Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(4):1002-1014. doi:10.7150/ijms.109161 This issue Cite
Review
Aberrant DNA Methylation in Esophageal Squamous Cell Carcinoma and its Clinical Implications in Systemic Chemotherapy
1. Department of Health Prevention and Care, Beijing Hospital, National Center of Gerontology, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, No. 1 Dahua Road, Beijing 100730, China.
2. Department of Oncology, Beijing Hospital, National Center of Gerontology, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, No. 1 Dahua Road, Beijing, 100730, China.
3. Graduate School of Peking Union Medical College, Chinese Academy of Medical Sciences, No. 5 Dong Dan San Tiao, Beijing, 100005, China.
4. Department of Thoracic Surgery, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Panjiayuan Nanli No.17, Beijing 100021, China.
Received 2024-12-21; Accepted 2025-1-25; Published 2025-2-3
Abstract
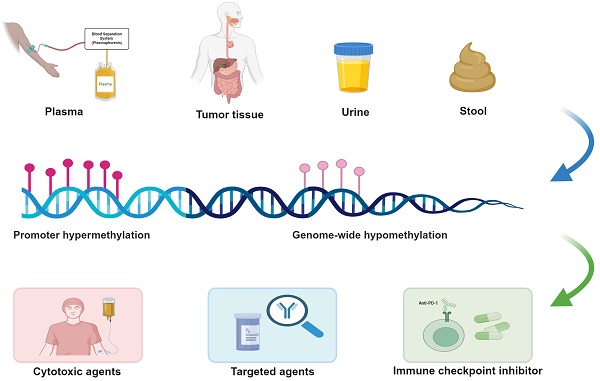
Esophageal cancer is a significant global health concern, with esophageal squamous cell carcinoma being the predominant subtype in high-incidence regions like China. Despite advances in multidisciplinary treatments, the prognosis for ESCC remains poor, with systemic chemotherapy facing the challenge of drug resistance. Epigenetic alterations, particularly DNA methylation, play a crucial role in ESCC carcinogenesis and therapeutic response. Aberrant DNA methylations, including global hypomethylation and promoter-specific hyper-methylation, disrupt critical pathways such as cell cycle regulation, apoptosis, and DNA repair, contributing to chemoresistance. Several studies have identified methylation markers that predict treatment response, particularly for chemotherapy, targeted therapy and immunotherapy, such as p16 and GPX3 for cisplatin, MTHFR for 5-FU, CHFR for paclitaxel. DNA methyltransferase inhibitors and other epigenetic therapies are being explored to reverse these methylation changes and enhance therapeutic efficacy. However, the clinical utility of these markers remains limited due to the lack of large-scale validation and concerns over off-target effects. This review aims to summarize all aberrant methylation alterations in ESCC and the clinical implications of aberrantly methylated candidate genes identified in ESCC systemic chemotherapy, with the goal of further understanding the underlying molecular mechanisms, refining methylation-targeting therapies, and integrating them with conventional treatments to improve patient outcomes.
Keywords: esophageal squamous cell carcinoma, DNA methylation, systemic chemotherapy, therapeutic response, epigenetic therapy
Introduction
Esophageal cancer ranks as the 11th most common newly diagnosed cancer and is the 7th leading cause of cancer mortality worldwide, with approximately 511,000 new cases and 445,000 deaths reported in 2022 according to the latest global cancer statistics [1]. Esophageal cancer (EC) is histologically classified into esophageal squamous cell carcinoma (ESCC), esophageal adenocarcinoma (EAC) and other subtypes [2]. ESCC and EAC have significant differences in many features such as geographic distribution, etiologies, pathogenesis, physiological and molecular mechanisms [3-6]. Cigarette smoking, alcoholic abuse and inappropriate dietary pattern (e.g. nutritional deficiencies, nitrosamines, betel quid chewing, pickled vegetables and hot food/beverages) are the major risk factors for ESCC [5, 7]. Meanwhile EAC accounts for nearly two-thirds of esophageal cancer cases in high-income countries with excess body weight, gastroesophageal reflux disease, and Barrett's esophagus as the major risk factors [7]. Esophageal squamous cell carcinoma is the predominant subtype in China, accounting for approximately 90 percent of esophageal cancer and ranking the 7th in incidence and 5th in mortality [8]. Multidisciplinary treatments including surgery, radiotherapy, chemotherapy, molecular-targeting therapy and immunotherapy have improved overall survival of esophageal cancer patients, nonetheless the prognosis is still poor with overall 5-year survival ranging from 15% to 25% [9, 10].
Systemic chemotherapy which mainly consists of cytotoxic agents, molecular-targeted agents and immune checkpoint inhibitors play an essential role in the treatment of ESCC patients. Cytotoxic agents, such as cisplatin (DDP) and 5-fluorouracil (5-FU), have been used as the standard treatment for decades [11]. However, their limited efficacy due to resistance and inevitable adverse effects significantly impact the long-term survival of ESCC patients. In terms of immunotherapy, the Food and Drug Administration (FDA) has approved the PD-1 inhibitor pembrolizumab for second-line treatment of PD-L1 positive, advanced or metastatic ESCC [12, 13]. Based on phase 2 and phase 3 clinical trials, nivolumab as another PD-1 inhibitor was found to outperform conventional chemotherapy irrespective of PD-L1 status [14, 15]. Multiple molecular-targeted agents against EGFR, HER-2, VEGFR, c-Met and other oncogenes were also investigated in ESCC treatment. As vital candidates, anti-EGFR inhibitors such as cetuximab and nimotuzumab have some promising results in clinical trials [16, 17]. However, most molecular-targeted agents haven't demonstrated clinical utility in randomized phase 3 trials of ESCC so far.
Negative results from multiple systemic chemotherapy clinical trials suggest the importance of identifying predictive biomarkers of responses to specific agents. Nevertheless, definitive predictive biomarkers to determine right patient candidates remain unclear for conventional chemotherapy, targeted therapy and immunotherapy in patients with ESCC. Recently intriguing findings have emerged that both genetic and epigenetic alterations equally contribute to ESCC with collusion between them, which provides novel theories and understandings about therapeutic effect evaluation, potential therapeutic targets and prognostic prediction in ESCC patients [18].
DNA methylation, along with other epigenetic alterations such as histone modification, chromatin remodeling and the expression of microRNAs, plays an important role in transcriptional regulation, thus interfering driver genes expression [19, 20]. The best-understood epigenetic mechanism is DNA methylation, which mainly involves global hypomethylation and specific gene promoter hypermethylation especially of tumor suppressor genes (TSGs). Sufficient evidence has suggested that TSGs transcriptional silence by promoter hypermethylation and increased chromosome instability by global hypomethylation were critical steps in ESCC carcinogenesis [21]. Besides, epigenetic alterations of upstream transcription factors have been proved to cause tumor suppressor network silence as well. Silenced genes with aberrant methylation occurred in almost every significant molecular pathway associated with systemic chemotherapy, such as cell cycle regulation, cell apoptotic, DNA damage repair, cell adhesion and immune response regulation pathways [22].
DNA methylation has a crucial role in esophageal carcinogenesis, which was indicated in previous researches. More specifically, aberrant methylated genes have been detected in fields of early screening, diagnosis, prognosis and chemo-sensitivity prediction. Promising predictive markers have great potentiality to improve limited therapeutic effect and poor prognosis in ESCC. In this review, we aim to summarize the molecular mechanisms of aberrant methylated candidate genes in ESCC systemic chemotherapy and their clinical applications on biomarkers of chemotherapy response and therapeutic potentials.
Aberrantly methylated genes in ESCC
Numerous DNA methylation profiling researches have revealed that, similar to other cancers, ESCC genome exhibits widespread regions of hypomethylation and focal regions of hypermethylation, when compared with adjacent normal tissues or noncancer controls [23, 24]. Although the general patterns of hyper- and hypomethylation are similar, the distribution of specific methylation sites differs from that observed in other related cancers (e.g. head and neck squamous cell carcinoma). Large-scale genomic and epigenomic data further confirm that ESCC and EAC exhibit nearly mutually exclusive sets of driver genes and significantly distinct epigenetic alterations. Hence, a focused summary of aberrant methylation alterations in ESCC is both crucial and indispensable.
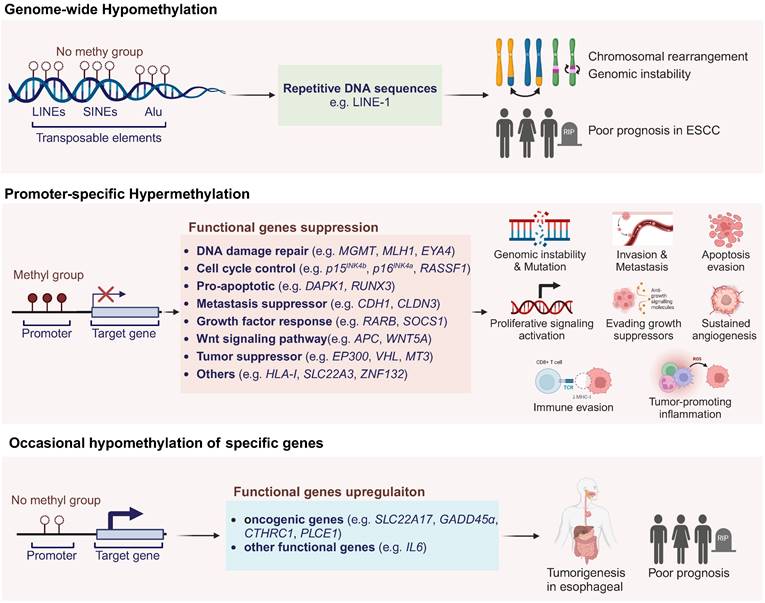
As illustrated in Figure 1, these alterations in the epigenetic landscape, characterized by global genomic DNA hypomethylation and specific genes promoter hypermethylation, both contribute to the pathogenesis and development of ESCC via distinct mechanisms [25]. Besides those, the occasional hypomethylation of specific genes has also seen some research progress, which will be detailed in the following sections.
Aberrant DNA methylation patterns in ESCC. Promoter hypermethylation silencing tumor suppressor genes and hypomethylation of repetitive DNA sequences lead to genomic instability and tumorigenesis. Occasional hypomethylation of specific genes were also reported to contribute to ESCC development. Abbreviations: LINEs, long interspersed nuclear elements; SINEs, short interspersed nuclear elements; LINE-1, long interspersed nuclear element-1; ESCC, esophageal squamous cell carcinoma; TCR, T-cell receptor; MHC-I, major histocompatibility complex class I; ROS, reactive oxygen species. Created in BioRender. Li, YK. (2025) https://BioRender.com/z74h803 (accessed on 25 January 2025).
Global genomic DNA hypomethylation
Feinberg et.al [26] reported global genomic DNA hypomethylation in tumor tissues as early as 1983. Gaudet et.al [27] managed to induce tumorigenesis in mice by genomic hypomethylation, which validated the correlation between genomic hypomethylation and tumorigenesis. However, the underlying mechanism of global genomic DNA hypomethylation in ESCC is still equivocal. Current studies showed that DNA hypomethylations, especially those located in long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs) and other highly repetitive DNA sequences like Alu elements, could destabilize chromosome and cause rearrangements [28, 29]. LINE-1 elements constitute nearly 17% of human genome as important parts, so methylation of LINE-1 indicates the global DNA methylation level [21]. Variable studies showed that LINE-1 methylation in ESCC ranged from 25% to 92%, and low methylation level of LINE-1 was indicated to correlated significantly with poor prognosis [30, 31]. Global genomic DNA hypomethylation could still influence tumorigenesis through some other mechanisms that require further investigations [25].
Promoter-specific hypermethylation in ESCC
DNA methylation has become the best-studied epigenetic field due to the stable characteristic and credible detection techniques. Previously published studies have shown that as much as hundreds of genes have been found to be silenced by promoter hypermethylation. Numbers of evidences have suggested that the promoter hypermethylation of tumor suppressor genes is the critical step in carcinogenesis of ESCC [32]. In the Table 1, we summarized all reported TSGs in ESCC which play significant roles in signal pathways including cell-cycle regulation, apoptosis control, cell adhesion, metastasis, DNA damage repair, growth factor response, etc. The other functional genes which are silenced by promoter hypermethylation, accompanied with detailed gene information and corresponding methylation frequency in tumor tissues, were also listed as below.
Rare hypomethylation of specific genes in ESCC
According to the comprehensive analysis of the DNA methylation profile in ESCC, only 12.4% out of 26081 differentially methylated regions exhibited hypomethylation while 87.6% showed hypermethylation [106]. In ESCC carcinogenesis, the phenomenon of occasional hypomethylation of specific genes has been a subject of intense research [38, 106]. Cui et al. found that the hypomethylation of NGALR (SLC22A17) gene contributes to its overexpression in ESCC, implicating its role in tumorigenesis [107]. GADD45α, involved in DNA repair, is also overexpressed due to promoter hypomethylation in ESCC [108]. The DNA polymerase iota (Polι) gene, overexpressed in ESCC, is linked to hypomethylation, potentially driving genomic instability [109]. The IGF2 gene is influenced by hypomethylation at its DMR0, correlating with poor prognosis [110]. Similarly, the OCT1 and CTHRC1 genes are upregulated in ESCC due to hypomethylation, activating MAPK/MEK/ERK pathway and facilitating tumor progression [111, 112]. The hypomethylation-associated upregulation of PLCE1 has been linked to tumorigenesis, tumor angiogenesis and poor prognosis in ESCC [113]. The IGFBP7 gene shows lower promoter methylation, with its unmethylated state correlating with increased globulin levels and reflux in ESCC patients [114]. Notably, the gene CDKN1C, a putative tumor suppressor, exhibits diminished expression associated with the loss of methylation at the differentially methylated region (DMR)-LIT1, rather than its own promoter CpG methylation [115]. Lastly, the interleukin 6 (IL6) gene, which is known to activate the JAK/STAT3 pathway, is implicated in ESCC and influenced by hypomethylation [45]. These findings underscore the complex interplay between hypomethylation and the dysregulation of key genes in ESCC pathogenesis.
Clinical implications of aberrantly methylated genes in systemic chemotherapy
DNA methylation and therapeutic response prediction in ESCC
As mentioned in the above review, DNA methylation has been implicated in silencing apoptotic genes, disrupting DNA repair mechanisms, dysregulating cell cycle and even enhancing drug efflux, all of which contribute to chemotherapy resistance. Notably, these specific genes epigenetically modified by DNA hypermethylation regulate cancer response to systemic chemotherapy, making them potential predictive markers for therapy response, as shown in Figure 2.
In 2006, Hamilton et al. firstly identified a combined 9-gene panel (p16, REPRIMO, p57, p73, RUNX-3, CHFR, MGMT, TIMP-3, and HPP1) to predict adjuvant chemotherapy (DDP and 5-FU) responses based on methylation levels from 12 ESCC patients and 23 EAC patients [116]. Among these genes, REPRIMO and p16, key regulators of the cell cycle, exhibited significantly higher methylation rates in esophageal cancer patients non-responsive to chemoradiation, and promoter methylation of both genes impairs the ability to arrest at the G2/M checkpoint, where cisplatin exerts their effects, leading to chemoresistance [116, 117]. The study conducted by Chang et al. in 2017 stands out for establishing a six-CpG panel, including IFNGR2, KCNK4, NOTCH4, NPY, PAX6, and SOX17, which predicts treatment response to chemoradiation (DDP, 5-FU and concurrent radiotherapy). This panel's risk score equation, derived from the methylation status of selected genes in 91 ESCC patients, demonstrated a remarkable power in discriminating poor responders from good responders, with an AUC of 0.930, underscoring its potential in personalized medicine [118]. Iwabu et al. identified FGF5 as an oncogenic growth factor and its methylation as a predictive marker for definitive chemoradiotherapy (dCRT) sensitivity from 117 ESCC patients, with low methylation linked to poor response (sensitivity 45%, specificity 90%), highlighting its potential clinical utility. FGF5 could be induced by dCRT in ESCC when it is unmethylated, supporting cell survival and leading to resistance [119]. In a comprehensive study by Salta et al. to identify DNA methylation-based biomarkers for early detection and prediction of response to therapy in ESCC patients, the methylation of GPX3 has been identified as a potential marker for predicting responses to therapy. GPX3 encodes glutathione peroxidase, which mitigates oxidative damage. Methylation-induced inactivation, along with compensatory overexpression of other antioxidants and increased genomic instability, further enhances cisplatin resistance in cancer cells [120]. Based on molecular profiles from 36 ESCC patients in 2021, de Klerk et al. revealed that TP63 amplification and TFPI2 gene promoter methylation are associated with unfavorable responses to neoadjuvant chemoradiotherapy which consists of carboplatin, paclitaxel and concurrent radiotherapy [121]. Huang et al. demonstrated that PON3 hypermethylation downregulates its expression, contributing to multidrug resistance in ESCC, although the underlying mechanisms remain to be further elucidated [122]. The study by Min et al. sheds light on the epigenetic evolution of acquired resistance to combination therapy (cisplatin and verapamil) in ESCC. SLC8A3 was identified as a potential multidrug resistance gene from 6 ESCC patients' WGBS results, whose promoter descending methylation dynamics were associated with resistance to chemotherapy [123].
Hypermethylated functional genes in esophageal squamous cell carcinoma
| Classification | Year | Gene Symbol | Description | Major functions | Location | Methylation rate in tumor (%) | References |
|---|---|---|---|---|---|---|---|
| DNA damage repair genes | 1998 | FHIT | Fragile Histidine Triad Diadenosine Triphosphatase | DNA damage response, cell-cycle regulation | 3p14.2 | 14-33, 50 | [33, 34] |
| 2003 | MGMT | O-6-Methylguanine-DNA Methyltransferase | DNA damage repair | 10q26 | 27-72 | [35, 36] | |
| 2005 | MLH1 | MutL Homolog 1 | DNA mismatch repair, cell-cycle regulation | 3p22.3 | 3-62 | [37-39] | |
| 2005 | MSH2 | MutS Homolog 2 | DNA mismatch repair, cell-cycle regulation | 2p21 | 29-32 | [40] | |
| 2006 | BRCA1 | BRCA1 DNA Repair Associated | DNA damage repair, cell-cycle regulation, transcriptional activator | 17q21.31 | 26-28 | [38] | |
| 2015 | HIC1 | HIC ZBTB Transcriptional Repressor 1 | DNA damage response, apoptosis, Wnt signaling pathway, tumor suppressor | 17p13.3 | 84.2 | [41] | |
| 2018 | EYA4 | EYA Transcriptional Coactivator and Phosphatase 4 | DNA damage repair, migration and invasion, epithelial-mesenchymal transition, AKT signaling pathway, tumor suppressor | 6q23.2 | 78-85.7 | [42] | |
| Cell cycle control genes | 1999 | CDKN2A (p16INK4a) | Cyclin Dependent Kinase Inhibitor 2A | Cell-cycle regulation, tumor suppressor | 9p21 | 12-88 | [35, 37, 43-45] |
| 1999 | CDKN2B (p15INK4b) | Cyclin Dependent Kinase Inhibitor 2B | Cell-cycle regulation | 9p21 | 13-18 | [43] | |
| 2003 | RASSF1 | Ras Association Domain Family Member 1 | Cell-cycle regulation, apoptosis | 3p21.3 | 14-53 | [34] | |
| 2015 | CHFR | Checkpoint With Forkhead and Ring Finger Domains | Cell cycle and apoptosis, tumor suppressor | 12q24.33 | 45 | [46] | |
| Pro-apoptotic genes | 2005 | UCHL1 (PGP9.5) | Ubiquitin C-Terminal Hydrolase L1 | Ubiquitin regulation, cell growth inhibition, apoptosis, tumor suppressor | 4p13 | 42 | [47] |
| 2006 | DAPK1 | Death Associated Protein Kinase 1 | Cell survival, apoptosis, and autophagy | 9q21.33 | 26-38 | [38, 44] | |
| 2010 | ZNF382 | Zinc Finger Protein 382 | Pro-apoptotic transcription factor, tumor suppressor | 19q13.12 | 89 | [48] | |
| 2011 | RUNX3 | RUNX Family Transcription Factor 3 | Cellular growth and differentiation, Notch and TGF-β signaling pathway, tumor suppressor | 1p36.11 | 35.8-51.4 | [49, 50] | |
| Metastasis suppressor genes | 2003 | CADM1 (TSLC1) | Cell Adhesion Molecule 1 | Migration and invasion, cell proliferation, tumor suppressor | 11q23.3 | 50 | [51] |
| 2004 | CDH1 (E-cadherin) | Cadherin 1 | Cell adhesion, proliferation, metastasis, epithelial-mesenchymal transition | 16q22.1 | 14-61 | [38, 44, 52] | |
| 2004 | CDH13 | Cadherin 13 | Cell adhesion and recognition, tumor growth, cell cycle, tumor suppressor | 16q23.3 | 14-39.4 | [53] | |
| 2006 | PCDH10 | Protocadherin 10 | Cell-cell interaction, cell migration, tumor suppressor | 4q28.3 | 51-81 | [54] | |
| 2006 | CLDN3 | Claudin 3 | Cell-cell adhesion, cell tight junctions | 7q11.23 | 69 | [35] | |
| 2008 | DCC | DCC Netrin 1 Receptor | Cell adhesion, cell proliferation and differentiation, apoptosis, tumor suppressor | 18q21.2 | 74 | [55] | |
| 2010 | UPK1A | Uroplakin 1A | Cell motility, metastasis, tetraspanin superfamily, tumor suppressor | 19q13.12 | 62 | [56] | |
| 2011 | CLDN4 | Claudin 4 | Cell-cell adhesion, cell tight junctions | 7q11.23 | 41.7 | [57] | |
| 2012 | CDH11 | Cadherin 11 | Cell adhesion, migration and invasion, Wnt/β-catenin signaling pathway | 16q21 | 93 | [58] | |
| Growth factor response-related genes | 2003 | RARB | Retinoic Acid Receptor Beta | Growth factor response, cellular growth and differentiation | 3p24 | 25, 39, 67-70 | [34, 35, 44] |
| 2005 | RBP1 | Retinol Binding Protein 1 | Retinoid signaling | 3q23 | 17.9-31 | [35, 59] | |
| 2005 | RARRES1 (TIG1) | Retinoic Acid Receptor Responder 1 | Retinoid signaling | 3q25.32 | 17.9 | [59] | |
| 2007 | CRABP1 | Cellular Retinoic Acid Binding Protein 1 | Cell cycle, proliferation, tumor suppressor | 15q25.1 | 52.8 | [60] | |
| 2011 | SOCS1 | Suppressor Of Cytokine Signaling 1 | Negative regulator of JAK/STAT pathway and interferon signaling, tumor suppressor | 16p13.13 | 45.3 | [61] | |
| Wnt signaling related genes | 2007 | APC | APC Regulator of WNT Signaling Pathway | Wnt signaling pathway, cell polarity and chromosome segregation | 5q21-q22 | 27-46 | [37] |
| 2007 | SFRP1 | Secreted Frizzled Related Protein 1 | Wnt signaling modulator | 8p11.21 | 29.6-64.3 | [37, 44, 50] | |
| 2007 | SFRP2 | Secreted Frizzled Related Protein 2 | Wnt signaling modulator | 4q31.3 | 19.6 | [37] | |
| 2007 | WIF1 | WNT Inhibitory Factor 1 | Wnt signaling pathway inhibitor | 12q14.3 | 27-35 | [44, 62] | |
| 2011 | DKK3 | Dickkopf WNT Signaling Pathway Inhibitor 3 | Wnt signaling inhibitor, tumor suppressor | 11p15.3 | 37.4 | [50] | |
| 2012 | SOX17 | SRY-Box Transcription Factor 17 | Cell proliferation, Wnt signaling pathway, transcription regulator | 8q11.23 | 65 | [63] | |
| Genes with tumor suppressive functions | 2003 | ECRG4 | ECRG4 Augurin Precursor | Cell proliferation, cell cycle, senescence, tumor suppressor | 2q12.2 | 80 | [64] |
| 2003 | PRSS3 (Trypsinogen 4) | Serine Protease 3 | Proteolytic activity, tumor suppressor | 9p13.3 | 50 | [65] | |
| 2003 | VHL | Von Hippel-Lindau Tumor Suppressor | Ubiquitin ligase component, tumor suppressor | 3p25.3 | 36 | [34] | |
| 2005 | MT3 | Metallothionein 3 | Cellular growth inhibition, intracellular metal homeostasis | 16q13 | 52 | [66] | |
| 2006 | GRIN2B (NMDAR2B) | Glutamate Ionotropic Receptor NMDA Type Subunit 2B | Apoptosis, signal transduction, tumor suppressor | 12p13.1 | 95 | [67] | |
| 2007 | DLC1 | DLC1 Rho GTPase Activating Protein | Cytoskeleton organization, Ras-mediated signaling pathways, cell adhesion, cell cycle, tumor suppressor | 8p22 | 51 | [68] | |
| 2007 | CDX2 | Caudal Type Homeobox 2 | Cellular growth and differentiation, transcription factor | 13q12.2 | 49 | [69] | |
| 2007 | PLCD1 | Phospholipase C Delta 1 | Phospholipase, cell proliferation, cell cycle, migration and invasion, tumor suppressor | 3p22.2 | 38 | [70] | |
| 2007 | ADAMTS18 | ADAM Metallopeptidase with Thrombospondin Type 1 Motif 18 | Tumor microenvironment modulator, cell proliferation, tumor suppressor | 16q23 | 52-88 | [71] | |
| 2007 | EP300 | E1A Binding Protein P300 | Transcription regulator activity, tumor invasion and metastasis | 22q13.2 | 42 | [72] | |
| 2008 | GNG7 | G Protein Subunit Gamma 7 | Signal transduction, tumor suppressor | 19p13.3 | 33.3 | [73] | |
| 2008 | SST | Somatostatin | Somatostatin hormone, tumor suppressor | 3q27.3 | 53.8 | [74] | |
| 2008 | HOPX | HOP Homeobox | Tumor suppressor, serum response factor-related pathway | 4q12 | 50 | [75] | |
| 2008 | ITGA4 | Integrin Subunit Alpha 4 | Cell attachment to ECM, signal transduction, tumor suppressor | 2q31.3 | 21 | [44] | |
| 2008 | IRF8 | Interferon Regulatory Factor 8 | Transcriptional factor, IFN-γ signaling pathway, immune response, cellular growth and differentiation, tumor suppressor | 16q24.1 | 58 | [76] | |
| 2008 | ENG | Endoglin | TGF-β signaling pathway, angiogenesis, tumor suppressor | 9q34.11 | 46.2 | [77] | |
| 2009 | KAT2B (PCAF) | Lysine Acetyltransferase 2B | Transcription regulator activity, histone acetylation, cell cycle, tumor suppressor | 3p24.3 | 70 | [78] | |
| 2010 | NEFH | Neurofilament Heavy Chain | Mitochondrial function, glycolysis, Akt/β-catenin pathway, tumor suppressor | 22q12.2 | 65 | [79] | |
| 2011 | HSPB2 | Heat Shock Protein Family B (Small) Member 2 | Stress response and resistance, heat shock protein activity, tumor suppressor | 11q23.1 | 95.7 | [80] | |
| 2012 | PTK6 | Protein Tyrosine Kinase 6 | Cell proliferation, cellular differentiation, apoptosis, migration and invasion, tumor suppressor | 20q13.33 | 40 | [81] | |
| 2012 | TFPI2 | Tissue Factor Pathway Inhibitor 2 | Proteolytic activity, ECM degradation, tumor suppressor | 7q21.3 | 67 | [82] | |
| 2012 | RAB25 | RAB25, Member RAS Oncogene Family | Cell survival, migration, invasion and angiogenesis, MAPK/ERK signaling pathway, tumor suppressor | 1q22 | 75-100 | [83] | |
| 2013 | DIRAS1 | DIRAS Family GTPase 1 | Apoptosis, cell motility, ERK and p38 MAPK signaling pathways, tumor suppressor | 19p13.3 | 40 | [84] | |
| 2014 | DACH1 | Dachshund Family Transcription Factor 1 | TGF-β signaling, transcription factor, tumor suppressor | 13q21.33 | 61.5 | [85] | |
| 2014 | RASSF10 | Ras Association Domain Family Member 10 | Cell proliferation, cell cycle regulation, microtubule stability, tumor suppressor | 11p15.3 | 44.3 | [86] | |
| 2014 | ADAMTS8 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 8 | Cell proliferation, apoptosis, EGFR-MEK-ERK signaling pathway, tumor suppressor | 11q24.3 | 22 | [87] | |
| 2014 | SPINT2 | Serine Peptidase Inhibitor, Kunitz Type 2 | Cell proliferation, apoptosis, tumor suppressor | 19q13.2 | 52.08 | [88] | |
| 2015 | RASSF5A | Ras Association Domain Family Member 5A | Apoptosis, tumor suppressor | 1q32.1 | 47.1 | [89] | |
| 2017 | BIN1 | Bridging Integrator 1 | Cell proliferation, tumor suppressor | 2q14.3 | 62 | [90] | |
| 2017 | PAX1 | Paired Box 1 | Transcription factor, tumor suppressor | 20p11.22 | 96-100 | [91, 92] | |
| 2018 | RHCG | Rh Family C Glycoprotein | Ammonium transporter, invasion, metastasis, NF-κB signaling pathway, tumor suppressor | 15q26.1 | 86.4 | [93] | |
| 2019 | CHL1 | Cell Adhesion Molecule L1 Like | Cell proliferation, cell cycle, metastasis, AKT signaling pathway, tumor suppressor | 3p26.3 | 59.0 | [94] | |
| 2019 | SEMA3B | Semaphorin 3B | Cell proliferation, invasion, tumor suppressor | 3p21.31 | 54.3 | [95] | |
| 2024 | PREX2 | Phosphatidylinositol-3,4,5-Trisphosphate Dependent Rac Exchange Factor 2 | Migration, invasion, tumor suppressor | 8q13.2 | 14.6 | [96] | |
| Other functional genes | 2006 | GATA4 | GATA Binding Protein 4 | Zinc-finger transcription factor | 8p23.1 | 61 | [97] |
| 2006 | GATA5 | GATA Binding Protein 5 | Zinc-finger transcription factor | 20q13.33 | 32 | [97] | |
| 2006 | MT1G | Metallothionein 1G | Cellular stress response, metal metabolism, detoxification | 16q13 | 62 | [35] | |
| 2007 | TAC1 | Tachykinin Precursor 1 | Neurotransmitter | 7q21.3 | 50 | [98] | |
| 2007 | CACNA1G | Calcium Voltage-Gated Channel Subunit Alpha1 G | Calcium-dependent process, hormone or neurotransmitter release, cell proliferation, cell motility and cell death | 17q21.33 | 23.2 | [37] | |
| 2008 | SCGB3A1 (HIN-1) | Secretoglobin Family 3A Member 1 | Signal transduction, cytokine activity, cellular growth | 5q35.3 | 50-72 | [99] | |
| 2011 | HLA-I | Major Histocompatibility Complex, Class I | Immune response, immune evasion, metastasis | 6p21-22 | 70.1 | [100] | |
| 2011 | PTX3 | Pentraxin 3 | Innate immune defense, inflammatory reactions, angiogenesis | 3q25.32 | 85 | [101] | |
| 2011 | GPX3 | Glutathione Peroxidase 3 | Antioxidative defense | 5q33.1 | 54.8-71.4 | [102] | |
| 2017 | PAX5 | Paired Box 5 | Cell proliferation, cell cycle, chemosensitivity, transcription factor | 9p13.2 | 85.9 | [103] | |
| 2017 | ZNF582 | Zinc Finger Protein 582 | Transcriptional regulator | 19q13.43 | 85.7-93.2 | [91, 92] | |
| 2018 | ZNF132 | Zinc Finger Protein 132 | Cell proliferation, apoptosis, migration, transcription factor | 19q13.43 | 70.8 | [104] | |
| 2018 | SLC22A3 | Solute Carrier Family 22 Member 3 | Antioxidative defense, DNA damage repair | 6q25.3 | NA | [105] | |
| 2019 | SOX1 | SRY-Box Transcription Factor 1 | Transcription factor | 13q34 | 89.2 | [92] |
Overview of DNA methylation markers in ESCC systemic chemotherapy, including cytotoxic agents, targeted agents and immune checkpoint inhibitors. Abbreviations: PI3K, phospho-inositol-3 kinase; ATR, ataxia telangiectasia and Rad3-related protein kinase. Created in BioRender. Li, YK. (2025) https://BioRender.com/d45q992 (accessed on 25 January 2025).
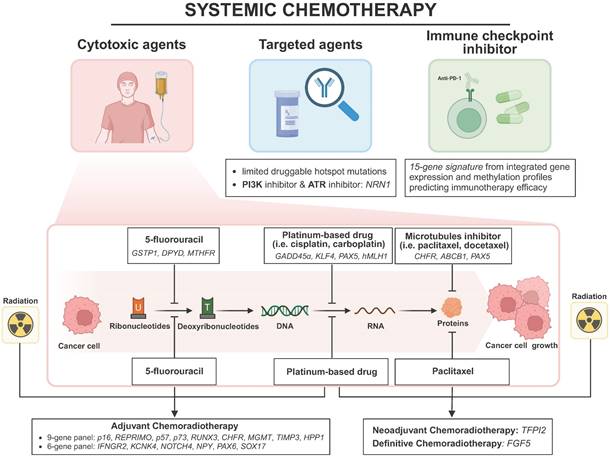
Other findings about methylation markers in predicting platinum-based drug efficacy are as follows. Wang et al. linked GADD45α overexpression to promoter hypomethylation, affecting cisplatin sensitivity through the disruption of apoptotic pathways [108]. Lin et al. demonstrated that long-term cisplatin exposure induces OCT1 methylation, causing cisplatin resistance [124]. Both KLF4 and PAX5 gene methylation were discovered to predict cisplatin sensitivity and clinical outcomes. KLF enhances cisplatin sensitivity in ESCC cells through apoptosis induction and cell cycle arrest, whereas PAX5 contributes to cisplatin resistance by regulating GLUT1, a known chemoresistance factor in cancer cells [103, 125]. Cao et al. confirmed the role of hMLH1 promoter methylation in cisplatin resistance through mismatch repair deficiency, suggesting demethylation treatment may enhance conventional chemotherapy efficacy [126].
About the response to 5-fluorouracil, GSTP1 encoding a detoxifying enzyme was found to be methylated by DNMTs which were recruited by the long noncoding RNAs LINC01270 and LINC01419, thereby reducing the chemosensitivity of ESCC to 5-FU [127, 128]. Meanwhile, dihydropyrimidine dehydrogenase (DPYD) as a 5-FU degrading enzyme, was repressed by LINC00261 overexpression via the methylation-dependent manner, ultimately enhancing the 5-FU response in ESCC [129]. Moreover, MTHFR methylation mediated by lncRNA HOTAIR inhibits 5-FU sensitivity by regulating the systemic exposure of ESCC cells to 5-FU, suggesting that targeting HOTAIR or MTHFR could potentially overcome chemoresistance [130].
As for the efficacy of microtubule inhibitors, Yun et al. reported that CHFR methylation sensitizes ESCC cells to docetaxel and paclitaxel. This occurs because the CHFR protein's role at the prophase checkpoint, where it delays mitotic entry in response to microtubule poisons, thereby attenuating the cytotoxic efficacy of microtubule inhibitors [46]. Sumarpo et al. found that ABCB1 gene amplification and promoter demethylation contribute to the acquisition of taxane resistance in ESCC cell lines [131]. Additionally, Zhang et al. revealed PAX5 as a tumor suppressor gene regulated by promoter methylation, sensitizing ESCC cell lines to docetaxel and 5-FU by promoting p53 signaling activity [132].
With regards to molecular-targeted therapies, few studies on ESCC exist, most of which target EGFR and limited druggable hotspot mutations. Du et al. found that NRN1 methylation is a sensitive marker for PI3K-Akt-mTOR and ATR inhibitors, implying that demethylation of NRN1 could enhance the efficacy of these inhibitors in ESCC [133].
Recent multiple randomized studies have shown that immune checkpoint inhibitors (ICIs), especially PD-1/PD-L1 inhibitors, exhibit antitumor activity in advanced ESCC [134, 135]. Nonetheless, the predictive value of existing maker for immunotherapy like PD-L1 status in ESCC is unsatisfactory [136]. Identifying sensitive and solid predictive markers is essential to improve response and prognosis. Zheng et al. integrated gene expression and DNA methylation profiles to characterize two molecular subtypes of ESCC associated with distinct immune-related pathways and clinical outcomes. The 15-gene expression signature could predict the response rate to immunotherapy [137].
Researches on methylation biomarkers for predicting therapeutic response in ESCC are still ongoing. The studies mentioned above contribute to stratifying ESCC patients for personalized therapy and provide new strategies for overcoming chemotherapeutic drug resistance.
DNA methylation as a therapeutic target in ESCC
DNA methylations differ from genetic alterations by being reversible, making them promising targets for therapeutic intervention [138]. DNA methyltransferases (DNMTs), including DNMT1, DNMT3A, and DNMT3B, catalyze DNA methylation process. Nucleoside analogs such as 5-azacytidine and its derivative 5-aza-2'-deoxycytidine (decitabine) inhibit DNMT activity and are currently approved for treating myelodysplastic syndrome (MDS), significantly improving patient survival. Theoretically, DNMT inhibitors can reverse the hypermethylation-mediated silencing of tumor suppressor or other functional genes. Consequently, multiple preclinical and clinical trials are carried out to assess the efficacy of these agents in solid tumors, including ESCC [139, 140]. To be noted, in a phase Ib/II study by Chen et al. low-dose decitabine-primed chemoimmunotherapy re-sensitized resistant cancer cells and showing promising safety and efficacy in ESCC patients, suggesting DNMT inhibitor as a potential therapeutic agent in relapsed/refractory ESCC patients [140]. Two independent researches both highlighted that low-dose decitabine epigenetically upregulates MAGE-A antigens, enhancing T cell-mediated tumor recognition and antigen-specific responses. These findings suggest that combining decitabine with immunotherapy may be a promising strategy for treating advanced ESCC [141, 142].
Recently Chang et al. demonstrated that Nutlin-3, a murine double min 2 (MDM2) small molecule inhibitor, could function as a novel DNMT inhibitor via enhancing p53 and RB expression, making it a promising DNMT inhibitor for sensitizing chemoradiation-resistant ESCC to therapy [143]. Besides that, natural compounds have demonstrated potential as DNMT inhibitors. Fang et al. reported that (-)-epigallocatechin-3-gallate (EGCG), a major green tea extract, inhibits DNMT1 activity, reversing methylation and restoring the expression of tumor suppressor genes such as CDKN2A, RARB, MGMT, and MLH1 in ESCC cell lines [144]. Similarly, genistein, a soy-derived isoflavone, was found to reverse the hypermethylation of TSGs via a direct inhibition of DNA methyltransferase, suggesting these natural extracts could contribute to the chemoprevention of ESCC [145]. Huang et al. indicated that black raspberries (BRBs) could reduce DNMT1 and DNMT3b mRNA levels, thereby demethylating Sfrp4, inhibiting WNT signaling and suppressing esophageal tumorigenesis in rats, a model akin to human ESCC. This suggests that BRBs may prevent the aberrant DNA methylation in ESCC development, offering a natural approach to target epigenetic alterations in cancer prevention [146].
Nevertheless, currently available DNMT inhibitors are not sufficiently selective, leading to off-target adverse effects including global DNA hypomethylation, oncogene activation and increased genomic instability. Therefore, further researches are needed to develop targeted therapies that specifically reverse the hypermethylation of tumor suppressor genes, thereby improving therapeutic efficacy.
In addition to DNMTs, TET1 as a CpG demethylase was reported to have the catalytic domain to inhibit the CpG methylation of TSG promoters and reactive their expression [147]. Interestingly, TET1 underwent promoter CpG methylation-mediated silencing in human cancers including ESCC, leading to elevated methylation levels in tumor cells via a DNA methylation feedback loop, which suggests targeting TET1 as a therapeutic target is worth further research. UHRF1, a multidomain nuclear protein that recruits DNMT1 to maintain DNA methylation has also been identified as a potential therapeutic target in ESCC patients, particularly those with elevated UHRF1 expression [148].
Regarding the repurposing of non-cancer drugs, there have been some interesting findings. In a pre-clinical trial, Liu et al. showed that the nonsteroidal anti-inflammatory drug celecoxib demethylated TSGs and boosted apoptosis, highlighting its potential as an epigenetic therapy [149]. The study by Wang et al. revealed that metformin, a first-line drug for diabetes, could counteract nicotine-upregulated CHRNA7 expression in ESCC by enhancing promoter hypermethylation, thereby inhibiting JAK2/STAT3/SOX2 signaling pathway and tumor progression. The finding underscores the therapeutic potential of metformin in modulating epigenetic changes and offers a novel strategy for ESCC treatment, especially in smoking patients [150].
Discussion and Perspectives
Despite continuous advancements and improvements in multidisciplinary treatments, esophageal squamous cell carcinoma remains one of the most lethal malignancies in China and worldwide. DNA methylation, a major component of epigenetic modifications, plays a crucial role in the initiation and progression of ESCC. Compared to message RNA and protein, DNA is more stable biochemically, and tumor-derived DNA can be readily obtained from patient specimens including plasma, stool, sputum and urine after being released into the circulation. Moreover, techniques to detect DNA methylation like pyrosequencing, methylated DNA immunoprecipitation sequencing (MeDIP-Seq) and methylation-specific PCR (MSP), provide significant technical advantages. Taken together, the detection of aberrantly methylated genes can be exploited as potent predictive methods for clinical translation.
The first part of this review provides a comprehensive summary of all aberrantly methylated genes identified in ESCC to date and their associated characteristics. Previous studies and reviews have repeatedly highlighted the clinical applications of DNA methylation biomarkers in ESCC, particularly for early screening, early diagnosis and prognostic prediction. However, given the limited efficacy of systemic chemotherapy in ESCC and lack of reliable predictive biomarkers and novel therapeutic targets, there is a pressing need for further exploration. From the perspectives of significance and necessity, the latter part of this review meticulously discusses the predictive value of aberrant DNA methylation for therapeutic efficacy in systemic treatments and its potential as a promising therapeutic target.
Through extensive review of the literature, we have found that the major obstacle preventing DNA methylation markers from becoming truly useful biomarkers in ESCC is the lack of suitable large-scale, prospective clinical trial validation. In the future, we will focus on more large-sample prospective cohort studies and even initiate large-scale investigator-driven clinical trials based on our promising preliminary findings to identify and verify DNA methylation markers that can genuinely improve treatment outcomes and prognosis in clinical settings. Besides that, we have observed that epigenetic patterns like DNA methylation, exhibit considerable plasticity and undergo dynamic changes during tumor progression or under therapeutic selection pressure. Therefore, it is essential to adopt a more dynamic perspective to evaluate DNA methylation markers, paying close attention to their temporal changes. By flexibly interpreting thresholds and incorporating these dynamic processes, we can achieve more precise treatment and timely therapeutic adjustments.
ESCC exhibits a relatively high proportion of methylation abnormalities, and epigenetic therapies targeting these aberrations hold substantial potential, with multiple clinical studies currently underway. It is important to note, however, that epigenetic therapies generally lack specificity, leading to considerable off-target effects and toxicity. As a result, most methylation-targeting drugs are currently administrated at low doses and are used in combination with conventional chemotherapy agents. Future research needs to develop more specific methylation-targeting drugs to enhance both efficacy and safety, as well as exploring the synergistic effects of these drugs with other conventional therapies, with the aim of significantly improving treatment outcomes and reversing drug resistance. It is anticipated that large-scale clinical trials in ESCC along with further exploration of the regulatory mechanisms of DNA methylation will discover more effective predictors of therapeutic response and identify novel therapeutic targets for eventual clinical application.
Conclusion
This review underscores the pivotal role of DNA methylation in ESCC, particularly its involvement in systemic chemotherapy. The key findings highlight the identification of numerous aberrantly methylated genes that disrupt critical pathways in ESCC. Clinically, DNA methylation markers hold promise for predicting therapeutic responses, though their utility is hindered by insufficient validation and off-target effects. Future research should prioritize large-scale validation of methylation markers through prospective trials and the development of more targeted epigenetic therapies. Integrating dynamic changes in methylation during treatment could enhance precision medicine. Further exploration of methylation mechanisms may reveal new therapeutic targets and improve clinical outcomes in ESCC.
Abbreviations
ESCC: Esophageal squamous cell carcinoma; EAC: Esophageal adenocarcinoma; DDP: Cisplatin; 5-FU: 5-fluorouracil; FDA: Food and drug administration; TSG: Tumor suppressor gene; LINEs: Long interspersed nuclear elements; SINEs: Short interspersed nuclear elements; IL6: Interleukin 6; dCRT: Definitive chemoradiotherapy; WGBS: Whole genome bisulfite sequencing; DNMTs: DNA methyltransferases; DPYD: dihydropyrimidine dehydrogenase; ATR: Ataxia Telangiectasia and Rad3-related protein kinase; ICI: Immune checkpoint inhibitor; MeDIP-Seq: Methylated DNA immunoprecipitation sequencing; MSP: Methylation-specific polymerase chain reaction.
Acknowledgements
The authors thank their colleagues in the Department of Oncology, the Department of Health Prevention and Care, Beijing Hospital and the Department of Thoracic Surgery, Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, for supporting the review.
Funding
The authors received no specific funding for this work.
Author contributions
Z.L. and Y.Z. had the idea for the review. Z.L. and X.C. performed the literature search, data analysis and table preparation. YK.L. and Y.Z. designed and created illustrations. Z.L. and Y.Z. drafted the original manuscript. Y.Z. and Y.X. critically reviewed and revised the work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Laversanne M, Sung H. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-63
2. Arnold M, Soerjomataram I, Ferlay J. et al. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381-7
3. Lipenga T, Matumba L, Vidal A. et al. A concise review towards defining the exposome of oesophageal cancer in sub-Saharan Africa. Environ Int. 2021;157:106880
4. Codipilly DC, Qin Y, Dawsey SM. et al. Screening for esophageal squamous cell carcinoma: recent advances. Gastrointest Endosc. 2018;88:413-26
5. Sheikh M, Poustchi H, Pourshams A. et al. Individual and Combined Effects of Environmental Risk Factors for Esophageal Cancer Based on Results From the Golestan Cohort Study. Gastroenterology. 2019;156:1416-27
6. Li M, Park JY, Sheikh M. et al. Population-based investigation of common and deviating patterns of gastric cancer and oesophageal cancer incidence across populations and time. Gut. 2023;72:846-54
7. Blot W, Tarone R. Esophageal Cancer In: Thun MJ, Linet MS, Cerhan JR, Haiman C, Schottenfeld D, editors. Schottenfeld and Fraumeni Cancer Epidemiology and Prevention. New York, NY: Oxford University Press. 2018
8. Han B, Zheng R, Zeng H. et al. Cancer incidence and mortality in China, 2022. J Natl Cancer Cent. 2024;4:47-53
9. Li K, Leng X, He W. et al. Resected lymph nodes and survival of patients with esophageal squamous cell carcinoma: an observational study. Int J Surg. 2023;109:2001-9
10. Samson P, Robinson C, Bradley J. et al. Neoadjuvant Chemotherapy versus Chemoradiation Prior to Esophagectomy: Impact on Rate of Complete Pathologic Response and Survival in Esophageal Cancer Patients. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2016;11:2227-37
11. Kies MS, Rosen ST, Tsang T. et al. Cisplatin and 5-fluorouracil in the primary management of squamous esophageal cancer. Cancer. 1987;60:2156-60
12. Shah MA, Kojima T, Hochhauser D. et al. Efficacy and safety of pembrolizumab for heavily pretreated patients with advanced, metastatic adenocarcinoma or squamous cell carcinoma of the esophagus: the phase 2 KEYNOTE-180 study. JAMA oncology. 2019;5:546-50
13. Kojima T, Muro K, Francois E. et al. Pembrolizumab versus chemotherapy as second-line therapy for advanced esophageal cancer: Phase III KEYNOTE-181 study. American Society of Clinical Oncology. 2019;38:4138-4148
14. Kudo T, Hamamoto Y, Kato K. et al. Nivolumab treatment for oesophageal squamous-cell carcinoma: an open-label, multicentre, phase 2 trial. Lancet Oncol. 2017;18:631-9
15. Takahashi M, Kato K, Okada M. et al. Nivolumab versus chemotherapy in Japanese patients with advanced esophageal squamous cell carcinoma: a subgroup analysis of a multicenter, randomized, open-label, phase 3 trial (ATTRACTION-3). Esophagus. 2021;18:90-9
16. Ruhstaller T, Thuss-Patience P, Hayoz S. et al. Neoadjuvant chemotherapy followed by chemoradiation and surgery with and without cetuximab in patients with resectable esophageal cancer: a randomized, open-label, phase III trial (SAKK 75/08). Ann Oncol. 2018;29:1386-93
17. Han X, Lu N, Pan Y. et al. Nimotuzumab Combined with Chemotherapy is a Promising Treatment for Locally Advanced and Metastatic Esophageal Cancer. Medical science monitor: international medical journal of experimental and clinical research. 2017;23:412-8
18. Lin DC, Wang MR, Koeffler HP. Genomic and Epigenomic Aberrations in Esophageal Squamous Cell Carcinoma and Implications for Patients. Gastroenterology. 2018;154:374-89
19. You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell. 2012;22:9-20
20. Miremadi A, Oestergaard MZ, Pharoah PD. et al. Cancer genetics of epigenetic genes. Human molecular genetics. 2007;16:R28-R49
21. Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nature Reviews Genetics. 2009;10:691-703
22. Hogg SJ, Beavis PA, Dawson MA. et al. Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discov. 2020;19:776-800
23. Li Y, Liu B, Zhou X. et al. Genome-Scale Multimodal Analysis of Cell-Free DNA Whole-Methylome Sequencing for Noninvasive Esophageal Cancer Detection. JCO Precis Oncol. 2024;8:e2400111
24. Talukdar FR, Soares Lima SC, Khoueiry R. et al. Genome-Wide DNA Methylation Profiling of Esophageal Squamous Cell Carcinoma from Global High-Incidence Regions Identifies Crucial Genes and Potential Cancer Markers. Cancer research. 2021;81:2612-24
25. Cao W, Lee H, Wu W. et al. Multi-faceted epigenetic dysregulation of gene expression promotes esophageal squamous cell carcinoma. Nat Commun. 2020;11:3675
26. Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89-92
27. Gaudet F, Hodgson JG, Eden A. et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489-92
28. Kawano H, Saeki H, Kitao H. et al. Chromosomal instability associated with global DNA hypomethylation is associated with the initiation and progression of esophageal squamous cell carcinoma. Annals of surgical oncology. 2014;21(Suppl 4):S696-702
29. Baba Y, Watanabe M, Murata A. et al. LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:1114-24
30. Iwagami S, Baba Y, Watanabe M. et al. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Annals of surgery. 2013;257:449-55
31. Hoshimoto S, Takeuchi H, Ono S. et al. Genome-wide hypomethylation and specific tumor-related gene hypermethylation are associated with esophageal squamous cell carcinoma outcome. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2015;10:509-17
32. Li JS, Ying JM, Wang XW. et al. Promoter methylation of tumor suppressor genes in esophageal squamous cell carcinoma. Chinese journal of cancer. 2013;32:3-11
33. Tanaka H, Shimada Y, Harada H. et al. Methylation of the 5′ CpG island of the FHIT gene is closely associated with transcriptional inactivation in esophageal squamous cell carcinomas. Cancer research. 1998;58:3429-34
34. Kuroki T, Trapasso F, Yendamuri S. et al. Allele loss and promoter hypermethylation of VHL, RAR-β, RASSF1A, and FHIT tumor suppressor genes on chromosome 3p in esophageal squamous cell carcinoma. Cancer research. 2003;63:3724-8
35. Roth MJ, Abnet CC, Hu N. et al. p16, MGMT, RARbeta2, CLDN3, CRBP and MT1G gene methylation in esophageal squamous cell carcinoma and its precursor lesions. Oncology reports. 2006;15:1591-7
36. Zhang L, Lu W, Miao X. et al. Inactivation of DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relation to p53 mutations in esophageal squamous cell carcinoma. Carcinogenesis. 2003;24:1039-44
37. Ishii T, Murakami J, Notohara K. et al. Oesophageal squamous cell carcinoma may develop within a background of accumulating DNA methylation in normal and dysplastic mucosa. Gut. 2007;56:13-9
38. Guo M, Ren J, House MG. et al. Accumulation of promoter methylation suggests epigenetic progression in squamous cell carcinoma of the esophagus. Clin Cancer Res. 2006;12:4515-22
39. Tzao C, Hsu H-S, Sun G-H. et al. Promoter methylation of the hMLH1 gene and protein expression of human mutL homolog 1 and human mutS homolog 2 in resected esophageal squamous cell carcinoma. The Journal of thoracic and cardiovascular surgery. 2005;130:1371 e1-. e8
40. Zhang G, Ma C, Liu Q. et al. Detection of methylation of hMSH2 gene promoter region of esophageal cancer. Zhonghua zhong liu za zhi [Chinese journal of oncology]. 2005;27:541-3
41. Li P, Liu X, Dong ZM. et al. Epigenetic silencing of HIC1 promotes epithelial-mesenchymal transition and drives progression in esophageal squamous cell carcinoma. Oncotarget. 2015;6:38151-65
42. Luo M, Li Y, Shi X. et al. Aberrant methylation of EYA4 promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. Cancer science. 2018;109:1811-24
43. Xing EP, Nie Y, Song Y. et al. Mechanisms of inactivation of p14ARF, p15INK4b, and p16INK4a genes in human esophageal squamous cell carcinoma. Clin Cancer Res. 1999;5:2704-13
44. Lee EJ, Lee BB, Han J. et al. CpG island hypermethylation of E-cadherin (CDH1) and integrin alpha4 is associated with recurrence of early stage esophageal squamous cell carcinoma. International journal of cancer. 2008;123:2073-9
45. Lima SC, Hernandez-Vargas H, Simao T. et al. Identification of a DNA methylome signature of esophageal squamous cell carcinoma and potential epigenetic biomarkers. Epigenetics. 2011;6:1217-27
46. Yun T, Liu Y, Gao D. et al. Methylation of CHFR sensitizes esophageal squamous cell cancer to docetaxel and paclitaxel. Genes & cancer. 2015;6:38-48
47. Mandelker DL, Yamashita K, Tokumaru Y. et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer research. 2005;65:4963-8
48. Cheng Y, Geng H, Cheng SH. et al. KRAB zinc finger protein ZNF382 is a proapoptotic tumor suppressor that represses multiple oncogenes and is commonly silenced in multiple carcinomas. Cancer research. 2010;70:6516-26
49. Zheng Y, Zhang Y, Huang X. et al. Analysis of the RUNX3 gene methylation in serum DNA from esophagus squamous cell carcinoma, gastric and colorectal adenocarcinoma patients. Hepato-gastroenterology. 2011;58:2007-11
50. Liu JB, Qiang FL, Dong J. et al. Plasma DNA methylation of Wnt antagonists predicts recurrence of esophageal squamous cell carcinoma. World journal of gastroenterology. 2011;17:4917-21
51. Ito T, Shimada Y, Hashimoto Y. et al. Involvement of TSLC1 in progression of esophageal squamous cell carcinoma. Cancer research. 2003;63:6320-6
52. Takeno S, Noguchi T, Fumoto S. et al. E-cadherin expression in patients with esophageal squamous cell carcinoma. American journal of clinical pathology. 2004;122:78-84
53. Hibi K, Kodera Y, Ito K. et al. Methylation pattern of CDH13 gene in digestive tract cancers. British journal of cancer. 2004;91:1139-42
54. Ying J, Li H, Seng TJ. et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25:1070-80
55. Park HL, Kim MS, Yamashita K. et al. DCC promoter hypermethylation in esophageal squamous cell carcinoma. International journal of cancer. 2008;122:2498-502
56. Kong KL, Kwong DL, Fu L. et al. Characterization of a candidate tumor suppressor gene uroplakin 1A in esophageal squamous cell carcinoma. Cancer research. 2010;70:8832-41
57. Sung CO, Han SY, Kim SH. Low expression of claudin-4 is associated with poor prognosis in esophageal squamous cell carcinoma. Annals of surgical oncology. 2011;18:273-81
58. Li L, Ying J, Li H. et al. The human cadherin 11 is a pro-apoptotic tumor suppressor modulating cell stemness through Wnt/β-catenin signaling and silenced in common carcinomas. Oncogene. 2012;31:3901-12
59. Mizuiri H, Yoshida K, Toge T. et al. DNA methylation of genes linked to retinoid signaling in squamous cell carcinoma of the esophagus: DNA methylation of CRBP1 and TIG1 is associated with tumor stage. Cancer science. 2005;96:571-7
60. Tanaka K, Imoto I, Inoue J. et al. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene. 2007;26:6456-68
61. Hussain S, Singh N, Salam I. et al. Methylation-mediated gene silencing of suppressor of cytokine signaling-1 (SOCS-1) gene in esophageal squamous cell carcinoma patients of Kashmir valley. Journal of receptor and signal transduction research. 2011;31:147-56
62. Chan SL, Cui Y, van Hasselt A. et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Laboratory investigation; a journal of technical methods and pathology. 2007;87:644-50
63. Jia Y, Yang Y, Zhan Q. et al. Inhibition of SOX17 by microRNA 141 and methylation activates the WNT signaling pathway in esophageal cancer. The Journal of molecular diagnostics: JMD. 2012;14:577-85
64. Yue CM, Deng DJ, Bi MX. et al. Expression of ECRG4, a novel esophageal cancer-related gene, downregulated by CpG island hypermethylation in human esophageal squamous cell carcinoma. World journal of gastroenterology. 2003;9:1174-8
65. Yamashita K, Mimori K, Inoue H. et al. A tumor-suppressive role for trypsin in human cancer progression. Cancer research. 2003;63:6575-8
66. Smith E, Drew PA, Tian ZQ. et al. Metallothionien 3 expression is frequently down-regulated in oesophageal squamous cell carcinoma by DNA methylation. Molecular cancer. 2005;4:42
67. Kim MS, Yamashita K, Baek JH. et al. N-methyl-D-aspartate receptor type 2B is epigenetically inactivated and exhibits tumor-suppressive activity in human esophageal cancer. Cancer research. 2006;66:3409-18
68. Seng TJ, Low JS, Li H. et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934-44
69. Guo M, House MG, Suzuki H. et al. Epigenetic silencing of CDX2 is a feature of squamous esophageal cancer. International journal of cancer. 2007;121:1219-26
70. Fu L, Qin YR, Xie D. et al. Characterization of a novel tumor-suppressor gene PLC delta 1 at 3p22 in esophageal squamous cell carcinoma. Cancer research. 2007;67:10720-6
71. Jin H, Wang X, Ying J. et al. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene. 2007;26:7490-8
72. Zhang C, Li K, Wei L. et al. p300 expression repression by hypermethylation associated with tumour invasion and metastasis in oesophageal squamous cell carcinoma. Journal of clinical pathology. 2007;60:1249-53
73. Ohta M, Mimori K, Fukuyoshi Y. et al. Clinical significance of the reduced expression of G protein gamma 7 (GNG7) in oesophageal cancer. British journal of cancer. 2008;98:410-7
74. Jin Z, Mori Y, Hamilton JP. et al. Hypermethylation of the somatostatin promoter is a common, early event in human esophageal carcinogenesis. Cancer. 2008;112:43-9
75. Yamashita K, Kim MS, Park HL. et al. HOP/OB1/NECC1 promoter DNA is frequently hypermethylated and involved in tumorigenic ability in esophageal squamous cell carcinoma. Molecular cancer research: MCR. 2008;6:31-41
76. Lee KY, Geng H, Ng KM. et al. Epigenetic disruption of interferon-gamma response through silencing the tumor suppressor interferon regulatory factor 8 in nasopharyngeal, esophageal and multiple other carcinomas. Oncogene. 2008;27:5267-76
77. Wong VC, Chan PL, Bernabeu C. et al. Identification of an invasion and tumor-suppressing gene, Endoglin (ENG), silenced by both epigenetic inactivation and allelic loss in esophageal squamous cell carcinoma. International journal of cancer. 2008;123:2816-23
78. Zhu C, Qin YR, Xie D. et al. Characterization of tumor suppressive function of P300/CBP-associated factor at frequently deleted region 3p24 in esophageal squamous cell carcinoma. Oncogene. 2009;28:2821-8
79. Kim MS, Chang X, LeBron C. et al. Neurofilament heavy polypeptide regulates the Akt-beta-catenin pathway in human esophageal squamous cell carcinoma. PloS one. 2010;5:e9003
80. Chang X, Yamashita K, Sidransky D. et al. Promoter methylation of heat shock protein B2 in human esophageal squamous cell carcinoma. International journal of oncology. 2011;38:1129-35
81. Ma S, Bao JYJ, Kwan PS. et al. Identification of PTK6, via RNA sequencing analysis, as a suppressor of esophageal squamous cell carcinoma. Gastroenterology. 2012;143:675-86.e12
82. Jia Y, Yang Y, Brock MV. et al. Methylation of TFPI-2 is an early event of esophageal carcinogenesis. Epigenomics. 2012;4:135-46
83. Tong M, Chan KW, Bao JY. et al. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer research. 2012;72:6024-35
84. Zhu YH, Fu L, Chen L. et al. Downregulation of the novel tumor suppressor DIRAS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer research. 2013;73:2298-309
85. Wu L, Herman JG, Brock MV. et al. Silencing DACH1 promotes esophageal cancer growth by inhibiting TGF-beta signaling. PloS one. 2014;9:e95509
86. Lu D, Ma J, Zhan Q. et al. Epigenetic silencing of RASSF10 promotes tumor growth in esophageal squamous cell carcinoma. Discovery medicine. 2014;17:169-78
87. Choi GC, Li J, Wang Y. et al. The metalloprotease ADAMTS8 displays antitumor properties through antagonizing EGFR-MEK-ERK signaling and is silenced in carcinomas by CpG methylation. Molecular cancer research: MCR. 2014;12:228-38
88. Yue D, Fan Q, Chen X. et al. Epigenetic inactivation of SPINT2 is associated with tumor suppressive function in esophageal squamous cell carcinoma. Experimental cell research. 2014;322:149-58
89. Guo W, Wang C, Guo Y. et al. RASSF5A, a candidate tumor suppressor, is epigenetically inactivated in esophageal squamous cell carcinoma. Clinical & experimental metastasis. 2015;32:83-98
90. Wang X, Wang J, Jia Y. et al. Methylation decreases the Bin1 tumor suppressor in ESCC and restoration by decitabine inhibits the epithelial mesenchymal transition. Oncotarget. 2017;8:19661-73
91. Huang J, Wang G, Tang J. et al. DNA Methylation Status of PAX1 and ZNF582 in Esophageal Squamous Cell Carcinoma. International journal of environmental research and public health. 2017;14:216
92. Tang L, Liou YL, Wan ZR. et al. Aberrant DNA methylation of PAX1, SOX1 and ZNF582 genes as potential biomarkers for esophageal squamous cell carcinoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;120:109488
93. Ming XY, Zhang X, Cao TT. et al. RHCG Suppresses Tumorigenicity and Metastasis in Esophageal Squamous Cell Carcinoma via Inhibiting NF-κB Signaling and MMP1 Expression. Theranostics. 2018;8:185-98
94. Tang H, Jiang L, Zhu C. et al. Loss of cell adhesion molecule L1 like promotes tumor growth and metastasis in esophageal squamous cell carcinoma. Oncogene. 2019;38:3119-33
95. Dong Z, Liang X, Wu X. et al. Promoter hypermethylation-mediated downregulation of tumor suppressor gene SEMA3B and lncRNA SEMA3B-AS1 correlates with progression and prognosis of esophageal squamous cell carcinoma. Clinical & experimental metastasis. 2019;36:225-41
96. Zhou Y, Mo S, Cui H. et al. Immune-tumor interaction dictates spatially directed evolution of esophageal squamous cell carcinoma. Natl Sci Rev. 2024;11:nwae150
97. Guo M, House MG, Akiyama Y. et al. Hypermethylation of the GATA gene family in esophageal cancer. International journal of cancer. 2006;119:2078-83
98. Jin Z, Olaru A, Yang J. et al. Hypermethylation of tachykinin-1 is a potential biomarker in human esophageal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:6293-300
99. Guo M, Ren J, Brock MV. et al. Promoter methylation of HIN-1 in the progression to esophageal squamous cancer. Epigenetics. 2008;3:336-41
100. Qifeng S, Bo C, Xingtao J. et al. Methylation of the promoter of human leukocyte antigen class I in human esophageal squamous cell carcinoma and its histopathological characteristics. The Journal of thoracic and cardiovascular surgery. 2011;141:808-14
101. Wang JX, He YL, Zhu ST. et al. Aberrant methylation of the 3q25 tumor suppressor gene PTX3 in human esophageal squamous cell carcinoma. World journal of gastroenterology. 2011;17:4225-30
102. He Y, Wang Y, Li P. et al. Identification of GPX3 epigenetically silenced by CpG methylation in human esophageal squamous cell carcinoma. Digestive diseases and sciences. 2011;56:681-8
103. Kurimoto K, Hayashi M, Guerrero-Preston R. et al. PAX5 gene as a novel methylation marker that predicts both clinical outcome and cisplatin sensitivity in esophageal squamous cell carcinoma. Epigenetics. 2017;12:865-74
104. Jiang D, He Z, Wang C. et al. Epigenetic silencing of ZNF132 mediated by methylation-sensitive Sp1 binding promotes cancer progression in esophageal squamous cell carcinoma. Cell Death Dis. 2018;10:1
105. Xiong JX, Wang YS, Sheng J. et al. Epigenetic alterations of a novel antioxidant gene SLC22A3 predispose susceptible individuals to increased risk of esophageal cancer. Int J Biol Sci. 2018;14:1658-68
106. Chen C, Peng H, Huang X. et al. Genome-wide profiling of DNA methylation and gene expression in esophageal squamous cell carcinoma. Oncotarget. 2016;7:4507-21
107. Cui L, Xu LY, Shen ZY. et al. NGALR is overexpressed and regulated by hypomethylation in esophageal squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:7674-81
108. Wang B, Yin BL, He B. et al. Overexpression of DNA damage-induced 45 α gene contributes to esophageal squamous cell cancer by promoter hypomethylation. Journal of experimental & clinical cancer research: CR. 2012;31:11
109. Zhou J, Zhang S, Xie L. et al. Overexpression of DNA polymerase iota (Polι) in esophageal squamous cell carcinoma. Cancer science. 2012;103:1574-9
110. Murata A, Baba Y, Watanabe M. et al. IGF2 DMR0 methylation, loss of imprinting, and patient prognosis in esophageal squamous cell carcinoma. Annals of surgical oncology. 2014;21:1166-74
111. Wang C, Li Z, Shao F. et al. High expression of Collagen Triple Helix Repeat Containing 1 (CTHRC1) facilitates progression of oesophageal squamous cell carcinoma through MAPK/MEK/ERK/FRA-1 activation. Journal of experimental & clinical cancer research: CR. 2017;36:84
112. He W, Gong S, Wang X. et al. DNA methylation integratedly modulates the expression of Pit-Oct-Unt transcription factors in esophageal squamous cell carcinoma. Journal of Cancer. 2021;12:1634-43
113. Chen Y, Wang D, Peng H. et al. Epigenetically upregulated oncoprotein PLCE1 drives esophageal carcinoma angiogenesis and proliferation via activating the PI-PLCε-NF-κB signaling pathway and VEGF-C/ Bcl-2 expression. Molecular cancer. 2019;18:1
114. Kaya Z, Almalı N, Sahin ES. et al. Association of insulin-like growth factor binding protein-7 promoter methylation with esophageal cancer in peripheral blood. Molecular biology reports. 2022;49:3423-31
115. Soejima H, Nakagawachi T, Zhao W. et al. Silencing of imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at DMR-LIT1 in esophageal cancer. Oncogene. 2004;23:4380-8
116. Hamilton JP, Sato F, Greenwald BD. et al. Promoter methylation and response to chemotherapy and radiation in esophageal cancer. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2006;4:701-8
117. Hamilton JP, Sato F, Jin Z. et al. Reprimo methylation is a potential biomarker of Barrett's-Associated esophageal neoplastic progression. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:6637-42
118. Chang WL, Lai WW, Kuo IY. et al. A six-CpG panel with DNA methylation biomarkers predicting treatment response of chemoradiation in esophageal squamous cell carcinoma. Journal of gastroenterology. 2017;52:705-14
119. Iwabu J, Yamashita S, Takeshima H. et al. FGF5 methylation is a sensitivity marker of esophageal squamous cell carcinoma to definitive chemoradiotherapy. Scientific reports. 2019;9:13347
120. Salta S, Macedo-Silva C, Miranda-Gonçalves V. et al. A DNA methylation-based test for esophageal cancer detection. Biomark Res. 2020;8:68
121. de Klerk LK, Goedegebuure RSA, van Grieken NCT. et al. Molecular profiles of response to neoadjuvant chemoradiotherapy in oesophageal cancers to develop personalized treatment strategies. Mol Oncol. 2021;15:901-14
122. Huang D, Wang Y, He Y. et al. Paraoxonase 3 is involved in the multi-drug resistance of esophageal cancer. Cancer cell international. 2018;18:168
123. Min Q, Wang Y, Wu Q. et al. Genomic and epigenomic evolution of acquired resistance to combination therapy in esophageal squamous cell carcinoma. JCI Insight. 2021;6:e150203
124. Lin R, Li X, Li J. et al. Long-term cisplatin exposure promotes methylation of the OCT1 gene in human esophageal cancer cells. Digestive diseases and sciences. 2013;58:694-8
125. Chen C, Ma Z, Zhang H. et al. Krüppel-Like Factor 4 Enhances Sensitivity of Cisplatin to Esophageal Squamous Cell Carcinoma (ESCC) Cells. Medical science monitor: international medical journal of experimental and clinical research. 2017;23:3353-9
126. Cao Y, Chen Y, Huang Y. et al. In vitro study of human mutL homolog 1 hypermethylation in inducing drug resistance of esophageal carcinoma. Irish journal of medical science. 2017;186:257-63
127. Li N, Zhao Z, Miao F. et al. Silencing of long non-coding RNA LINC01270 inhibits esophageal cancer progression and enhances chemosensitivity to 5-fluorouracil by mediating GSTP1methylation. Cancer Gene Ther. 2021;28:471-85
128. Chen JL, Lin ZX, Qin YS. et al. Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation. Ther Adv Med Oncol. 2019;11:1758835919838958
129. Lin K, Jiang H, Zhuang SS. et al. Long noncoding RNA LINC00261 induces chemosensitization to 5-fluorouracil by mediating methylation-dependent repression of DPYD in human esophageal cancer. Faseb j. 2019;33:1972-88
130. Zhang S, Zheng F, Zhang L. et al. LncRNA HOTAIR-mediated MTHFR methylation inhibits 5-fluorouracil sensitivity in esophageal cancer cells. Journal of experimental & clinical cancer research: CR. 2020;39:131
131. Sumarpo A, Ito K, Saiki Y. et al. Genetic and epigenetic aberrations of ABCB1 synergistically boost the acquisition of taxane resistance in esophageal squamous cancer cells. Biochemical and biophysical research communications. 2020;526:586-91
132. Zhang W, Yan W, Qian N. et al. Paired box 5 increases the chemosensitivity of esophageal squamous cell cancer cells by promoting p53 signaling activity. Chin Med J (Engl). 2022;135:606-18
133. Du W, Gao A, Herman JG. et al. Methylation of NRN1 is a novel synthetic lethal marker of PI3K-Akt-mTOR and ATR inhibitors in esophageal cancer. Cancer science. 2021;112:2870-83
134. Sun JM, Shen L, Shah MA. et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): a randomised, placebo-controlled, phase 3 study. Lancet. 2021;398:759-71
135. Wang ZX, Cui C, Yao J. et al. Toripalimab plus chemotherapy in treatment-naïve, advanced esophageal squamous cell carcinoma (JUPITER-06): A multi-center phase 3 trial. Cancer Cell. 2022;40:277-88.e3
136. Yang H, Wang K, Wang T. et al. The Combination Options and Predictive Biomarkers of PD-1/PD-L1 Inhibitors in Esophageal Cancer. Frontiers in oncology. 2020;10:300
137. Zheng Y, Gao Q, Su X. et al. Genome-Wide DNA Methylation and Gene Expression Profiling Characterizes Molecular Subtypes of Esophagus Squamous Cell Carcinoma for Predicting Patient Survival and Immunotherapy Efficacy. Cancers. 2022;14:4970
138. Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442-60 e1
139. Schrump DS, Fischette MR, Nguyen DM. et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res. 2006;12:5777-85
140. Chen M, Nie J, Liu Y. et al. Phase Ib/II study of safety and efficacy of low-dose decitabine-primed chemoimmunotherapy in patients with drug-resistant relapsed/refractory alimentary tract cancer. International journal of cancer. 2018;143:1530-40
141. Shi X, Chen X, Fang B. et al. Decitabine enhances tumor recognition by T cells through upregulating the MAGE-A3 expression in esophageal carcinoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;112:108632
142. Liu WH, Sang MX, Hou SY. et al. Low-dose decitabine induces MAGE-A expression and inhibits invasion via suppression of NF-kB2 and MMP2 in Eca109 cells. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2014;68:745-50
143. Chang WL, Hsieh CH, Kuo IY. et al. Nutlin-3 acts as a DNA methyltransferase inhibitor to sensitize esophageal cancer to chemoradiation. Molecular carcinogenesis. 2023;62:277-87
144. Fang MZ, Wang Y, Ai N. et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer research. 2003;63:7563-70
145. Fang MZ, Chen D, Sun Y. et al. Reversal of hypermethylation and reactivation of p16INK4a, RARβ, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11:7033-41
146. Huang YW, Gu F, Dombkowski A. et al. Black raspberries demethylate Sfrp4, a WNT pathway antagonist, in rat esophageal squamous cell papilloma. Molecular carcinogenesis. 2016;55:1867-75
147. Li L, Li C, Mao H. et al. Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Scientific reports. 2016;6:26591
148. Ye J, Zhang Y, Liang W. et al. UHRF1 is an Independent Prognostic Factor and a Potential Therapeutic Target of Esophageal Squamous Cell Carcinoma. Journal of Cancer. 2017;8:4027-39
149. Liu JF, Li YS, Drew PA. et al. The effect of celecoxib on DNA methylation of CDH13, TFPI2, and FSTL1 in squamous cell carcinoma of the esophagus in vivo. Anti-cancer drugs. 2016;27:848-53
150. Wang L, Du L, Xiong X. et al. Repurposing dextromethorphan and metformin for treating nicotine-induced cancer by directly targeting CHRNA7 to inhibit JAK2/STAT3/SOX2 signaling. Oncogene. 2021;40:1974-87
Author contact
![]() Corresponding authors: Yang Zhou, Email: zhouyangac.cn, Tel.: +86-15210588950. Yan Xu, Email: 18618168070com, Tel.: +86-18618168070.
Corresponding authors: Yang Zhou, Email: zhouyangac.cn, Tel.: +86-15210588950. Yan Xu, Email: 18618168070com, Tel.: +86-18618168070.

 Global reach, higher impact
Global reach, higher impact