Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(8):1750-1761. doi:10.7150/ijms.110392 This issue Cite
Research Paper
Exploring the Relationship Between Gut Microbiota and Aortic Stenosis: Role of Inflammatory Proteins, Blood Metabolites, and Immune Cells
1. Department of Structural Heart Disease, National Center for Cardiovascular Disease, Fuwai Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
2. National Health Commission Key Laboratory of Cardiovascular Regeneration Medicine, Fuwai Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
3. State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
4. Key Laboratory of Innovative Cardiovascular Devices, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
5. National Clinical Research Center for Cardiovascular Diseases, Fuwai Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
6. College of Life Sciences, Hunan Normal University, Changsha, China.
* These authors contributed equally to the study and are considered co-corresponding authors.
Received 2025-1-13; Accepted 2025-2-27; Published 2025-3-10
Abstract
Background: Aortic stenosis is the most prevalent valvular heart disease in high-income population, and there are currently no medical therapies to slow the disease progression. Given that gut microbiota influences the immune system, lipid metabolism, and inflammation, there may be a potential link between gut microbiota and AS.
Aims: We aimed to examine the causal effects of gut microbiota on AS and to investigate the mediating roles of inflammatory proteins, blood metabolites, and immune cells.
Methods: Bidirectional Mendelian randomization analysis was performed to assess the causal relationships between gut microbiota, inflammatory proteins, blood metabolites, immune cells, and AS. Two-step Mendelian randomization was utilized to explore direct and indirect effects. The data were derived from genome-wide association study summary statistics available in public databases.
Results: The study identified nine gut microbial features (six microbial taxa and three pathways), four inflammatory proteins, 91 blood metabolites, and four immune cell traits associated with AS. However, no significant mediating roles were found for inflammatory proteins, blood metabolites, and immune cells in the causal pathway between gut microbiota and AS.
Conclusion: This study revealed novel causal associations between gut microbial features, inflammatory proteins, blood metabolites, and immune cell traits with AS. These findings offer new insights into the pathophysiology of AS and provide potential targets for therapeutic approaches.
Keywords: Aortic stenosis, Gut microbiota, Inflammatory protein, Blood metabolite, Immune cell, Mendelian randomization analysis
Introduction
Aortic stenosis (AS) is the most prevalent valvular heart disease in Europe and North America [1-3]. Although surgical aortic valve replacement and transcatheter aortic valve implantation are effective for severe AS [1,4], no medical therapies currently exist to slow disease progression. Investigations have identified several modifiable risk factors for AS, such as hypertension [5], obesity [6,7], diabetes [8], dyslipidaemia [9-11], which may give a few hints for future pharmaceutical development. Immune and inflammatory mechanisms were also found to play important roles in the progress of AS [12]. Meanwhile, the critical role of gut microbiota in modulating immune system [13], lipid metabolism [14], and inflammation [15] has been verified by many studies. Several studies have explored the connection between gut microbiota and AS [16]. Elevated acylcarnitine, choline and TMAO levels, which were closely linked to gut microbial metabolism, have been reported in AS patients [17,18]. Liu et al. [19] found distinct gut microbial profiles in AS compared to coronary artery disease. Nevertheless, no study explored the potential causal relationship between gut microbiota and AS at the genetic level.
Mendelian randomization (MR) is a potent epidemiological and genetic research method for exploring the causal links between risk factors and disease outcomes. It relies on Mendelian genetics principles, which dictate that alleles are dispersed at random during gamete formation, mirroring the randomization process in randomized controlled trials. This helps address the issues of reverse causality and confounding commonly seen in observational research [20,21].
In this study, we performed a comprehensive MR analysis to investigate the potential causal effects of gut microbiota on AS, inflammatory proteins on AS, blood metabolites on AS, immune cells on AS, respectively. Then we explored whether inflammatory proteins, blood metabolites, or immune cells serve as mediators in the relationship between gut microbiota and AS.
Materials and Methods
Study design
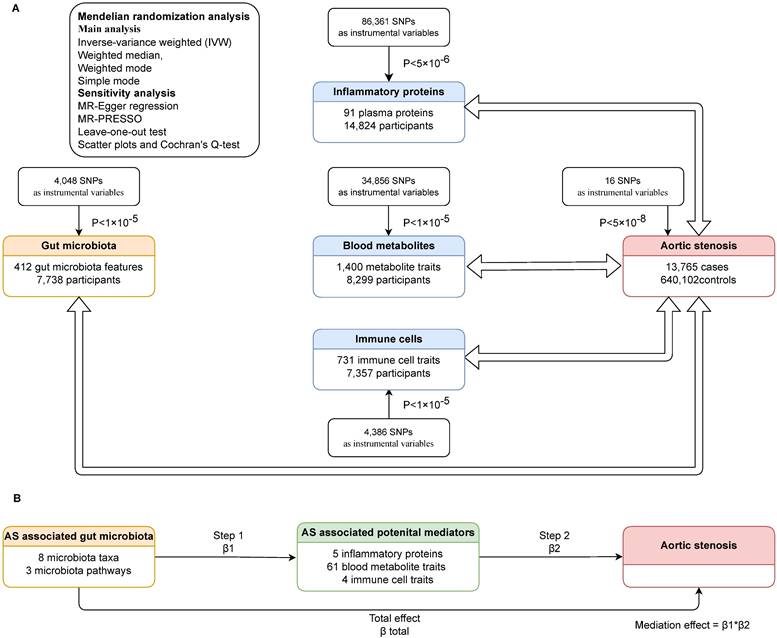
We carried out bidirectional Mendelian randomization to explore the links between gut microbiota and AS, as well as between inflammatory proteins and AS, blood metabolites and AS, immune cells and AS. Then we assessed whether gut microbiota had a causal impact on potential mediators (inflammatory proteins, blood metabolites and immune cells associated with AS), and if so, we adopted two-step Mendelian randomization (TSMR) to determine the direct and indirect effects of the gut microbiota and potential mediators on AS. In TSMR analysis, we included gut microbiota and potential mediators that have been demonstrated significant associated with AS, using only genetic instrumental variables (IVs) not used in step one, to evaluate the effect of the potential mediator on AS. The mediation effect was calculated only when the direction of the total effect (beta of the gut microbiota-AS association) aligned with the direction of the effect through the mediator (beta of the gut microbiota-potential mediator association × beta of the potential mediator-AS association). The study design is illustrated in the flowchart (Figure 1).
Data sources
All genome-wide association study (GWAS) summary statistics utilized in this study were sourced from public databases and were accessible, as listed in Table S1. The original studies have already obtained ethics and institutional review board approvals and informed consent was acquired from the participants or caregivers.
Data for AS were sourced from a GWAS meta-analysis of 10 cohorts, involving 13,765 cases and 640,102 controls of European ancestry and revealing 11,591,806 variants [22]. Gut microbiota data were derived from a GWAS study conducted by Esteban et al. [23], who analyzed metagenomic sequencing data of 207 microbial taxa and 205 pathways representing microbial composition and function in 7,738 people from the northern Netherlands. The data of inflammation-related proteins were obtained from the study performed by Zhao et al. [24], who carried out a genome-wide protein quantitative trait locus study of 91 plasma proteins in 14,824 participants. Blood metabolites data came from a GWAS study by Yiheng Chen et al. [25], encompassing 1,091 metabolites and 309 metabolite ratios in 8,299 individuals from the Canadian Longitudinal Study on Aging (CLSA) cohort, with 850 metabolites categorized into eight superpathways [25]. Immune cell traits data were extracted from the GWAS Catalog (accession numbers from GCST0001391 to GCST0002121). The original GWAS study [26] involved 3,757 European individuals and investigated the impact of approximately 22 million SNPs on 731 immunophenotypes.
Instrumental variable selection
Candidate IVs for gut microbiota, immune cell traits and blood metabolites were selected based on the genome-wide significance threshold P < 1×10-5, consistent with previous studies [26-28]. To increase the number of IVs for inflammatory proteins, a threshold of P < 5×10⁻⁶ was applied. IVs related to AS were chosen at the conventional GWAS threshold of P < 5×10⁻⁸.Then we clumped all those IVs using conventional thresholds of 10 Mb and r2<0.001 to identify independent IVs free from linkage disequilibrium, utilizing the 1,000 Genomes European reference panel. F statistics were calculated to assess the strength of each SNP as an IV. Weak instruments (F statistic <10) were excluded from analyses to reduce weak instrument bias [29].
Statistical analysis
The inverse-variance weighted (IVW) method was primarily used in the analysis to evaluate causal relationships between exposures and outcomes [30]. Weighted median, weighted mode, and simple mode were applied to test the robustness of the findings. Results were presented as beta (β) value with standard error for the continuous outcomes and odds ratio (OR) with a 95% confidence interval (CI) for the binary outcomes. To address multiple testing, false discovery rate (FDR) correction of P-values was applied using the Benjamini-Hochberg method for all IVW results to control Type I errors. Significant results were those with the FDR-adjusted P-values < 0.05.
In the sensitivity analysis, pleiotropy and heterogeneity were examined. MR-Egger regression method was utilized as the main estimation to account for potential pleiotropy [31]. MR-PRESSO (MR Pleiotropy RESidual Sum and Outlier) was employed to identify and correct for horizontal pleiotropy by removing outliers [32]. Leave-one-out test was used to estimate the potential pleiotropic effects of single SNPs. Scatter plots and Cochran's Q-test were used to estimate heterogeneity, and P < 0.05 indicates the presence of heterogeneity.
For mediation estimation, we adopted TSMR to identify the direct and indirect effects of the gut microbiota and candidate mediators on AS. The proportion mediated by a candidate mediator was calculated as the estimated effect of the gut microbiota on the candidate mediator multiplied by the estimated effect of the candidate mediator on AS.
The study design. (A)Bidirectional mendelian randomization between gut microbiota and AS, inflammatory proteins and AS, blood metabolites and AS, immune cells and AS. (B)TSMR to decompose the direct and indirect effects of the gut microbiota and potential mediators on AS. AS: aortic stenosis. TSMR: two-step Mendelian randomization. MR: mendelian randomization. SNP: single nucleotide polymorphism. β1, β2, β total: the causal effect of exposure on outcome in each step.
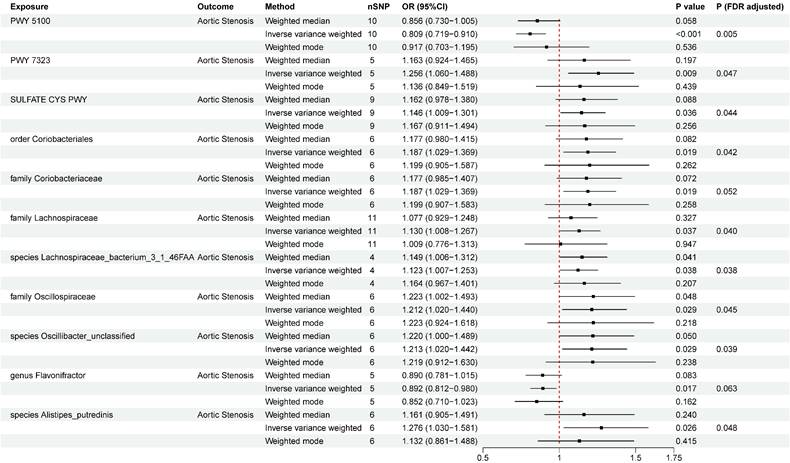
Two-sample Mendelian randomization estimations showing the effect of gut microbiota on aortic stenosis. Only the results of the IVW, weighted median, and weighted mode analysis methods were shown in the figure, and the results of other methods can be found in Table S7. nSNP: total number of instrumental variables used for analysis. PWY 5100: pyruvate fermentation to acetate and lactate II. PWY 7323: superpathway of GDP mannose derived O antigen building blocks biosynthesis. SULFATE CYS PWY: superpathway of sulfate assimilation and cysteine biosynthesis. P (FDR adjusted) < 0.05 was considered statistically significant.
All analyses were conducted using the R platform (version 4.2.1). The statistical analyses and data visualizations were performed with the 'TwoSampleMR', 'Mendelian Randomization', 'MRPRESSO' packages.
Results
Instrument variables included in analysis
A total of 4,048, 86,361, 34,856, and 4,386 SNPs were selected as IVs for gut microbiota, inflammatory proteins, blood metabolites, and immune cells, respectively. When AS was used as exposure factors, 16 SNPs were selected according to the selection criteria mentioned above. Detailed information of these IVs was shown in Table S2-S6.
The causal relationship between gut microbiota and AS
A total of eight microbial taxa (including three families, one genus, one order, and three species) and three pathways were identified associated with AS at the nominal significance level of 0.05 (Figure 2). After FDR correction, six microbial taxa (including two families, one order, and three species) and three pathways still remained significant. PWY 5100 (pyruvate fermentation to acetate and lactate II) (OR = 0.809, 95%CI = 0.719~0.910 , P FDR= 0.005) was associated with an decreased risk of AS, while order Coriobacteriales (OR=1.187, 95%CI =1.029~1.369, P FDR= 0.042), family Coriobacteriaceae (OR=1.187, 95%CI =1.029~1.369, P FDR= 0.052), family Lachnospiraceae (OR = 1.130, 95%CI = 1.008~1.267 , P FDR= 0.040), species Lachnospiraceae bacterium 3_1_46FAA (OR=1.123,95%CI=1.007~1.253, P FDR= 0.038), family Oscillospiraceae (OR = 1.212, 95%CI = 1.020~1.440, P FDR= 0.045), species Oscillibacter_unclassified (OR=1.213,95%CI =1.020~1.442, P FDR= 0.039), species Alistipes_putredinis (OR=1.276,95%CI =1.030~1.581, P FDR = 0.048), PWY 7323 (superpathway of GDP-mannose-derived O-antigen building blocks biosynthesis) (OR = 1.256, 95%CI = 1.060~1.488, P FDR= 0.047), and SULFATE CYS PWY (superpathway of sulfate assimilation and cysteine biosynthesis) (OR = 1.146, 95%CI = 1.009~1.301, P FDR= 0.044) were at risk of increasing AS. The associations remained consistent in sensitivity analyses, with no heterogeneity or pleiotropy was observed in the primary analysis (Table S7). To test reverse causality, we conducted a reverse MR analysis and found no evidence of associations of genetic liability to AS with identified gut microbiota was identified (Table S8). The results of “leave-one-out” analysis confirmed the reliability of the MR analysis (Figure S1). The scatter plots illustrated the overall effect of gut microbiota on AS (Figure S2).
The causal relationship between inflammatory proteins and AS
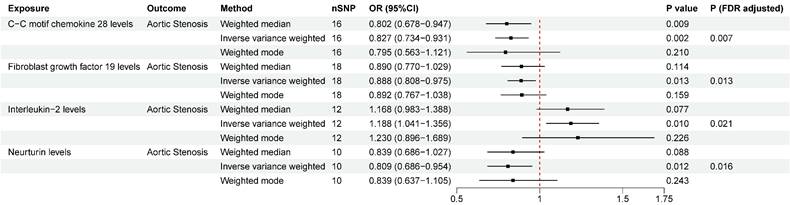
We detected significant associations of four inflammatory proteins with AS both at the nominal significance level of 0.05 and after FDR correction. (Figure. 3). C-C motif chemokine ligand 28 (CCL-28) (OR = 0.827, 95%CI = 0.734~0.931, P FDR= 0.007), fibroblast growth factor 19 (FGF-19) (OR = 0.888, 95%CI = 0.808~0.975, P FDR= 0.013), and neurtuin (OR = 0.809, 95%CI = 0.686~0.954, P FDR= 0.016) were associated with a lower risk of AS. While interleukin-2 (IL-2) (OR = 1.188, 95%CI = 1.041~1.356, P FDR= 0.021) was observed to increase the risk of AS. No significant heterogeneity or horizontal pleiotropy was detected based on Cochrane's Q, MR-Egger, and MR-PRESSO tests. In the reverse MR analysis, we found no evidence linking genetic liability to AS with the levels of identified inflammatory proteins (Table S10). The results of “leave-one-out” analysis proved that MR analysis confirmed the reliability of the MR analysis (Figure S3). The scatter plots demonstrated the overall effect of inflammatory proteins on AS (Figure S4).
The causal relationship between blood metabolites and AS
A total of 91 blood metabolites has significant causal associations with AS (Table S11), both at the nominal significance level of 0.05 and after FDR correction. Heterogeneity was observed in 25 metabolites by Cochran's Q-test and 16 metabolites were detected to have horizontal pleiotropy by MR-Egger, and these metabolites were excluded in following analysis. Additionally, six blood metabolites were excluded in the subsequent analysis due to having reverse causal relationship with AS (Table S12). The most significant causal effects were observed between seven blood metabolic traits and AS (Figure. 4). 1-stearoyl-GPE (18:0) (OR =1.162, 95%CI = 1.083~1.246, P FDR= 5.03×10-4), 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4) (OR = 1.171, 95%CI = 1.119~1.227, P FDR= 1.13×10-9), 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4) (OR = 1.155, 95%CI = 1.082~1.233, P FDR= 3.57×10-4), 1-oleoyl-2-arachidonoyl-GPE (18:1/20:4) (OR = 1.163, 95%CI = 1.091~1.238, P FDR= 8.88×10-5), 1-stearoyl-2-arachidonoyl-gpc (18:0/20:4) (OR = 1.096, 95%CI = 1.050~1.145, P FDR= 4.21×10-4), and Linoleoyl-arachidonoyl-glycerol (18:2/20:4) [2] (OR = 1.150, 95%CI = 1.074-1.232, P FDR= 8.53×10-4) were associated with an increased risk of AS. Oleoyl-linoleoyl-glycerol (18:1 to 18:2) [2] to linoleoyl-arachidonoyl-glycerol (18:2 to 20:4) [1] ratio (OR = 0.870, 95%CI = 0.823~0.918, P FDR= 2.37×10-5) was associated with a decreased risk of AS. Heterogeneity was detected in 1-stearoyl-GPE (18:0), 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4), 1-oleoyl-2-arachidonoyl-GPE (18:1/20:4), and Linoleoyl-arachidonoyl-glycerol (18:2/20:4) [2], while no obvious horizontal pleiotropy was found. We found no evidence linking genetic liability to AS with the identified 61 blood metabolites in the reverse MR analysis (Table S12). The results of “leave-one-out” analysis confirmed the reliability of the MR analysis (Figure S5). The scatter plots illustrated the overall effect of blood metabolites on AS (Figure S6).
The causal relationship between immune cells and AS
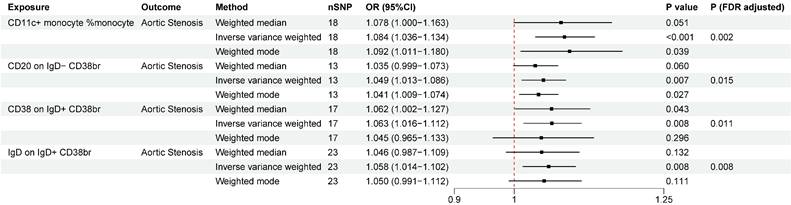
A total of four immunophenotypes were positively correlated with AS both at the nominal significance level of 0.05 and after FDR correction (Figure. 5), including CD11c+ monocyte %monocyte (cDC panel, OR = 1.084, 95%CI = 1.036 ~ 1.134 , P FDR= 0.002), CD20 on IgD- CD38br (B cell panel, OR = 1.049, 95%CI = 1.013 ~ 1.086, P FDR= 0.015), CD38 on IgD+ CD38br (B cell panel, OR = 1.063, 95%CI = 1.016 ~ 1.112, P FDR= 0.011), IgD on IgD+ CD38br (B cell panel, OR = 1.058, 95%CI = 1.014 ~ 1.102, P FDR= 0.008). Across these outcomes, the B-cell panel emerged as the most significantly associated factor with AS. No pleiotropy was detected by MR-PRESSO for the immune cells analyzed in the primary analysis. In pleiotropy analysis, the P-value of CD20 on IgD- CD38br was below 0.05, while P-values of the other three immune cells were greater than 0.05. The reverse MR analysis indicated no reverse causal relationship between immune cells and AS (Table S14). The results of “leave-one-out” analysis confirmed the reliability of the MR analysis (Figure S7). The scatter plots demonstrated the overall effect of immune cells on AS (Figure S8).
The mediation effect of inflammatory proteins, blood metabolites and immune cells in the causal relationship between gut microbiota and AS
It is widely recognized that gut microbiota plays a significant role in the development of various diseases by influencing metabolism and immune function. We performed MR analysis to determine the causal effect of the AS-associated microbial features on the potential mediator (inflammatory proteins, blood metabolites and immune cells) and followed by TSMR.
Two-sample Mendelian randomization estimations showing the effect of inflammatory proteins on aortic stenosis. Only the results of the IVW, weighted median, and weighted mode analysis methods were shown in the figure, and the results of other methods can be found in Table S9. nSNP: total number of instrumental variables used for analysis. P (FDR adjusted) < 0.05 was considered statistically significant.
Two-sample Mendelian randomization estimations showing the effect of blood metabolites on aortic stenosis. Only the results of the IVW, weighted median, and weighted mode analysis methods were shown in the figure, and the results of other methods can be found in Table S11. nSNP: total number of instrumental variables used for analysis. P (FDR adjusted) < 0.05 was considered statistically significant.

Two-sample mendelian randomization estimations showing the effect of immune cells on aortic stenosis. Only the results of the IVW, weighted median, and weighted mode analysis methods were shown in the figure, and the results of other methods can be found in Table S13. nSNP: total number of instrumental variables used for analysis. P (FDR adjusted) < 0.05 was considered statistically significant.
Two gut microbiota-inflammatory proteins-AS combinations were tested, indicating that higher relative abundance of family Oscillospiraceae (OR = 0.841, 95%CI = 0.747 ~ 0.948, P = 0.005) and species Oscillibacter_unclassified (OR = 0.842, 95%CI = 0.747 ~ 0.949, P = 0.005) were associated with decreased blood neurturin concentration (Table S15). However, no significant mediation effect was detected (mediation effect of family Oscillospiraceae = 0.0365(-0.00403, 0.077), P = 0.078; mediation effect of species Oscillibacter_unclassified = 0.0366(-0.00393, 0.0772), P = 0.077).
We explored the potential meditating roles of 61 blood metabolites, among which 27 metabolites may increase the risk of AS and the other 34 were negatively correlated with AS. In order to detect more potential meditating roles of blood metabolites, we included a total of 61 metabolites which were shown related to AS at significant level of 0.05. When evaluating the causal effects of GM on blood metabolites, 31 blood metabolites were related to 10 gut microbiota taxa or pathways (Table S16), generating 46 potential gut microbiota-metabolites-AS combinations. In the following mediation analysis, only 10 combinations (involving nine blood metabolites and four gut microbiotas and two pathways) were included based on the standard mentioned above. However, our results indicated that blood metabolites were not mediators in the pathway from gut microbiotas and AS (Table S17).
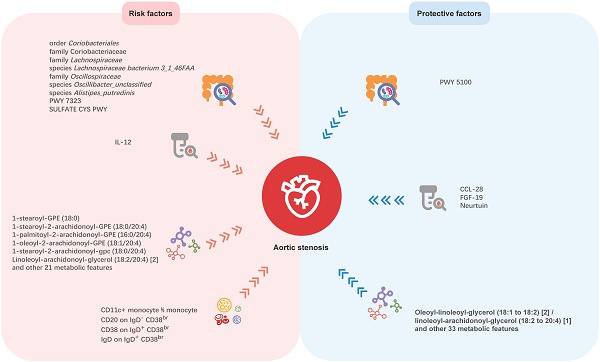
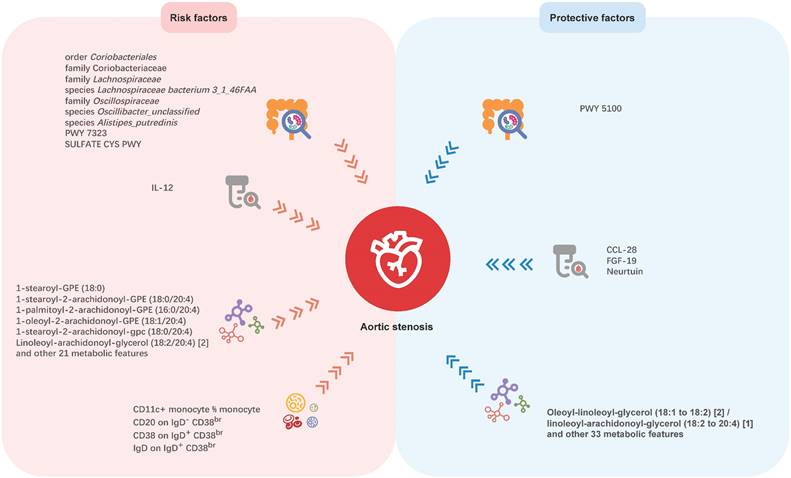
Mendelian randomization analyses show causal effects of gut microbial features, inflammatory proteins, blood metabolites, and immune cell traits on Aortic stenosis. PWY 5100: pyruvate fermentation to acetate and lactate II; PWY 7323: superpathway of GDP-mannose-derived O-antigen building blocks biosynthesis; SULFATE CYS PWY: superpathway of sulfate assimilation and cysteine biosynthesis; CCL-28: C-C motif chemokine ligand 28; FGF-19: fibroblast growth factor 19; IL-12: interleukin-2.
In MR analysis of gut microbiota and immune cells, all gut microbiota associated with AS were unrelated with immune cells associated with AS (Table S18), which means the effect of gut microbiota on AS may not mediated by immune cells.
Discussion
Although previous studies have revealed the association between gut microbiota and AS, the causal relationship remains unclear, due to the intrinsic defects of the observational study. Thus, our study aimed to investigate the causal links between genetically predicted gut microbiota characteristics and AS. In this large-scale and comprehensive MR study, we explored causal relationships between 412 gut microbial features and AS, 91 inflammation-related proteins with AS, 1,400 blood metabolites and AS, and 731 immune cell traits and AS. The potential mediation roles of inflammatory proteins, blood metabolites and immune cells were also been tested in the causal relationship between gut microbiota and AS. We found nine microbial features, four inflammatory proteins, 91 blood metabolites, and four immune cell traits having causal associations with AS, which may offer new insights for future investigations (Figure. 6). But no mediating roles of inflammatory proteins, blood metabolites, and immune cells was detected in the causal association between gut microbiota and AS.
In the present study, most of the AS-related microbiotas are from order Clostridiales, class Clostridia in phylum Firmicutes. Family Oscillospiraceae and species Oscillibacter_unclassified are members of the family Oscillospiraceae in order Clostridiales and are positively related to AS. It is noteworthy that the genus Oscillospira, an important component of human gut microbiota which has attracted attention from researchers, is also a member of family Oscillospiraceae, but is classified into family Ruminococcaceae. The genus Oscillospira is capable of producing various short-chain fatty acids dominated by butyrate [33,34], and has been proved to have protective effect of many diseases, especially obesity [32,33] and obesity-related chronic inflammatory and metabolic diseases[33,37,38]. This may be attributed to its negative correlation with chronic inflammation [39]. But there are some evidences that it is positively associated to some central nervous system disorders [37,38]and degenerative diseases [33,42-44]. Therefore, it is reasonable to assume that different species in family Oscillospiraceae may play varying roles in human health. In the original GWAS study for gut microbial traits [23], only two menmbers of family Oscillospiraceae were included (genus Oscillibacter and species Oscillibacter_unclassified), and genus Oscillospira was not included. Therefore, the associations between family Oscillospiraceae, genus Oscillospira and AS still remain to be explored and more research is required to reveal the underlying mechanisms. Although family Coriobacteriaceae did not show significant relation with AS, order Coriobacteriales seems to be a risk factor for AS, suggesting other family members in order Coriobacteriales may be related to AS. Family Lachnospiraceae and species Lachnospiraceae bacterium_3_1_46FAA may increases the risk of AS. Family Lachnospiraceae has been determined to be associated with diabetes, obesity, liver diseases, kidney diseases, as well as inflammatory bowel disease, by altering glucose and lipid metabolism, modulating immune system and inflammation [45-47]. Limited study has been carried out on the function of species Lachnospiraceae bacterium_3_1_46FAA. Further studies are needed to reveal if and how it is implicated in AS development. Taken together, these microbiotas have not been reported yet and may serve as potential targets for the treatment of AS. Prevotella copri, a member of Bacteroidetes, seemed to play roles in immunity and inflammation, and has been correlated with atherosclerosis, hypertension, and heart failure. Liu et al. [19] performed 16S rRNA sequencing on fecal samples and indicated that gut microbiota in patients with cardiac valve calcification, which was characterized by increased Prevotella copri, was different from both healthy controls and patients with coronary artery disease. Interestingly, our study did not identify a significant causal correlation between Prevotella copri and AS. This discrepancy may be partly attributed to the small sample size and study design employed by Liu et al [19], which included only 19 patients with cardiac valve calcification and did not distinguish aortic valve calcification from the calcification of other cardiac valves. Besides, cross-sectional studies can only reveal associations and are not capable of establishing causality. Therefore, longitudinal studies with a lager sample size are needed to validate these associations. Additionally, it should be noted that species Alistipes_putredinis (a member of genus Alistipes, family Rikenellaceae, order Bacteroidales, class Bacteroidetes, phylum Bacterodetes) was determined to be a risk factor of AS. The genus Alistipes has drawn increasing interests of researchers and been linked with cardiovascular diseases such as atrial fibrillation, congestive heart failure, and atherosclerosis cardiovascular disease [48]. Although there are accumulating evidences, the involvement of Alistipes in cardiovascular diseases is still contradictory [49], and our study may bring some new insights. Altogether, our study highlights the potential of microbiome-targeting therapy for AS prevention and treatment. But when we interpret the influence of gut microbiota on AS, it should be noted that the gut microbiota itself may be influenced by a variety of factors, such as dietary habits, medication use, and comorbid conditions, and it is also in a state of dynamic change. Besides, other traditional factors which has been widely accepted as risk factors of AS, including older age, male sex, hypertension, smoking, diabetes, and elevated serum lipoprotein(a) levels, still need to be prioritized when we discuss the etiology of AS.
Aortic valve interstitial cells (VICs), playing a crucial role in the development of AS, are a heterogeneous population of cells including fibroblasts and smooth muscle cells [12,50]. Cytokines have been demonstrated to influence the differentiation of VICs, therefore we explored the link between inflammatory proteins and AS. As a result, CCL-28, FGF-19 and neurturin were detected to be protective factors, while IL-2 was linked to increased risk of AS. In previous studies, CCL-28, FGF-19 and IL-2 were mostly investigated in immune disorders and tumor treatment [51-54], and neurturin was mainly discussed in muscular, neurodegenerative and psychiatric disorders [55,56]. Here we revealed their potential effects on AS for the first time.
Concerning the relationship between blood metabolites and AS, 1-stearoyl-2-arachidonoyl-gpc (18:0/20:4) and 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4) are both glycerophospholipids generated by the amalgamation of stearic acid and arachidonic acid (AA), constituting different phosphoryl groups. These glycerophospholipids have been found to be related with ischemic heart diseases and left ventricular diastolic dysfunction, and inversely associated with Intestinimonas_massilliensis [57]. However, clinical study revealing the relationship between specific gut microbiota and the glycerophospholipids and glycerolipids associated with AS are relatively lacking, and more evidence comes from MR study. An MR study [58] suggested that genus Eubacterium nodatum was negatively associated with 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4), 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4), and 1-oleoyl-2-arachidonoyl-GPE (18:1/20:4). Genus Holdemanella and genus Peptococcus were also showed to be related with the level of 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4) [59]. Regrettably, we could not verify these relationships because the primary GWAS study [23] we used did not include the microbiota mentioned above. The oleoyl-linoleoyl-glycerol (18:1 to 18:2) [2] to linoleoyl-arachidonoyl-glycerol (18:2 to 20:4) [1] ratio was associated with a decreased risk of AS, but the oleoyl-linoleoyl-glycerol (18:1 to 18:2) [2] level was not associated with AS (P FDR > 0.05). To be noted, the genetic features of linoleoyl-arachidonoyl-glycerol (18:2 to 20:4) [1] level were not included in the original GWAS study. Therefore, it seems that linoleoyl-arachidonoyl-glycerol (18:2 to 20:4) [1] level may be a risk factor of AS. All the 7 metabolites which showed most significant causal effects on AS are composed of AA. Studies have unveiled that AA could be metabolized by three distinct enzyme systems: cyclooxygenases (COXs, also known as PGG/H synthases), lipoxygenases, and cytochrome P450 enzymes (ω-hydroxylases and epoxygenases), producing a wide range of biologically active fatty acid mediators [60], which also have been verified to be important in the development and progression of cardiovascular diseases [61]. This investigation postulates that these glycerolipids may contribute to the development of AS. Regretfully, there is currently short of clinical research concerning these metabolites, highlighting the need for further exploration in the future. Trimethylamine N-oxide (TMAO), a well-studied gut microbiota metabolite, has been found to accelerate the progression of AS and was associated with adverse outcome after transcatheter aortic valve implantation [18,62]. Some studies also highlighted the roles of tryptophan derivatives, especially indoxyl sulfate and serotonin, in osteogenic differentiation and inflammation of AS[16]. We also included TMAO, some tryptophan derivatives, and some bile acid derivatives in our analysis. However, none of these metabolites was found to be associated with AS at the genetic level. As we know, these metabolites are highly influenced by dietary components and dependent on hepatic metabolism. Therefore, the increase of these metabolites in patients with AS may not be determined by genetics, but rather influenced by diet or other metabolic factors.
Our study found that the risk of AS increased with an elevation in the proportion of CD11c+ monocyte. CD11c+ monocyte mostly consists of intermediate monocytes and nonclassical monocytes [63], comprising only 10% of total blood monocytes, yet secreting high levels of proinflammatory cytokines [64-66]. They have also been found to be related with high body mass index (BMI), hyperlipidemia and cardiovascular diseases [65,67-70], including atherosclerosis and coronary artery diseases. Our study adds new evidence of the possibly crucial role of these monocytes in the development of AS. Within B cell panel, our analysis detected a significant relationship between CD38 and IgD on IgD+ CD38br B-cells and AS, as well as a causal relation between CD20 on IgD- CD38br B-cells and AS. CD38 is expressed on peripheral B cells during early differentiation and activation, and has been verified to be linked with multiple B-cell-related diseases [71]. IgD is expressed on the surface of mature B cells after they have encountered their specific antigen for the first time, and orchestrates a surveillance system at the interface of immunity and inflammation [72,73]. CD20 is regarded as an effective therapeutic target for a majority of B-cell malignancies, but the precise physiological role and function in disease progress remain unclear [74]. However, it should be noted that only immune cells in peripheral blood were included in the original GWAS study. Concerning the participation of immune cells in aortic valve diseases, the aortic valve is inhabited by tissue-resident macrophages, mast cells, dendritic cells and T cells [75-77], and T cells were recognized as important inflammatory drivers of progressive calcific aortic valve disease [78,79]. Monocytes can also be recruited after aortic valve endothelial injury [80]. But the role of B cells in the development of AS has received less attention.
Our study had some limitations. Firstly, this study only included European populations. Variations in genetic factors, environmental exposures, and lifestyles across different populations could impact the relationship between gut microbiota, metabolites, and AS, limiting the generalizability of the findings to other ethnic or regional groups. Secondly, the analysis did not completely account for potential confounders, such as comorbidities and antibiotics, which could change the intestinal microbiota composition and alter metabolite profiles, influencing the link between gut microbiota and AS. Additionally, the Mendelian randomization approach itself has limitations. It can only analyze association between exposures and outcomes at genetic level, but genetic influences may be compensated for by other biological processes over time, potentially weakening the observed genetic effects. Overreliance on MR may reinforce genetic determinism, overshadowing the importance of environmental and lifestyle factors in disease etiology. Combining MR with other methodologies, such as randomized controlled trials and mechanistic studies, can provide a more comprehensive understanding of causal relationships and inform clinical and public health interventions. Future studies are warranted to address these limitations, explore mechanisms behind gut microbiota-AS relationship and offer new perspectives on AS prevention and treatment.
Conclusion
Using Mendelian randomization analysis, we identified several gut microbiota, inflammatory proteins, blood metabolites, and immune cell traits genetically associated with AS. However, no significant mediating effects have been identified for the gut bacteria-mediated inflammatory proteins, blood metabolites or immune cell traits associated with AS. These findings provide new insights into the pathophysiology of AS and establish a theoretical basis for future therapeutic approaches.
Abbreviations
AS: aortic stenosis; MR: Mendelian randomization; TSMR: two-step Mendelian randomization; IV: instrumental variables; GWAS: genome-wide association study; IVW: inverse-variance weighted; OR: odds ratio; CI: confidence interval; FDR: false discovery rate; MR-PRESSO: MR Pleiotropy RESidual Sum and Outlier; CCL-28: C-C motif chemokine ligand 28; FGF-19: fibroblast growth factor 19; IL-2: interleukin-2; VIC: valve interstitial cells; TMAO: Trimethylamine N-oxide; SNP: single nucleotide polymorphism.
Supplementary Material
Supplementary figures.
Supplementary tables.
Acknowledgements
We acknowledge the high-quality data of original GWAS studies involved in this study.
Funding
This work was funded by: National Key R&D Program of China (2022YFC2503400); National Clinical Research Center for Cardiovascular Diseases, Fuwai Hospital, Chinese Academy of Medical Sciences (NCRC2020009); National High Level Hospital Clinical Research Funding (2022-GSP-GG-18); CAMS Innovation Fund for Medical Sciences (2021-I2M-1-065).
Author contributions
Pan X contributed to the study design, supervision and preparation. Jing F and Zhou J conducted the formal analysis and drafted the paper. Zhang F, Zhao G and Fang F reviewed and revised the manuscript. All authors read and approved the final manuscript.
Data availability
All data used in this study were obtained from publicly released GWAS. The data of gut microbial features can be obtained in this website (https://dutchmicrobiomeproject.molgeniscloud.org/). The data for aortic stenosis can be obtained in this website (https://doi.org/10. 5281/zenodo.7505361). The data for inflammatory proteins can be obtained in this website (https://www.phpc.cam.ac.uk/ceu/proteins). The data for blood metabolites can obtained in this website (https://doi.org/10.1038/s41588-022-01270-1). The data for immune cell traits can obtained in this website (https://www.ebi.ac.uk/gwas/studies/).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Vahanian A, Beyersdorf F, Praz F. et al. 2021 ESC/EACTS guidelines for the management of valvular heart disease. Eur Heart J. 2022;43(7):561-632
2. Iung B, Delgado V, Rosenhek R. et al. Contemporary Presentation and Management of Valvular Heart Disease: The EURObservational Research Programme Valvular Heart Disease II Survey. Circulation. 2019;140(14):1156-1169
3. Andell P, Li X, Martinsson A. et al. Epidemiology of valvular heart disease in a Swedish nationwide hospital-based register study. Heart. 2017;103(21):1696-1703
4. Otto CM, Nishimura RA, Bonow RO. et al. 2020 ACC/AHA guideline for the management of patients with valvular heart disease. J Thorac Cardiovasc Surg. 2021;162(2):e183-e353
5. Nazarzadeh M, Pinho-Gomes AC, Smith Byrne K. et al. Systolic Blood Pressure and Risk of Valvular Heart Disease: A Mendelian Randomization Study. JAMA Cardiol. 2019;4(8):788-795
6. Kaltoft M, Langsted A, Nordestgaard BG. Obesity as a Causal Risk Factor for Aortic Valve Stenosis. J Am Coll Cardiol. 2020;75(2):163-176
7. Larsson SC, Bäck M, Rees JMB, Mason AM, Burgess S. Body mass index and body composition in relation to 14 cardiovascular conditions in UK Biobank: a Mendelian randomization study. Eur Heart J. 2020;41(2):221-226
8. Larsson SC, Wallin A, Håkansson N, Stackelberg O, Bäck M, Wolk A. Type 1 and type 2 diabetes mellitus and incidence of seven cardiovascular diseases. Int J Cardiol. 2018;262:66-70
9. Kjeldsen EW, Thomassen JQ, Rasmussen KL, Nordestgaard BG, Tybjærg-Hansen A, Frikke-Schmidt R. Cardiovascular risk factors and aortic valve stenosis: towards 10-year absolute risk charts for primary prevention. Eur J Prev Cardiol. Published online May 22, 2024:zwae177.
10. Kamstrup PR, Tybjærg-Hansen A, Nordestgaard BG. Elevated lipoprotein(a) and risk of aortic valve stenosis in the general population. J Am Coll Cardiol. 2014;63(5):470-477
11. Smith JG, Luk K, Schulz CA. et al. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. JAMA. 2014;312(17):1764-1771
12. Moncla LHM, Briend M, Bossé Y, Mathieu P. Calcific aortic valve disease: mechanisms, prevention and treatment. Nat Rev Cardiol. 2023;20(8):546-559
13. Rizzetto L, Fava F, Tuohy KM, Selmi C. Connecting the immune system, systemic chronic inflammation and the gut microbiome: The role of sex. J Autoimmun. 2018;92:12-34
14. Brown EM, Clardy J, Xavier RJ. Gut microbiome lipid metabolism and its impact on host physiology. Cell Host Microbe. 2023;31(2):173-186
15. Schirmer M, Smeekens SP, Vlamakis H. et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167(4):1125-1136.e8
16. Chong-Nguyen C, Yilmaz B, Coles B. et al. A scoping review evaluating the current state of gut microbiota and its metabolites in valvular heart disease physiopathology. Eur J Clin Invest. Published online January 10, 2025:e14381.
17. Bengel P, Elkenani M, Beuthner BE. et al. Metabolomic profiling in patients with different hemodynamic subtypes of severe aortic valve stenosis. Biomolecules. 2023;13(1):95
18. Guo Y, Xu S, Zhan H. et al. Trimethylamine N-oxide levels are associated with severe aortic stenosis and predict long-term adverse outcome. J Clin Med. 2023;12(2):407
19. Liu Z, Li J, Liu H. et al. The intestinal microbiota associated with cardiac valve calcification differs from that of coronary artery disease. Atherosclerosis. 2019;284:121-128
20. Carter AR, Sanderson E, Hammerton G. et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol. 2021;36(5):465-478
21. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. The BMJ. 2018;362:k601
22. Yu Chen H, Dina C, Small AM. et al. Dyslipidemia, inflammation, calcification, and adiposity in aortic stenosis: a genome-wide study. Eur Heart J. 2023;44(21):1927-1939
23. Lopera-Maya EA, Kurilshikov A, Van Der Graaf A. et al. Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch Microbiome Project. Nat Genet. 2022;54(2):143-151
24. Zhao JH, Stacey D, Eriksson N. et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol. 2023;24(9):1540-1551
25. Chen Y, Lu T, Pettersson-Kymmer U. et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat Genet. 2023;55(1):44-53
26. Orrù V, Steri M, Sidore C. et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. 2020;52(10):1036-1045
27. Sanna S, van Zuydam NR, Mahajan A. et al. Causal relationships between gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51(4):600-605
28. Liu X, Tong X, Zou Y. et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet. 2022;54(1):52-61
29. Burgess S, Thompson SG, CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755-764
30. Slob EAW, Burgess S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44(4):313-329
31. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377-389
32. Wang Q, Dai H, Hou T. et al. Dissecting Causal Relationships Between Gut Microbiota, Blood Metabolites, and Stroke: A Mendelian Randomization Study. J Stroke. 2023;25(3):350-360
33. Yang J, Li Y, Wen Z, Liu W, Meng L, Huang H. Oscillospira - a candidate for the next-generation probiotics. Gut Microbes. 2021;13(1):1987783
34. Konikoff T, Gophna U. Oscillospira: A central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016;24(7):523-524
35. Ng M, Fleming T, Robinson M. et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet Lond Engl. 2014;384(9945):766-781
36. Goodrich JK, Waters JL, Poole AC. et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789-799
37. Del Chierico F, Nobili V, Vernocchi P. et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatol Baltim Md. 2017;65(2):451-464
38. Stanislawski MA, Lozupone CA, Wagner BD. et al. Gut microbiota in adolescents and the association with fatty liver: the EPOCH study. Pediatr Res. 2018;84(2):219-227
39. Shintouo CM, Mets T, Beckwee D. et al. Is inflammageing influenced by the microbiota in the aged gut? A systematic review. Exp Gerontol. 2020;141:111079
40. Zhang F, Yue L, Fang X. et al. Altered gut microbiota in Parkinson's disease patients/healthy spouses and its association with clinical features. Parkinsonism Relat Disord. 2020;81:84-88
41. Zhai Q, Cen S, Jiang J, Zhao J, Zhang H, Chen W. Disturbance of trace element and gut microbiota profiles as indicators of autism spectrum disorder: A pilot study of Chinese children. Environ Res. 2019;171:501-509
42. Keren N, Konikoff FM, Paitan Y. et al. Interactions between the intestinal microbiota and bile acids in gallstones patients. Environ Microbiol Rep. 2015;7(6):874-880
43. Zhang J, Luo D, Lin Z. et al. Dysbiosis of gut microbiota in adult idiopathic membranous nephropathy with nephrotic syndrome. Microb Pathog. 2020;147:104359
44. Parthasarathy G, Chen J, Chen X. et al. Relationship Between Microbiota of the Colonic Mucosa vs Feces and Symptoms, Colonic Transit, and Methane Production in Female Patients With Chronic Constipation. Gastroenterology. 2016;150(2):367
45. Kameyama K, Itoh K. Intestinal colonization by a Lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microbes Environ. 2014;29(4):427-430
46. Zhai B, Zhang C, Sheng Y. et al. Hypoglycemic and hypolipidemic effect of S-allyl-cysteine sulfoxide (alliin) in DIO mice. Sci Rep. 2018;8(1):3527
47. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022-1023
48. Jie Z, Xia H, Zhong SL. et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8(1):845
49. Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The genus alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11:906
50. Xu K, Xie S, Huang Y. et al. Cell-Type Transcriptome Atlas of Human Aortic Valves Reveal Cell Heterogeneity and Endothelial to Mesenchymal Transition Involved in Calcific Aortic Valve Disease. Arterioscler Thromb Vasc Biol. 2020;40(12):2910-2921
51. Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3(2):133-146
52. Mohan T, Deng L, Wang BZ. CCL28 chemokine: An anchoring point bridging innate and adaptive immunity. Int Immunopharmacol. 2017;51:165-170
53. Gadaleta RM, Moschetta A. Metabolic Messengers: fibroblast growth factor 15/19. Nat Metab. 2019;1(6):588-594
54. Ogawa H, Iimura M, Eckmann L, Kagnoff MF. Regulated production of the chemokine CCL28 in human colon epithelium. Am J Physiol Gastrointest Liver Physiol. 2004;287(5):G1062-1069
55. Correia JC, Kelahmetoglu Y, Jannig PR. et al. Muscle-secreted neurturin couples myofiber oxidative metabolism and slow motor neuron identity. Cell Metab. 2021;33(11):2215-2230.e8
56. Creedon DJ, Tansey MG, Baloh RH. et al. Neurturin shares receptors and signal transduction pathways with glial cell line-derived neurotrophic factor in sympathetic neurons. Proc Natl Acad Sci. 1997;94(13):7018-7023
57. Luo K, Taryn A, Moon EH. et al. Gut microbiota, blood metabolites, and left ventricular diastolic dysfunction in US hispanics/latinos. Microbiome. 2024;12:85
58. Yan J. et al. Gut microbiota's role in glioblastoma risk, with a focus on the mediating role of metabolites. Front Neurol. 2024;15:1386885
59. Xie X, Ren W, Zhou W. et al. Genetic prediction of the effect of gut microbiota on uveitis via blood metabolites: A mediated mendelian randomization investigation. Medicine (Baltimore). 2024;103(50):e40922
60. Wang B, Wu L, Chen J. et al. Metabolism pathways of arachidonic acids: mechanisms and potential therapeutic targets. Signal Transduct Target Ther. 2021;6(1):94
61. Marklund M, Wu JHY, Imamura F. et al. Biomarkers of dietary omega-6 fatty acids and incident cardiovascular disease and mortality. Circulation. 2019;139(21):2422-2436
62. Xiong Z, Li J, Huang R. et al. The gut microbe-derived metabolite trimethylamine-N-oxide induces aortic valve fibrosis via PERK/ATF-4 and IRE-1α/XBP-1s signaling in vitro and in vivo. Atherosclerosis. 2024;391:117431
63. Loke P, Niewold TB. By CyTOF: Heterogeneity of Human Monocytes. Arterioscler Thromb Vasc Biol. 2017;37(8):1423-1424
64. Belge KU, Dayyani F, Horelt A. et al. The proinflammatory CD14+CD16+DR++ monocytes are a major source of TNF. J Immunol Baltim Md 1950. 2002;168(7):3536-3542
65. Ziegler-Heitbrock L. The CD14+ CD16+ blood monocytes: their role in infection and inflammation. J Leukoc Biol. 2007;81(3):584-592
66. Mukherjee R, Kanti Barman P, Kumar Thatoi P, Tripathy R, Kumar Das B, Ravindran B. Non-classical monocytes display inflammatory features: Validation in sepsis and systemic lupus erythematous. Sci Rep. 2015;5:13886
67. Rogacev KS, Ulrich C, Blömer L. et al. Monocyte heterogeneity in obesity and subclinical atherosclerosis. Eur Heart J. 2010;31(3):369-376
68. Ozaki Y, Imanishi T, Taruya A. et al. Circulating CD14+CD16+ monocyte subsets as biomarkers of the severity of coronary artery disease in patients with stable angina pectoris. Circ J Off J Jpn Circ Soc. 2012;76(10):2412-2418
69. Tacke F, Randolph GJ. Migratory fate and differentiation of blood monocyte subsets. Immunobiology. 2006;211(6-8):609-618
70. Poitou C, Dalmas E, Renovato M. et al. CD14dimCD16+ and CD14+CD16+ monocytes in obesity and during weight loss: relationships with fat mass and subclinical atherosclerosis. Arterioscler Thromb Vasc Biol. 2011;31(10):2322-2330
71. Zeng F, Zhang J, Jin X. et al. Effect of CD38 on B-cell function and its role in the diagnosis and treatment of B-cell-related diseases. J Cell Physiol. 2022;237(7):2796-2807
72. Bautista D, Vásquez C, Ayala-Ramírez P. et al. Differential expression of IgM and IgD discriminates two subpopulations of human circulating IgM+IgD+CD27+ B cells that differ phenotypically, functionally, and genetically. Front Immunol. 2020;11:736
73. Chen K, Xu W, Wilson M. et al. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat Immunol. 2009;10(8):889-898
74. Pavlasova G, Mraz M. The regulation and function of CD20: an “enigma” of B-cell biology and targeted therapy. Haematologica. 2020;105(6):1494-1506
75. Hulin A, Hortells L, Gomez-Stallons MV. et al. Maturation of heart valve cell populations during postnatal remodeling. Dev Camb Engl. 2019;146(12):dev173047
76. Helske S, Syväranta S, Kupari M. et al. Possible role for mast cell-derived cathepsin G in the adverse remodelling of stenotic aortic valves. Eur Heart J. 2006;27(12):1495-1504
77. Kim AJ, Xu N, Yutzey KE. Macrophage lineages in heart valve development and disease. Cardiovasc Res. 2021;117(3):663-673
78. Guauque-Olarte S, Droit A, Tremblay-Marchand J. et al. RNA expression profile of calcified bicuspid, tricuspid, and normal human aortic valves by RNA sequencing. Physiol Genomics. 2016;48(10):749-761
79. Raddatz MA, Madhur MS, Merryman WD. Adaptive immune cells in calcific aortic valve disease. Am J Physiol Heart Circ Physiol. 2019;317(1):H141-H155
80. Lavine KJ, Epelman S, Uchida K. et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A. 2014;111(45):16029-16034
Author contact
![]() Corresponding authors: Xiangbin Pan, Fang Fang, and Guangzhi Zhao, Department of Structural Heart Disease, Fuwai Hospital, No.167 North Lishi Road, Xicheng District, Beijing 100037, China; E-mail: panxiangbinorg, fangfang_ffcom, guangzhilalacom; Tel: +86(10)88396655; Fax: +86(10)88396655
Corresponding authors: Xiangbin Pan, Fang Fang, and Guangzhi Zhao, Department of Structural Heart Disease, Fuwai Hospital, No.167 North Lishi Road, Xicheng District, Beijing 100037, China; E-mail: panxiangbinorg, fangfang_ffcom, guangzhilalacom; Tel: +86(10)88396655; Fax: +86(10)88396655

 Global reach, higher impact
Global reach, higher impact