapy in Japan. Eur J Gastroenterol Hepatol. 2003 ;15:27-33
18. Furuta T, Shirai N, Xiao F. et al. Polymorphism of interleukin-1B affects the eradication rates of Helicobacter pylori by triple therapy. Clin Gastroenterol Hepatol. 2004 ;2:22-30
19. Hamajima N, Matsuo K, Saito T. et al. Interleukin 1 polymorphisms, lifestyle factors, and Helicobacter pylori infection. Jpn J Cancer Res. 2001 ;92:383-9
20. Beales I.L, Calam J. Interleukin 1 beta and tumour necrosis factor alpha inhibit acid secretion in cultured rabbit parietal cells by multiple pathways. Gut. 1998 ;42:227-34
21. Hamajima N, Shibata A, Katsuda N. et al. Subjects with TNF-A-857TT and -1031TT genotypes showed the highest Helicobacter pylori seropositive rate compared with those with other genotypes. Gastric Cancer. 2003 ;6:230-6
22. Hamajima N, Saito T, Matsuo K. et al. Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping. Jpn J Cancer Res. 2000 ;91:865-8
23. Atsuta Y, Kawase H, Hamajima N. et al. Use of duplex PCR-CTPP methods for CYP2E1 RsaI/IL-2 T-330G and IL-1B C-31T/TNF-A T-1031C polymorphisms. Mol Diagn. 2005 ;9:89-94
24. Sugimoto M, Furuta T, Shirai N. et al. Poor metabolizer genotype status of CYP2C19 is a risk factor for developing gastric cancer in Japanese patients with Helicobacter pylori infection. Aliment Pharmacol Ther. 2005 ;22:1033-40
25. Sapone A, Vaira D, Trespidi S. et al. The clinical role of cytochrome P450 genotypes in Helicobacter pylori management. Am J Gastroenterol. 2003 ;98:1010-15
26. Timm R, Kaiser R, Lotsch J. et al. Association of cyclophosphamide pharmacokinetics to polymorphic cytochrome P450 2C19. Pharmacogenomics J. 2005 ;5:365-73
27. Hamajima N, Ito H, Matsuo K. et al. Helicobacter pylori seropositivity, the interleukin 1B polymorphism, and smoking among first-visit outpatients. Asian Pac J Cancer Prev. 2002 ;3:23-8
28. Zambon CF, Basso D, Navaglia F. et al. Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine. 2005 ;29:141-52
29. Skoog T, van't Hooft FM, Kallin B. et al. A common functional polymorphism (C-->A substitution at position -863) in the promoter region of the tumour necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet. 1999 ;8:1443-9
30. Kamada T, Haruma K, Komoto K. et al. Effect of smoking and histological gastritis severity on the rate of H. pylori eradication with omeprazole, amoxicillin and clarithromycin. Helicobacter. 1999 ;4:204-10
31. Perri F, Villani M.R, Festa V. et al. Predictors of failure of Helicobacter pylori eradication with the standard “Maastricht triple therapy. Aliment Pharmacol Ther. 2001 ;15:1009-23
32. Matsuo K, Hamajima N, Ikehara Y. et al. Smoking and polymorphisms of fucosyltransferase gene Le affect success of H. pylori eradication with lansoprazole, amoxicillin, and clarithromycin. Epidemiol Infect. 2003 ;130:227-33
33. Saitoh T, Fukushima Y, Otsuka H. et al. Effects of rabeprazole, lansoprazole and omeprazole on intragastric pH in CYP2C19 extensive metabolizers. Aliment Pharmacol Ther. 2002 ;16:1811-17
Author contact
![]() Correspondence to: Yasuyuki Goto, M.D., Ph.D., Department of Preventive Medicine / Biostatistics and Medical Decision Making, Nagoya University Graduate School of Medicine, 65 Tsurumai-cho, Showa-ku, Nagoya 466-8550 Japan, TEL:+81-52-744-2132, FAX:+81-52-744-2971, e-mail: y-gotonagoya-u.ac.jp
Correspondence to: Yasuyuki Goto, M.D., Ph.D., Department of Preventive Medicine / Biostatistics and Medical Decision Making, Nagoya University Graduate School of Medicine, 65 Tsurumai-cho, Showa-ku, Nagoya 466-8550 Japan, TEL:+81-52-744-2132, FAX:+81-52-744-2971, e-mail: y-gotonagoya-u.ac.jp

 Global reach, higher impact
Global reach, higher impact