Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(8):1875-1884. doi:10.7150/ijms.108527 This issue Cite
Research Paper
Gene Signature-Based Prognostic Model for Acute Myeloid Leukemia: The Role of BATF, EGR1, PD-1, PD-L1, and TIM-3
1. Key Laboratory for Regenerative Medicine of Ministry of Education, Institute of Hematology, School of Medicine, Jinan University, Guangzhou 510632, China.
2. Department of Hematology, First Affiliated Hospital, Jinan University, Guangzhou 510632, China.
3. Department of Hematology, Guangzhou First People's Hospital, South China University of Technology, Guangzhou 510180, China.
4. Central People's Hospital of Zhanjiang, Zhanjiang, China.
5. Zhanjiang Key Laboratory of Leukemia Pathogenesis and Targeted Therapy Research, Zhanjiang, China.
# Yupei Zhang and Zhixi Chen contributed equally to this work.
Received 2024-12-10; Accepted 2025-3-5; Published 2025-3-19
Abstract
Background: Acute myeloid leukemia (AML) is a malignancy of hematopoietic stem and progenitor cells, with T cell exhaustion linked to poor outcomes. Our previous research has shown that basic leucine zipper ATF-like transcription factor (BATF) and early growth response 1 (EGR1) play a role in chimeric antigen receptor T (CAR-T) cell exhaustion during AML tumor elimination. However, the roles of BATF and EGR1 and their association with immune checkpoint genes in AML prognosis remain underexplored.
Methods: Bone marrow (BM) samples from 92 newly diagnosed AML patients at our clinical center (JUN-dataset) were analyzed to detect the expression levels of BATF, EGR1, programmed cell death 1 (PD-1), programmed death-ligand 1 (PD-L1), T cell immunoglobulin and mucin domain-containing protein 3 (TIM3) together with conducting a prognostic assessment. Our findings were validated using RNA sequencing data from 155 AML patients from the TCGA database and 199 AML patients from the Beat-AML database.
Results: High BATF expression correlated with poor overall survival (OS) (P = 0.030), whereas high EGR1 expression indicated a favorable prognosis (P = 0.040). Patients with high BATF and low EGR1 expression had worst outcomes (P < 0.001). Among those receiving allogenic hematopoietic stem cell transplantation (allo-HSCT), high BATF expression was linked to shorter OS (P = 0.004). Moreover, a prognostic model incorporating BATF, EGR1, PD-1, PD-L1, and TIM-3 calculated a risk score, with high-risk patients demonstrating significantly shorter OS than low-risk patients in both total AML patients and allo-HSCT recipients (P < 0.001). Similar results were found in both the TCGA and Beat-AML datasets.
Conclusions: We establish a prognostic model based on BATF, EGR1, PD-1, PD-L1, and TIM-3 expression that effectively predicts survival outcomes for AML patients and allo-HSCT recipients. This model may provide valuable insights for prognosis assessment and treatment strategies.
Keywords: BATF, EGR1, allogenic hematopoietic stem cell transplantation, acute myeloid leukemia, T cell exhaustion
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous malignant disease characterized by abnormal proliferation and differentiation blockade of myeloid hematopoietic stem and progenitor cells [1]. Despite ongoing improvements in treatment strategies over recent years, overall outcomes for AML patients have improved modestly with refractoriness and high relapse rates remaining major clinical challenges [2-6]. The 5-year survival rate for AML patients remains below 35% with patients over 60 years of age having a 5-year survival rate of less than 20% [7-9]. With the rapid development and widespread application of next-generation sequencing (NGS) technologies, molecular biology has become an essential foundation for diagnosis, treatment decision-making, and prognosis assessment. The inclusion of genes such as TP53, FLT3, and NPM1 in risk evaluation systems has improved the accuracy of risk stratification [10-12]. However, approximately 40-50% of patients in the current risk stratification system are classified as intermediate-risk, where significant variability in prognosis exists. Existing stratification criteria struggle to accurately predict treatment response and long-term prognosis [13-16]. To optimize the current risk stratification system, more precise predictive models are needed. Therefore, it is worth exploring novel biomarkers to assess prognosis and identify new therapeutic targets.
Continuous stimulation by tumor antigens can alter the phenotype and function of T cells, ultimately driving them into an exhausted state. This state is characterized by the upregulation of inhibitory receptors, such as programmed cell death 1 (PD-1), programmed death-ligand 1 (PD-L1), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), and T cell immunoglobulin and mucin domain-containing protein 3 (TIM3), impaired cell proliferation and cytokine secretion, compromised immune memory function, and metabolic dysregulation. Together, these changes weaken the immune response against tumors and adversely affect patient prognosis [17, 18]. Increasing data have shown that T cell exhaustion is a key factor contributing to poor prognosis in AML patients [19, 20]. In our previous research, we observed that the expression levels of these inhibitory receptor genes are closely correlated with the prognosis of patients with hematologic malignancies [21, 22]. However, the correlation between immune checkpoint genes and genes that regulate T cell function remains unknown.
Basic leucine zipper ATF-like transcription factor (BATF) is an important transcription factor that is involved in immune regulation, cancer initiation and progression, and inflammation, and it plays a key regulatory role in the exhaustion of effector T cells [23, 24]. High BATF expression is thought to inhibit the cytotoxic function of chimeric antigen receptor T (CAR-T) cells, promoting their transition into an exhausted state and ultimately weakening their antitumor efficacy [25-27]. While the transcription factor early growth response 1 (EGR1), plays an important role in T cell differentiation and activation, upregulation of EGR1 can enhance T cell activity and immune responsiveness, suggesting that modulating this gene may be a potential approach for boosting T cell functionality and delaying exhaustion [28, 29]. In our previous research on CAR-T cell-mediated elimination of AML tumor cells, we found that BATF downregulation and EGR1 upregulation are closely associated with reduced CAR-T cell exhaustion and enhanced cell functionality [30].
Based on these findings, further investigation into the roles of key factors such as BATF and EGR1 in the prognosis of AML patients, as well as their relationships with immune checkpoint molecules, may provide new insight into the molecular mechanisms underlying T cell exhaustion. In this study, we first analyzed bone marrow (BM) samples collected from 92 AML patients at our clinical center, examining the association between the BATF and EGR1 expression levels and patient prognosis with a particular focus on outcomes for patients receiving allogenic hematopoietic stem cell transplantation (allo-HSCT). Additionally, we developed a predictive model that integrates BATF, EGR1, and immune checkpoint gene (PD-1, PD-L1, and TIM-3) expression to evaluate the combined impact of these factors on prognosis. We subsequently validated the generalizability of our findings using two publicly available datasets (TCGA and Beat-AML).
Materials and methods
BM samples
Bone marrow samples from 92 newly diagnosed AML patients were collected at our clinical center from January 1, 2013 to December 31, 2023, forming the JUN-dataset. Inclusion criteria: new diagnosis of AML based on the Chinese guidelines for the diagnosis and treatment of Acute Myeloid Leukemia [31]. Corresponding clinical characteristics, including gender, age, treatment regimen, survival time, and survival status, are detailed in Table S1. Follow-up was completed on August 31, 2024. The OS time was defined as the time from the date of diagnosis to death or the last follow-up. This study was conducted in accordance with the Helsinki Declaration and approved by the Ethics Committee of the First Affiliated Hospital of Jinan University. All participants signed informed consent forms.
Publicly available datasets
The TCGA dataset, which includes RNA-seq data and clinical information from 155 AML samples, was downloaded from the UCSC Xena website (https://xenabrowser.net/datapages/) [32, 33]. The Beat-AML dataset, comprising RNA-seq data and clinical information from 199 AML samples, was obtained from the Beat-AML database (http://www.vizome.org/aml/) [34]. The RNA-seq data were presented as log2(RPKM + 1), and the clinical information encompassed age, gender, survival time, survival status, white blood cell count, and treatment (Table S1).
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from mononuclear cells from BM samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The RNA was reverse transcribed into complementary DNA (cDNA) using a reverse transcription kit (Promega Corporation, Madison, Wisconsin, USA) [35, 36]. The relative expression levels of BATF, EGR1, PD-1, PD-L1, and TIM-3 were detected by qRT-PCR using a qRT-PCR kit (TIANGEN, Beijing, China). The qRT-PCR reaction procedures were as follows: preincubation, 95°C for 10 min and amplification, 95°C for 10 sec and 60°C for 20 sec for a total of 40 cycles. β-actin was selected as an internal control [37]. The qRT-PCR primer sequences are shown in Table S2. The results are expressed as 2^(-ΔCT).
Statistical analysis
Statistical analyses were performed using SPSS (version 25.0, IBM) and R (version 4.4.1, https://www.r-project.org/). The high or low level of gene expression data was determined by the optimal cutoff point, which is determined by the X-tile software (version 3.6.1) [38, 39]. Differences in Kaplan-Meier curves were compared by utilizing the log-rank test via the R package "survival". Univariate and multivariate Cox regression analyses were performed using the R packages " survminer" and "survival". The area under the curve (AUC) in receiver operating characteristic (ROC) curves was determined by the R package "pROC". A p-value < 0.05 was considered statistically significant.
Results
High BATF or low EGR1 expression is associated with poor OS in AML patients
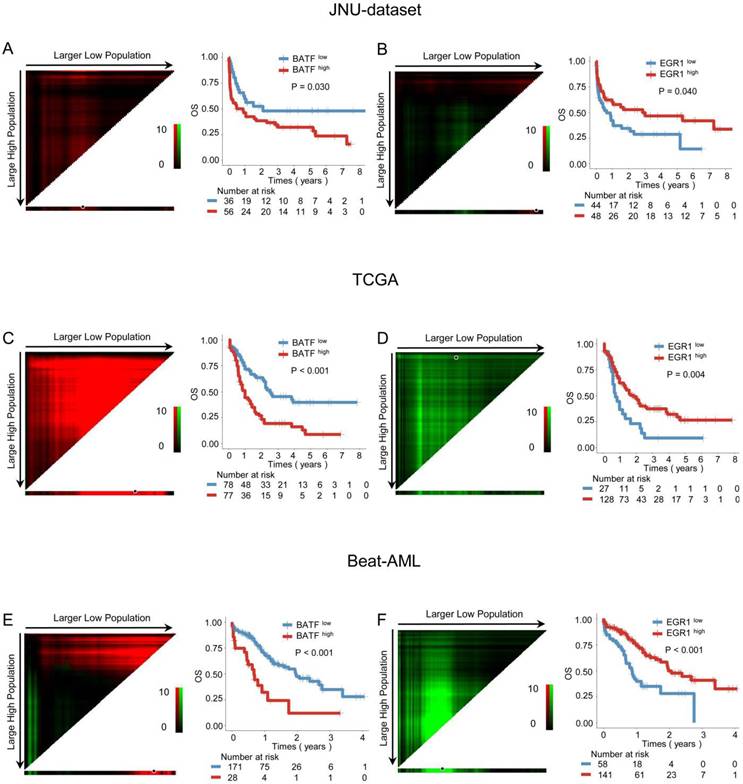
Our previous research found that a BRD4 inhibitor could reverse the exhaustion of CAR-T cells in killing AML cells by downregulating BATF and upregulating EGR1. Therefore, we aim to further investigate the impact of the BATF and EGR1 expression levels on prognosis for AML patients. Kaplan-Meier analysis demonstrated that high BATF expression was associated with poorer OS (3-year OS: 37.50% vs. 52.78%, P = 0.030) (Figure 1A), while high EGR1 expression was also linked to longer OS (3-year OS: 52.08% vs. 34.09%, P = 0.040) (Figure 1B). Furthermore, when age, gender, white blood cell counts at diagnosis, treatment choice, BATF expression, and EGR1 expression were included in univariate and multivariate Cox regression for survival analysis, the results indicated that BATF and EGR1 are independent prognostic predictors of OS (HR = 2.656, 95% CI: 1.460-4.832, P = 0.001; HR = 2.092, 95% CI: 1.198-3.655, P = 0.009) (Table S3).
The above results were further verified in AML patient cohorts from database. In the TCGA cohort, AML patients with higher BATF expression had poorer prognosis (3-year OS: 27.27% vs. 56.41%, P < 0.001) (Figure 1C), Conversely, higher EGR1 expression was linked to better prognosis (3-year OS: 46.88% vs. 18.52%, P = 0.004) in AML patients (Figure 1D). Similarly, in the Beat-AML cohort, high BATF expression was also correlated with poor outcome (3-year OS: 35.71% vs. 63.16%, P < 0.001) (Figure 1E), while high expression of EGR1 was associated with good outcome (3-year OS: 66.67% vs. 39.66%, P < 0.001) (Figure 1F). Next, we conducted univariate and multivariate Cox regression analyses including age, sex, white blood cell counts at diagnosis, treatment choice, BATF expression, and EGR1 expression in the TCGA and Beat-AML cohorts separately. The results demonstrated that BATF and EGR1 are an also independent prognostic factors for OS (TCGA: HR = 2.287, 95% CI: 1.474-3.550, P < 0.001; HR = 2.227, 95% CI: 1.357-3.657, P = 0.002; Beat-AML: HR = 3.282, 95% CI: 1.855-5.810, P < 0.001; HR = 2.506, 95% CI: 1.585-3.961, P < 0.001) (Table S4).
AML patients with high BATF expression together with low EGR1 together have the poorest prognosis
To investigate the role of BATF and EGR1 co-expression in predicting OS for AML patients, we conducted a combined group analysis. In the JUN-dataset cohort, AML patients with high BATF expression and low EGR1 expression had the poorest outcomes (3-year OS: 26.92% vs. 44.90% and 64.71%, P = 0.02) (Figure 2A). In the TCGA cohort, high BATF expression together with low EGR1 expression was associated with the poorest OS (3-year OS: 6.67% vs. 32.43% and 60.60%, P < 0.001) (Figure 2B). Similarly, in the Beat-AML cohort, patients with high BATF expression and low EGR1 expression had the shortest OS (3-year OS: 18.18% vs. 46.15% and 69.92%, P < 0.001) (Figure 2C). Through univariate and multivariate Cox regression analyses, high BATF and low EGR1 expression was able to predict poor OS of AML patients in the JUN-dataset (HR = 6.295, 95% CI: 2.431-16.301, P < 0.001) (Figures 2D, E). Similar results were also found in the TCGA and Beat-AML cohorts (TCGA: HR = 5.365, 95% CI: 2.720-10.583, P < 0.001; Beat-AML: HR = 5.638, 95% CI: 2.607-12.191, P < 0.001) (Table S5).
High BATF expression was associated with poor OS in AML patients receiving allo-HSCT
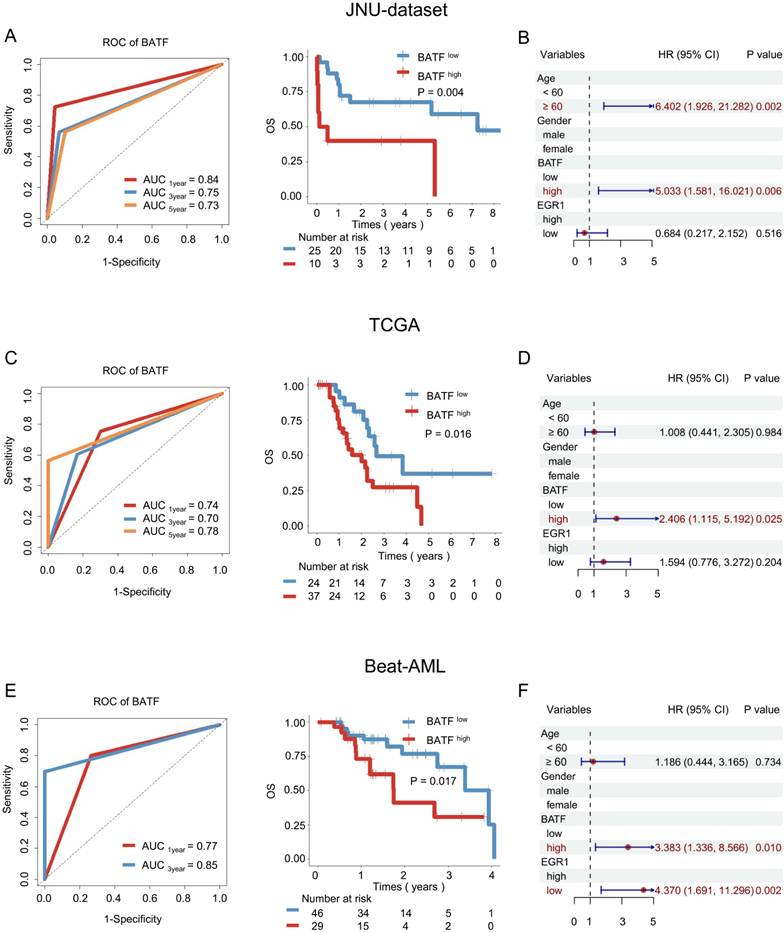
To further elucidate the potential of the BATF and EGR1 gene expression levels as prognostic biomarkers for AML patients undergoing allo-HSCT, we conducted an in-depth analysis of a cohort of allo-HSCT recipients. In the JUN-dataset, ROC analysis indicated that BATF has strong prognostic predictive power (AUC3year = 0.75) (Figure 3A). Kaplan-Meier analysis demonstrated that high BATF expression indicates poorer prognosis (3-year OS: 40.00% vs. 68.00%, P = 0.004) (Figure 3A). Furthermore, when age, gender, BATF expression, and EGR1 expression were included in univariate and multivariate Cox regression for survival analysis, BATF emerged as an independent prognostic factor for OS (HR = 5.033, 95%CI: 1.581-16.021, P = 0.006) (Figure 3B and Figure S1B).
Prognostic analysis of BATF and EGR1 expression in AML patients. A-B: Correlation between the expression levels of BATF or EGR1 and overall survival (OS) in the JUN-dataset. (left panel) The optimal cut-point for BATF or EGR1 was determined by X-tile software (version 3.6.1), which was shown as the highest pixel. (right panel) Kaplan-Meier curve was plotted according to the optimal cut-point. Blue and red indicate low and high expression of BATF or EGR1, which were plotted in Kaplan-Meier curves (top) with the number at risk AML patients (bottom). C-F: Relationship between the expression level of BATF or EGR1 and OS in the TCGA (C, D) and Beat-AML (E, F) datasets.
Co-expression of the BATF and EGR1 genes is associated with poor OS in AML patients. A-C: Kaplan-Meier curves are shown for different BATF and EGR1 combinations in the JUN-dataset (A). Kaplan-Meier curves are shown for different BATF and EGR1 combinations in the TCGA (B) and Beat-AML (C) datasets. According to the optimal cut-off value, the BATF and EGR1 genes were divided into low BATF expression and high EGR1 expression (red line), high BATF expression and low EGR1 expression (yellow line), low BATF expression and low EGR1 expression or high BATF expression and high EGR1 expression (blue line), which were plotted in Kaplan-Meier curves (top) with the number at risk AML patients (bottom). D-E: Univariate and multivariate Cox regression analysis of BATF and EGR1 co-expression in JUN-dataset.

To validate the above findings, further analysis was conducted using external databases. In the TCGA cohort of AML patients who received allo-HSCT, BATF demonstrated good prognostic predictive ability (AUC3year = 0.70) (Figure 3C), with high BATF expression associated with poorer survival (3-year OS: 27.27% vs. 56.41%, P = 0.016) (Figure 3C). Analysis of the Beat-AML cohort further confirmed the robust prognostic predictive power of BATF (AUC3year = 0.85) (Figure 3E), where high BATF expression was linked to worse OS (3-year OS: 35.71% vs. 63.16%, P = 0.017) (Figure 3E). When age, sex, BATF expression, and EGR1 expression were included in univariate and multivariate Cox regression for survival analysis, BATF was identified as an independent prognostic factor for OS (TCGA: HR = 2.406, 95% CI: 1.115-5.192, P = 0.025; Beat-AML: HR = 3.383, 95% CI: 1.336-8.566, P = 0.010) (Figures 3D, F and Figures S1D, F).
In the JUN-dataset, possibly due to the small sample size, Kaplan-Meier analysis indicated no statistically significant correlation between EGR1 expression and survival outcome for transplant patients (P > 0.05) (Figure S1A). In the Beat-AML cohort, patients expressing high levels of EGR1 demonstrated significantly improved survival outcome (P = 0.003) (Figure S1E). Analysis of the TCGA cohort showed that patients with elevated EGR1 expression exhibited a notable trend toward favorable prognosis, although this trend did not reach statistical significance (Figure S1C).
A model based on BATF, EGR1, and immune checkpoint gene expression can predict the prognosis of AML patients
It is known that upregulation of immune checkpoint factors is related to T cell exhaustion and may influence clinical outcome in AML. Therefore, we next sought to assess the relationship between the weighted combinations of BATF, EGR1, PD-1, PDL-1, TIM-3, and survival outcome. Multivariate Cox regression analysis was performed with these five genes.
BATF was associated with prognosis in AML patients undergoing allo-HSCT. A: ROC curve (left panel) and overall survival analysis (right panel) of BATF in the JUN-dataset. According to the optimal cut-off value, the BATF genes was divided into low BATF expression (blue line) and high BATF expression (red line), which were plotted in Kaplan-Meier curves (top) with the number at risk AML patients (bottom). B: Multivariate Cox regression analysis of allo-HSCT patients in the JUN-dataset. C: ROC curve (left panel) and overall survival analysis (right panel) of BATF in the TCGA dataset. D: Multivariate Cox regression analysis of allo-HSCT patients in the TCGA dataset. E: ROC curve (left panel) and overall survival analysis (right panel) of BATF in the Beat-AML dataset. F: Multivariate Cox regression analysis of allo-HSCT patients in the Beat-AML dataset.
A weighted combination of BATF, EGR1, and immune checkpoint gene expression levels is associated with AML patient prognosis. A: Radar plot showing the contribution of the BATF, EGR1, PD-1, PDL-1, and TIM-3 genes to OS in the JUN-dataset, which was determined by the coefficient β in the multivariate Cox regression model. Risk score = β1* (BATF expression) + β2* (EGR1 expression) + β3* (PD-1 expression) + β4* (PDL-1 expression) + β5* (TIM-3 expression). B: OS analysis of low-risk and high-risk score based on the combination of BATF, EGR1, PD-1, PDL-1, and TIM-3 in the JUN-dataset for total patients (left panel) and those receiving allo-HSCT (right panel). C-D: Overall survival (OS) analysis of low-risk and high-risk scores calculated based on the prognostic model in the TCGA (C) and Beat-AML (D) datasets for total patients (left panel) and those receiving allo-HSCT (right panel). According to the optimal cut-off value, the risk score was divided into a low risk score (blue line) and a high risk score (red line), which were plotted in Kaplan-Meier curves (top) with the number at risk AML patients (bottom).
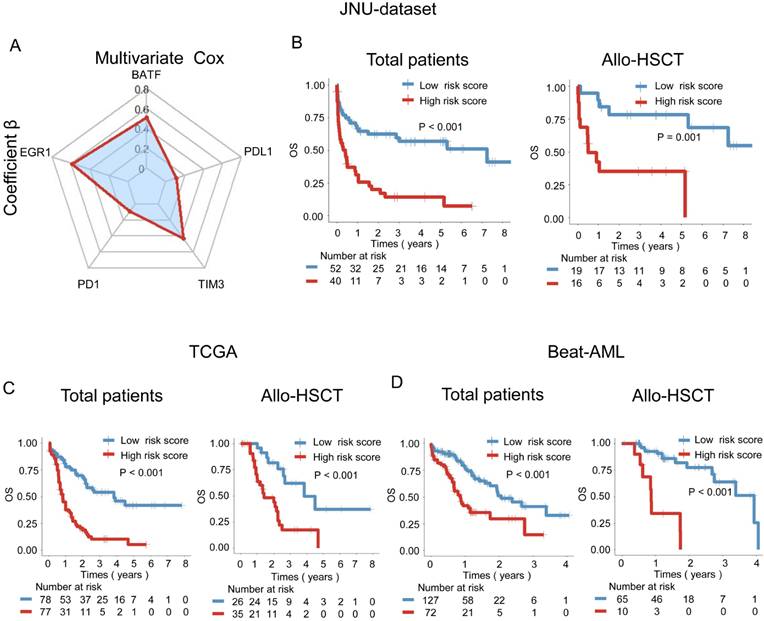
The following formula can be used to calculate the risk score of each patient according to the coefficients in our center's dataset: risk score = 0.60 × (BATF expression level) + 0.69 × (EGR1 expression level) + 0.10 × (PD-1 expression level) + 0.13 × (PDL-1 expression level) + 0.50 × (TIM-3 expression level) (Figure 4A). Importantly, patients with AML were divided into low-risk and high-risk score subgroups according to the optimal cut-point for the risk score. Patients with high-risk scores had significantly shorter OS than patients with a low-risk scores in the JUN-dataset (P < 0.001) (Figure 4B). Additionally, among patients who received allo-HSCT, those with high-risk scores had significantly poorer OS (P = 0.001) (Figure 4B). The results were further validated using the TCGA dataset, where high-risk patients also exhibited significantly poorer prognosis compared to low-risk patients (P < 0.001) (Figure 4C). Among patients who received allo-HSCT, those with high-risk scores had significantly shorter OS (P < 0.001) (Figure 4C). Similarly, in the Beat-AML cohort, high-risk patients demonstrated worse survival in both the overall patient population and the allo-HSCT subgroup (P < 0.001) (Figure 4D).
Discussion
T cell exhaustion, which leads to T cell dysfunction, is a significant factor contributing to the poor prognosis of AML patients [40, 41]. While the contribution of different exhaustion-related genes appears to be relatively different, defining the weight of single gene or the combined weight of such genes for T cell dysfunction as well as their association with prognosis for AML patients is worthwhile. Recent studies have identified BATF as a crucial regulator of T cell exhaustion, and the transcription factor EGR1 plays an important role in T cell differentiation and activation [27, 29]. Although there are a number of studies indicating that higher expression and co-expression of immune checkpoint genes such as PD-1, PD-L1, CTLA-4, and TIM-3, which induce T cell exhaustion, are associated with poor clinical outcomes for AML patients [42, 43] , it remains unclear whether alterations in T cell function genes alone or in cooperation with immune checkpoint factors contribute to T cell exhaustion and influence the clinical outcome of AML patients.
In this study, we first analyzed BM samples from 92 newly diagnosed AML patients at our center and observed high BATF expression and low EGR1 expression. Our previous study, indicated that the upregulation of BATF and the downregulation of EGR1 were closely associated with worsened CAR-T cell exhaustion and reduced cellular function. Additionally, several studies by other researchers have independently linked BATF or EGR1 to T cell or CAR-T cell functionality [44, 45]. Thus, we further investigated the association between alterations in the expression of both genes and clinical outcomes of AML patients. Indeed, we discovered that high BATF expression or low EGR1 expression alone was closely associated with poor OS. Moreover, combined analysis revealed that patients with high BATF and low EGR1 expression demonstrated significantly worse prognostic outcomes. This finding was also validated in two AML databases, suggesting that these genes may provide valuable insights for assessing AML prognosis.
It is recognized that one of the important influencing factors for overall survival is treatment strategy. Currently, allo-HSCT remains the only curative option for AML [46]. T cell dysfunction is one of the reasons for AML relapse after allo-HSCT, and restoring and maintaining normal T cell function are important approaches for eliminating minimal residual disease (MRD) and preventing disease relapse [47, 48]. Thus, alterations in T cell-regulating genes can serve as potential biomarkers evaluating the T cell immune state. Consequently, we conducted further analysis of an AML subgroup of patients who underwent allo-HSCT at our center. Significantly, BATF had strong prognostic predictive power with high expression of BATF associated with shorter survival for AML patients after allo-HSCT. As expected, high EGR1 expression was related to better prognosis in the allo-HSCT group, while these results were revealed from the database analysis. In our center's cohort, significant association couldn't observe between EGR1 expression and overall survival in the allo-HSCT group, the reason may be attributed to the relatively small sample size within this subgroup, which compromises to accurately identify any underlying correlations. Further extent of the cohort and confirmation of the results are needed. Overall, these findings may at least provide a useful reference for pre-transplant prognostic risk assessment for AML patients undergoing allo-HSCT.
The upregulation of immune checkpoint factors is known to be associated with T cell exhaustion and may impact the prognostic survival of AML patients. Our previous studies have also indicated that the expression levels of immune checkpoint genes were correlated with the prognosis of patients with hematological malignancies [21]. In this study, we further evaluated the relationship between the weighted combination of five genes (BATF, EGR1, PD-1, PD-L1, and TIM-3) and survival outcome. Using data from our center, we constructed a prognostic risk model, which indicated that high-risk score patients have significantly shorter survival times than low-risk score patients. Among patients receiving allo-HSCT, those with lower risk scores demonstrated improved survival outcomes. This finding was validated with two external databases. The impact of BATF and EGR1 on AML prognosis may be related to T cell exhaustion mechanisms, providing valuable insight for developing novel therapeutic targets aimed at T cell exhaustion. Overall, these findings highlight that the BATF and EGR1 expression levels are independent prognostic survival factors for AML patients. Elevated BATF expression is associated with poor prognosis in patients undergoing allo-HSCT. A prognostic model based on BATF, EGR1, and immune checkpoint gene expression effectively predicts prognosis for AML patients and those receiving allo-HSCT.
Nevertheless, the limitations of this study might include 1) as a single-center study conducted at our institution, it has a relatively small sample size and inherent selection bias and 2) further experiments are needed to elucidate the specific mechanisms by which BATF and EGR1 regulate immune checkpoint genes in AML patients. 3) In this study, we tried to find the predicted factor on gene expression levels. As post-transcriptional, translational regulation and protein modification occur, protein and gene expression levels often differ. More research is needed to explore how the proteins translated from these genes affect prognosis in AML patients.
Conclusions
We initially found that AML patients with high BATF expression or low EGR1 expression have shorter survival times. Combining previous findings of alterations in immune checkpoint factors in AML, we for the first time established a prognostic model based on BATF, EGR1, PD-1, PD-L1, and TIM-3 that effectively predicts survival outcomes for both AML patients and those receiving allo-HSCT. Our findings may offer valuable insights for prognosis assessment, monitoring disease progression, and informing treatment strategies.
Abbreviations
AML: Acute myeloid leukemia; BATF: basic leucine zipper ATF-like transcription factor; EGR1: early growth response 1; CAR-T: chimeric antigen receptor T; allo-HSCT: allogenic hematopoietic stem cell transplantation; PD-1: programmed cell death 1; PD-L1: programmed death-ligand 1; CTLA-4: cytotoxic T-lymphocyte-associated protein 4; TIM3: T cell immunoglobulin and mucin domain-containing protein 3; BM: bone marrow.
Supplementary Material
Supplementary figure and tables.
Acknowledgements
We would like to thank the contributors to the TCGA and the Beat-AML databases for the available data.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 82293632, 82293630 and 92474101) and Fundamental Research Funds for the Central Universities (No. 21623121).
Author contributions
YPZ and ZXC analyzed the data, performed the experiments, and wrote the manuscript. JMZ and SHC assisted with the data interpretation. LYZ and JC assisted with clinical sample collection. YQL, SNS, and CTC contributed to the concept development, study design, and editing of the manuscript. All authors read and approved the final manuscript.
Data availability
The TCGA and Beat-AML datasets used in this study was acquired from the UCSC Xena website (https://xenabrowser.net/datapages/) and the Beat-AML database (http://www.vizome.org/aml/). The datasets used and analyzed during this study are available from the corresponding author upon reasonable request.
Ethical statement
All bone marrow samples were collected in accordance with ethical requirements and regulations of the Ethics Committee of the First Affiliated Hospital of Jinan University. Informed consent was obtained from all subjects and studies were conducted under approval (No. 20200326).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Venugopal S, Sekeres MA. Contemporary Management of Acute Myeloid Leukemia: A Review. JAMA Oncol. 2024;10:1417-25
2. Thol F, Döhner H, Ganser A. How I treat refractory and relapsed acute myeloid leukemia. Blood. 2024;143:11-20
3. Haubner S, Mansilla-Soto J, Nataraj S, Kogel F, Chang Q, de Stanchina E. et al. Cooperative CAR targeting to selectively eliminate AML and minimize escape. Cancer Cell. 2023;41:1871-91
4. Xu J, Liu W, Fan F, Zhang B, Sun C, Hu Y. Advances in nano-immunotherapy for hematological malignancies. Exp Hematol Oncol. 2024;13:57
5. Tian C, Chen Z. Immune therapy: a new therapy for acute myeloid leukemia. Blood Sci. 2023;5:15-24
6. Wang C, Wang S. Targeting GPX4 degradation through disulfiram: A novel mechanism for ferroptosis induction in leukemia cells. Cell Investigation. 2025;1:100006
7. DiNardo CD, Erba HP, Freeman SD, Wei AH. Acute myeloid leukaemia. Lancet. 2023;401:2073-86
8. El Chaer F, Hourigan CS, Zeidan AM. How I treat AML incorporating the updated classifications and guidelines. Blood. 2023;141:2813-23
9. Shimony S, Stahl M, Stone RM. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol. 2023;98:502-26
10. Bhansali RS, Pratz KW, Lai C. Recent advances in targeted therapies in acute myeloid leukemia. J Hematol Oncol. 2023;16:29
11. Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H. et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345-77
12. Hou S, Liu J, Zhu Y. Multi-omics advances for molecular characterization, precision medicine, and prognostic implications in leukemia. Cell Investigation. 2025;1:100007
13. Song G-Y, Kim H-J, Kim T, Ahn SY, Jung S-H, Kim M. et al. Validation of the 2022 European LeukemiaNet risk stratification for acute myeloid leukemia. Sci Rep. 2024;14:8517
14. Lima AS, de Mello MR, Fernandes E, Bezerra MF, Oliveira MM, Duarte BK. et al. Clinical outcomes of patients with acute myeloid leukemia: evaluation of genetic and molecular findings in a real-life setting. Blood. 2015;126:1863-5
15. Chen X, Yuan L, Zhang Y, Wang F, Ma X, Fang J. et al. Advances towards genome-based acute myeloid leukemia classification: A comparative analysis of WHO-HAEM4R, WHO-HAEM5, and International Consensus Classification. Am J Hematol. 2024;99:824-35
16. Chen E, Jiao C, Yu J, Gong Y, Jin D, Ma X. et al. Assessment of 2022 European LeukemiaNet risk classification system in real-world cohort from China. Cancer Med. 2023;12:21615-26
17. Chow A, Perica K, Klebanoff CA, Wolchok JD. Clinical implications of T cell exhaustion for cancer immunotherapy. Nat Rev Clin Oncol. 2022;19:775-90
18. Sui S, Tian Y, Wang X, Zeng C, Luo OJ, Li Y. Single-cell RNA sequencing gene signatures for classifying and scoring exhausted CD8+ T cells in B-cell acute lymphoblastic leukaemia. Cell Prolif. 2024;57:e13583
19. Vadakekolathu J, Rutella S. Escape from T-cell-targeting immunotherapies in acute myeloid leukemia. Blood. 2024;143:2689-700
20. Zhang Z, Deng C, Zhu P, Yao D, Shi J, Zeng T. et al. Single-cell RNA-seq reveals a microenvironment and an exhaustion state of T/NK cells in acute myeloid leukemia. Cancer Sci. 2023;114:3873-83
21. Chen C, Liang C, Wang S, Chio CL, Zhang Y, Zeng C. et al. Expression patterns of immune checkpoints in acute myeloid leukemia. J Hematol Oncol. 2020;13:28
22. Wang P, Cai Q, Peng X, Dai Z, Liu J, Li W. et al. Increased co-expression of CTLA4/LAG3 predicted adverse clinical outcomes in patients with T-cell malignancies. Cell Investigation. 2025;1:100004
23. Trujillo-Ochoa JL, Kazemian M, Afzali B. The role of transcription factors in shaping regulatory T cell identity. Nat Rev Immunol. 2023;23:842-56
24. Topchyan P, Xin G, Chen Y, Zheng S, Burns R, Shen J. et al. Harnessing the IL-21-BATF Pathway in the CD8+ T Cell Anti-Tumor Response. Cancers. 2021;13:1263
25. Zhang X, Zhang C, Qiao M, Cheng C, Tang N, Lu S. et al. Depletion of BATF in CAR-T cells enhances antitumor activity by inducing resistance against exhaustion and formation of central memory cells. Cancer Cell. 2022;40:1407-22
26. Itahashi K, Irie T, Yuda J, Kumagai S, Tanegashima T, Lin Y-T. et al. BATF epigenetically and transcriptionally controls the activation program of regulatory T cells in human tumors. Sci Immunol. 2022;7:eabk0957
27. Li C, Liu Z, Wang Z, Yim WY, Huang Y, Chen Y. BATF and BATF3 deficiency alters CD8+ effector/exhausted T cells balance in skin transplantation. Mol Med. 2024;30:16
28. McMahon SB, Monroe JG. The role of early growth response gene 1 (egr-1) in regulation of the immune response. J Leukocyte Biol. 1996;60:159-66
29. Li T-T, Liu M-R, Pei D-S. Friend or foe, the role of EGR-1 in cancer. Med Oncol. 2019;37:7
30. Sui S, Zhong M, Zhong S, Peng X, Mao L, Chen C. et al. BRD4 inhibitor reduces exhaustion and blocks terminal differentiation in CAR-T cells by modulating BATF and EGR1. Biomark Res. 2024;12:124
31. [Chinese guidelines for diagnosis, treatment of adult acute myeloid leukemia (not APL) (2023)]. Zhonghua Xue Ye Xue Za Zhi. 2023; 44: 705-12.
32. Chen C, Xu L, Gao R, Wang S, Zhang Y, Wang C. et al. Transcriptome-Based Co-Expression of BRD4 and PD-1/PD-L1 Predicts Poor Overall Survival in Patients with Acute Myeloid Leukemia. Front Pharmacol. 2020;11:582955
33. Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675-8
34. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL. et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526-31
35. Chen C, Wang P, Mo W, Zhang Y, Zhou W, Deng T. et al. lncRNA-CCDC26, as a novel biomarker, predicts prognosis in acute myeloid leukemia. Oncol Lett. 2019;18:2203-11
36. Wang P, Zhang Y, Cai Q, Long Q, Pan S, Zhou W. et al. Optimal combination of immune checkpoint and senescence molecule predicts adverse outcomes in patients with acute myeloid leukemia. Ann Med. 2023;55:2201507
37. Zhong M, Chen C, Zhao W, Tan J, Chen J, Huang X. et al. High Co-Expression of PDCD1/TIGIT/CD47/KIR3DL2 in Bone Marrow Is Associated with Poor Prognosis for Patients with Myelodysplastic Syndrome. J Oncol. 2023;2023:1972127
38. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252-9
39. Wang P, Chen Y, Long Q, Li Q, Tian J, Liu T. et al. Increased coexpression of PD-L1 and TIM3/TIGIT is associated with poor overall survival of patients with esophageal squamous cell carcinoma. J Immunother Cancer. 2021;9:e002836
40. Baessler A, Vignali DAA. T Cell Exhaustion. Annu Rev Immunol. 2024;42:179-206
41. Gottschlich A, Thomas M, Grünmeier R, Lesch S, Rohrbacher L, Igl V. et al. Single-cell transcriptomic atlas-guided development of CAR-T cells for the treatment of acute myeloid leukemia. Nat Biotechnol. 2023;41:1618-32
42. Zhong M, Gao R, Zhao R, Huang Y, Chen C, Li K. et al. BET bromodomain inhibition rescues PD-1-mediated T-cell exhaustion in acute myeloid leukemia. Cell Death Dis. 2022;13:671
43. Zhou Y-J, Li G, Wang J, Liu M, Wang Z, Song Y. et al. PD-L1: expression regulation. Blood Sci. 2023;5:77-91
44. Jin R, Xu H, Zhou M, Lin F, Xu W, Xu A. EGR1 Mediated Reduction of Fibroblast Secreted-TGF-β1 Exacerbated CD8+ T Cell Inflammation and Migration in Vitiligo. Inflammation. 2024;47:503-12
45. Shan F, Cillo AR, Cardello C, Yuan DY, Kunning SR, Cui J. et al. Integrated BATF transcriptional network regulates suppressive intratumoral regulatory T cells. Sci Immunol. 2023;8:eadf6717
46. Chang Y-J, Pei X-Y, Huang X-J. Haematopoietic stem-cell transplantation in China in the era of targeted therapies: current advances, challenges, and future directions. Lancet Haematol. 2022;9:e919-e29
47. Notarantonio A-B, Bertrand A, Piucco R, Fievet G, Sartelet H, Boulangé L. et al. Highly immunosuppressive myeloid cells correlate with early relapse after allogeneic stem cell transplantation. Exp Hematol Oncol. 2024;13:50
48. Mathioudaki A, Wang X, Sedloev D, Huth R, Kamal A, Hundemer M. et al. The remission status of AML patients after allo-HCT is associated with a distinct single-cell bone marrow T-cell signature. Blood. 2024;143:1269-81
Author contact
![]() Corresponding authors: Cunte Chen (cuntechencom), Songnan Sui (suissncom), and Yangqiu Li (tyangqiuliedu.cn).
Corresponding authors: Cunte Chen (cuntechencom), Songnan Sui (suissncom), and Yangqiu Li (tyangqiuliedu.cn).

 Global reach, higher impact
Global reach, higher impact