Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(3):596-608. doi:10.7150/ijms.67840 This issue Cite
Research Paper
Loss-of-function Mutations K11E or E271K Lead to Novel Tumor Suppression, Implicate Nucleolar Helicase DDX24 Oncogenicity
1. Guangdong Provincial Key Laboratory of Biomedical Imaging and Guangdong Provincial Engineering Research Center of Molecular Imaging, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong Province 519000, China
2. Department of Ultrasound, The First Affiliated Hospital of Shenzhen University Health Science Center, Shenzhen Second People's Hospital, Shenzhen, Guangdong Province 518000, China
3. Department of Radiology, Zhongshan Affiliated Hospital, Guangzhou University of Chinese Medicine, Zhongshan, Guangdong Province 528400, China
4. Department of Radiology, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong Province 519000, China
5. Department of Ultrasound, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong Province 519000, China
6. Center of Oncology, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong Province 519000, China
7. Department of Interventional Medicine, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong Province 519000, China
* These authors contributed equally to this article.
Received 2021-10-8; Accepted 2022-2-17; Published 2022-3-14
Abstract
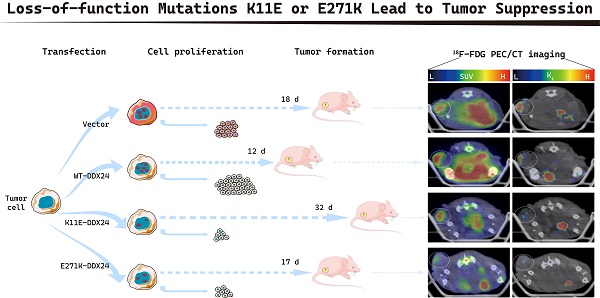
Purpose: Mutations (K11E or E271K) of DEAD-box RNA helicase 24 (DDX24) were related to multi-organ venous lymphatic malformation syndrome (MOVLD). However, the relationship between these mutations and DDX24-function still remains unknown. Understanding whether K11E and E271K cause “loss-of-function” or “gain-of-function” for DDX24 is significant for related diseases. DDX24 was reported to be related to tumors closely, thus this study aims to explore how K11E and E271K affect DDX24-function in tumor proliferation.
Methods: Cell lines stably expressing wild-type DDX24, K11E-DDX24, E271K-DDX24, along with vector only based on Chinese hamster ovary cells (CHO) and Balb/c tumor-bearing mice models were constructed. Then immunofluorescence staining, proliferation assay and colony formation assay in vitro and 18F-FDG PET/CT-scan were performed. Finally, the tumor tissues were collected to perform transcriptome sequencing to predict the potential mechanism.
Results: Contrasted with CHO-WT-DDX24, CHO-K11E-DDX24 or CHO-E271K-DDX24 showed a decreased number of nucleoli, a slower proliferation rate and a lower colony formation rate significantly. Moreover, mice, inoculated with CHO-K11E-DDX24 or CHO-E271K-DDX24 cells, showed lower tumor formation rate, slower tumor growth rate, better prognosis, reduced standard uptake value and Ki of glucose in subcutaneous tumors. Sequencing indicated CHO-K11E-DDX24 or CHO-E271K-DDX24 caused increasing expression of TNF or chemokines and alteration in immune-related signal pathways.
Conclusion: K11E or E271K mutation could lead to “loss-of-function” of DDX24 in cell proliferation and tumor bearing mice, which may be acted by non-specific immune killing to inhibit tumor growth.
Keywords: Nucleolar, mutation, loss-of-function, DDX24
Introduction
DEAD-box RNA helicases, named because of the characteristic Asp-Glu-Ala-Asp (DEAD) in the conserved motif, referred to as DDX, are the largest family in super family 2 helicases [1]. The helicase core region of DDX consists of two RecA-like domains with at least twelve conserved motifs, serving binding sites for ATP and RNA. The N-terminal and C-terminal domains, flanking the helicase core, confer diverse functions on DDX by interacting with other protein and RNAs (Figure S1A). The Q motif, together with motif I (or Walker A motif), motif II (or Walker B motif), and motif VI, is required for ATP binding and hydrolysis. Motifs Ia, Ib, Ic, IV, IVa and V are characterized less well but may be involved in RNA binding. In addition, motif III and Va may communicate ATPase and unwinding activities between ATP binding and RNA binding sites [2-4]. DDX mainly is participated in the regulation of RNA metabolism such as RNA export, RNA decay, RNA storage and ribosome biogenesis. Besides, DDX possesses a crucial role in transcriptional regulation of gene expression. Increasing researches suggested that DDX were highly multifunctional with positive character in carcinogenesis. For example, DDX27 was reported to enhance NF-κB signaling by increasing nucleophosmin-1 (NPM1) and NF-κB-p65 interaction in colorectal cancer [5]. DDX21 was reported as a positive regulator of polymerase-1 and potentiated cell growth in breast cancer [6]. DDX5 was regarded as a poor prognosis predictor for triple-negative breast cancer patients, because it promoted cancer cells proliferation by interacting with fos-related antigen 1 [7].
As a member of DDX family, DDX24 plays a critical role for the ribosome biogenesis [4]. In terms of immune function, previous studies showed that DDX24 inhibited the function of RIG-I-like receptors specifically to participate in the regulation of innate immune [8]. Moreover, DDX24 could promote the proliferation of HIV-1 virus [9]. In terms of tumor function, DDX24 was identified as a drug target in cancer therapeutics by screening of a shRNA library [10]. Yamauchi T, Shi D and their colleagues reported that the depletion of DDX24 might block p53 degradation by attenuating MDM2-mediated ubiquitylation or activating p300-p53 axis [11, 12]. In addition, previous study has figured out the point mutations of DDX24, K11E and E271K, were closely associated with multi-organ venous and lymphatic defect syndrome (MOVLD), in which DDX24 gene was confirmed to be involved in VEGF signaling and cell migration pathways [13]. The 11aa and 271aa sites are located at the amino domain and the core domain of DDX24, respectively. In addition, both 11aa and 271aa sites are highly conserved across distinct species (Figure S1B). Mutations in highly conserved regions are likely to produce important biological function changes [14, 15]. However, whether these mutations work as “loss of function” or “gain of function” remains unknown. DDX24 seemed to be a potential oncogene, thus our study here investigated how K11E and E271K affect the function of DDX24 in tumorigenesis in cell and in animal model.
DDX24 is generally expressed in human tissues and cells, therefore human cells were not suitable to build DDX24 point mutation model studies. Chinese hamster ovary cell (CHO) line, without detection of DDX24 expression, is an ideal cell to contrast wild-type and mutant DDX24. Moreover, CHO cells were widely used in the evaluation of gene functions by transfecting the target gene in vitro and in vivo, especially when long-term and stable gene expression was needed [16]. Thus, cell lines stably expressing wild-type DDX24, K11E-DDX24 and E271K-DDX24 based on CHO cells were constructed to explore the differences between wild-type DDX24 and mutant DDX24 in tumor proliferation. Here, we report our study regarding K11E or E271K DDX24 functions of oncogenicity using the CHO cells and tumor bearing mice model.
Methods
Cell lines and cell culture
Chinese hamster ovary (CHO)-K1 cell line was obtained from Institute of Biophysics, Chinese Academy of Sciences. CHO-K1 cells were cultured in DMEM/F12 medium with 10% fetal bovine serum and 1% penicillin-streptomycin at 37 ℃ in a humidified atmosphere containing 5% CO2. Human MHCC-97L (liver cancer), KYSE410 (esophagus squamous carcinoma), PC3 (prostate carcinoma) and HeLa (cervical cancer) cell lines were obtained from Sun Yat-sen University Medical Research Cell Banks and were cultured in complete DMEM or 1640 medium.
Establishment of monoclonal stably transfected cells
The following plasmids were synthesized by Yingrun Biotechnology Company (Hunan, China) and the sequence of corresponding primer was shown in Table S1. CHO-K1 cells were transfected with pmCherry-N1-WT-DDX24, pmCherry-N1-K11E-DDX24, pmCherry-N1-E271K-DDX24, along with pmCherry-N1 plasmid, respectively (Figure S2A, 2B) by using LipofectamineTM 3000 (L3000015, Thermo Fisher) and Opti-MEM® Reduced Serum Medium (31985070, Thermo Fisher), and then were cultured in a 96-well plate (100 cells/10mL) with medium containing 1500 μg/mL G418 (G8160-1, Solarbio). After two weeks, single cell clone with red-fluorescent was selected for expanding gradually to obtain a monoclonal cell line. Sequencing analyses of CHO-WT-DDX24, CHO-K11E-DDX24 and CHO-E271K-DDX24 cell lines (Figure S2C) showed that no mutations were observed in WT, while 31nt site mutation (A to G) was observed in CHO-K11E-DDX24 and 811nt site mutation (G to A) was observed in CHO-E271K-DDX24.
Quantitative real-time PCR
Total RNA was extracted from cells following the protocol of Total RNA Kit (R6834-02, Omega), and was reversely transcribed into cDNA using cDNA Synthesis Kit (M1681, Thermo Fisher). Transcript levels were detected by using SYBR Green Master Mix (A25742, Thermo Fisher) under a QuantStudio 7 Flex System (Thermo Fisher, USA). The following primers were used: DDX24, forward CCTGCCATCCGTGACAAACT, reverse AGGTGGTGCTTCGGTGTTAC; β-actin, forward CCTGGCACCCAGCACAAT, reverse GGGCCGGACTCGTCATACT (sequences from 5' to 3').
Western blot analysis
Total proteins from cells were extracted using RIPA lysis buffer (89900, Thermo Fisher) supplemental with Protease Inhibitor Cocktail (CW2200S, CWBIO). After quantification by BCA assay (P0010, Beyotime), equal amounts (30 μg) of proteins were electrophoresed on SDS-PAGE gels (M00661, GenScript) and then were transferred to PVDF membranes (ISEQ00010, Merck Millipore). Blots were incubated with the primary antibodies including DDX24 (ab70463, Abcam) or GADPH (60004, Proteintech). Bands were visualized under a Chemiluminescence Reaction System (Bio-Rad, USA) following manufacturer's recommendations.
Immunofluorescence (IF) staining
IF staining was performed to confirm the nucleolus location of DDX24 in the constructed cells. Cells were fixed with 4% paraformaldehyde (DF0135, Leagene) for 30 min, then were incubated with 0.5% Triton X-100 (1139ML500, Biofroxx) for 20 min and with 5% goat serum (16210064, GIBCO) at room temperature for 1 h in turn. Subsequently, cells were incubated with 5 μg/mL NPM1 antibody (32-5200, Invitrogen) overnight at 4 ℃ and then with secondary antibody conjugating Alexa Fluor® 488 (ab150125, Abcam) for 1 h at room temperature protected from light. At last, cells were antifaded by mounting medium with DAPI (H-1200, Vector) and were photographed under a Confocal Microscope (Zeiss, Germany). Cells were washed three times with PBST at every solution removal.
Cell proliferation assay
CHO-Vector, CHO-WT-DDX24, CHO-K11E-DDX24, CHO-E271K-DDX24 (800 cells/well), MHCC-97L, KYSE410, PC3 and HeLa cells (1200 cells/well) were inoculated in 96-well plates. After 24, 48, 72, 96 h cultured in an incubator, cell counting kit-8 (CCK-8) solution (CK04, Dojindo) was added to each well for 2 h. The absorbance at 450 nm was determined under a Microplate Reader (Synergy HTX, BioTek, USA) and then the cell growth curves were depicted basing on the OD values at indicated time points.
Colony formation assay
After CHO-Vector, CHO-WT-DDX24, CHO-K11E-DDX24, CHO-E271K-DDX24 cells were inoculated (1000 cells/well) in 6-well plates and were cultured in complete medium for 10 days, cells were fixed using 4% paraformaldehyde for 30 min and then were stained using crystal violet (G1062, Solarbio) for 30 min at room temperature. Then the colonies were pictured under the Chemiluminescence Reaction System (Bio-Rad, USA) and were counted by Image J (NIH).
Tumorigenicity assay in vivo
All animal experiments were performed according to the institutional ethical guidelines for animal experiments of The Fifth Affiliated Hospital of Sun Yat-sen University. Balb/c nude mice 4 ~ 6 weeks old were purchased from Medical Animal Experiment Center of Guangdong Province. Mice were randomly divided into four groups and then were injected subcutaneously with CHO-Vector, CHO-WT-DDX24, CHO-K11E-DDX24 and CHO-E271K-DDX24 cells at a dose of 5 x 106 in the left flanks, respectively. Tumor growth was monitored by measuring the volume with a vernier caliper. For the tumor-free survival curves, mice were considered tumor-free until tumors were visible or palpable (> 0.5 cm in diameter).
PET/CT 18F-FDG imaging
Mouse PET/CT scan was carried out when the tumor volume reached 200 mm3 using a NanoScan PET/CT system (Mediso, Hungary). Mice were anesthetized with isoflurane gas (2.5% for induction and 1.5% for maintenance). 18F-FDG (4.44 ~ 5.55 MBq) was administered intravenously via the tail vein. An emission scan of 60 min was acquired for all mice. The PET data were acquired in list mode and were reconstructed into 25 timeframes (10 x 3 s, 3 x 10 s, 4 x 60 s, 5 x 300 s, 3 x 600 s). The evaluation of dynamic PET data including standardized uptake value (SUV) was performed using the software of Carimas (Version 2.9) (Turku PET Center, Finland). The influx rate constant (Ki) of tumor was calculated using Patlak analysis by applying tumor time-activity curve (TAC) data to the Matlab software (Version 2014a) as we previously reported [17].
Immunohistochemistry (IHC) staining
The tumors were dissected and fixed in 10% buffered formalin. The tissue samples were paraffin-embedded and sectioned at 5 μm thickness. The slices were deparaffinized in xylene and rehydrated in graded alcohols. Briefly, after blocked with 5% goat serum for 60 min at room temperature, slices were incubated with primary antibodies (1: 200 dilution) anti-DDX24 (PA5-51721, Thermo Fisher) at 4 ℃ overnight, followed by the corresponding secondary antibody sheep anti-rabbit IgG polymer (MXB Biotechnologies, China) for 30 min at room temperature. Subsequently, the slices were treated with liquid 3,3′-diaminobenzidine (MXB Biotechnologies) and counterstained with hematoxylin. Finally, all the sections were scanned under a Pannoramic 250 Flash (3DHISTECH, Hungary).
RNA-sequencing (RNA-Seq)
To further explore the underlying molecular mechanisms of WT-DDX24 expression on promoting proliferation and K11E-DDX24 expression on suppressing tumorigenicity, the RNA-seq of tumors from Balb/c-nu mice of four groups, were used to estimate the transcriptome influence of wild-type or mutant DDX24 expressing cells on the surrounding microenvironment. Thus, 15 samples (4 replicates for Vector, 5 replicates for WT, 2 replicates for K11E and 4 replicates for E271K) were sequenced. Finally, the datasets were generated on BGIseq500 platform (BGI, Guangdong, China).
P value was corrected to Q value by differentially expressed genes (DEGs) sequencing [18]. Genes with fold change ≥ 2 and Q value ≤ 0.001 were screened as significantly DEGs. To guarantee the contrast, we selected their overlapped DEGs from our datasets. To further identify related pathways and genes, we performed both Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and Gene Ontology (GO) term enrichment. Moreover, Gene Set Enrichment Analysis (GSEA) was also performed on all genes in each group to avoid screening genes for false negatives at thresholds. In addition, clustered heatmaps of gene expression were obtained by running the R language.
Statistical analysis
Statistical analyses were performed through GraphPad Prism (Version 8). Quantitative data were shown as mean ± standard deviation from at least three independent experiments. If data were normally distributed, then unpaired T test or One-way ANOVA tests (Tukey multiple comparisons test, Dunnett multiple comparisons test or Holm-Sidak multiple comparisons test) was used for p value calculation. Otherwise, Kruskal Wallis test was used for multiple comparisons. Chi-square (and Fisher exact) test was used for tumor formation rate comparisons. Log-rank (Mantel-Cox) test was used for Kaplan-Meier tumor-free survival comparisons. P value, adjusted by test, less than 0.05 was considered statistically significant, where * indicated p < 0.05, ** indicated p < 0.01, *** indicated p < 0.001, respectively.
Results
The DDX24 expression was verified in stably transfected cell lines
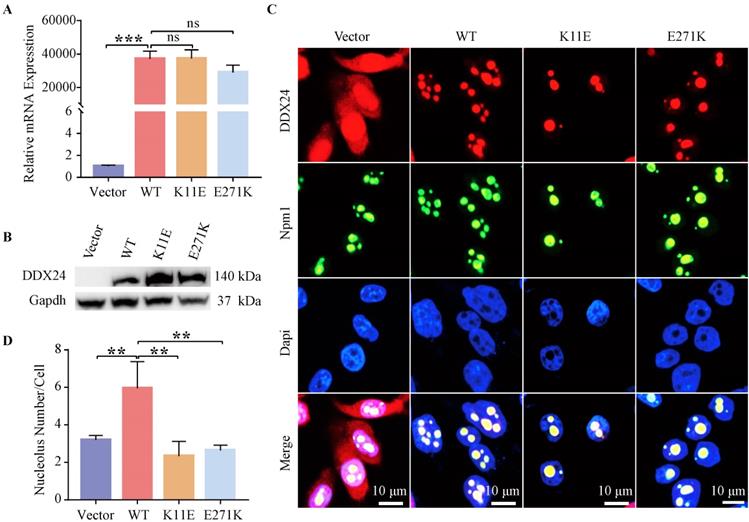
Constructed cell lines exhibited high transcriptional level (up to 20,000 times) of DDX24 except CHO-Vector, as tested by qPCR (Figure 1A). Western blot further corroborated DDX24 expression in the constructed cells compared to the empty plasmid (Figure 1B). The 11aa and 271aa sites of DDX24 in stably transfected cell lines were also verified by using cell-sequencing (Figure S1C).
The number of nucleoli in CHO-K11E-DDX24 or CHO-E271K-DDX24 cells was decreased
IF staining showed wild-type and point mutant DDX24 were both co-located with the nucleolar protein Npm1 (Figure 1C). Besides, the number of nucleoli was measured (Figure 1D), which revealed the number of nucleoli both decreased notably in CHO-K11E-DDX24 (2.33 ± 0.78) (p = 0.0014) and CHO-E271K-DDX24 (2.94 ± 0.59) (p = 0.017) compared with CHO-WT-DDX24 (5.95 ± 1.42).
The proliferation and colony formation in CHO-K11E-DDX24 and CHO-E271K-DDX24 were suppressed
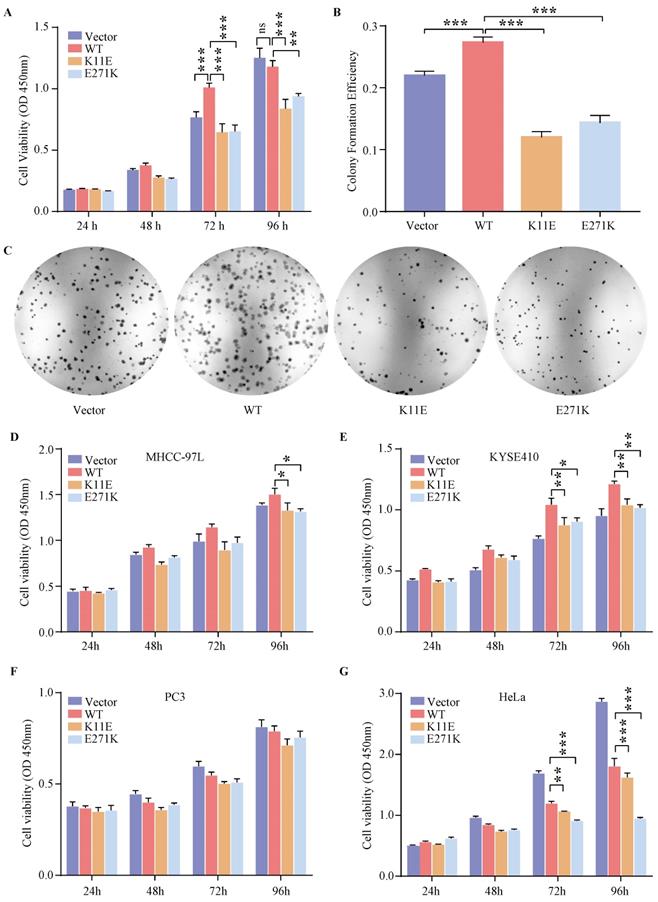
Expression of WT-DDX24 enhanced the growth of cells at 72 h compared with control vector cells (p < 0.0001) (Figure 2A). However, expression of K11E-DDX24 suppressed the growth of cells at 72 and 96 h (p < 0.0001). Likewise, expression of E271K-DDX24 inhibited the cell growth at 72 and 96 h (p < 0.0001 and p = 0.006 respectively). Furthermore, colony formation assay revealed that WT-DDX24 increased the colony forming efficiency (27.30 ± 0.75%, p < 0.0001). While the colony formation rates of K11E and E271K were 11.97 ± 0.77% and 14.3 ± 1.00% respectively, which were significantly lower than that of the vector cells (21.97 ± 0.57%, p < 0.0001) (Figure 2B, 2C). In addition, after knocking down DDX24 in CHO-WT-DDX24, cell proliferation and colony formation were inhibited significantly (Figure S3). The effects of point mutation and knockdown were consistent.
Detection of DDX24 expression in constructed CHO cell lines. (A) Relative mRNA levels of DDX24 in constructed CHO cell lines were detected by quantitative real-time PCR. P values were calculated with Tukey multiple comparisons test. ***, p < 0.001; ns, no significance. (B) Detection of DDX24 expression in the constructed CHO cell lines by Western blot. (C) Representative images demonstrating the nucleolar localization of DDX24 (co-locating with Npm1) in the cell lines under a confocal microscope. Nuclei were stained with Dapi (blue). Scale bars, 10 μm. (D) Quantitative analysis of nucleoli number in cell lines. P values were calculated with Dunnett multiple comparisons test. **, p < 0.01; Vector vs. WT, p = 0.0024; WT vs. K11E, p = 0.0014; WT vs. E271K, p = 0.0017.
The expression of DDX24 affected the proliferation in human tumor cells
Based on the cancer genome atlas project, we found the expression status of DDX24 in liver hepatocellular carcinoma (LIHC) and esophageal carcinoma (ESCA) was higher than that in the adjacent normal tissues, while the expression level of DDX24 in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) and prostate adenocarcinoma (PRAD) was lower than that in the adjacent normal tissues (Figure S4A). As shown in Figure S4B, high expression DDX24 was linked with poor prognosis of overall survival for LIHC (p = 0.021). However, low expression of DDX24 was related to poor overall survival prognosis for CESC (p = 0.03) and ESCA (p = 0.0059) (Figure S4C). Additionally, the genetic alteration status of DDX24 in LIHC, ESCA, CESC and PRAD was low (< 4%) (Figure S4D) and neither K11E nor E271K mutation was observed in above cancers (Figure S4E). Transient transfection (verified by qPCR in Figure S5) and CCK8 assay in MHCC-97L, KYSE410, PC3 and HeLa cells were performed. Expression of K11E-DDX24 or E271K-DDX24 suppressed the growth of MHCC-97L cells at 96 h (p = 0.0410 and p = 0.0192 respectively) compared with WT-DDX24 (Figure 2D). In KYSE410 cells, expression of K11E-DDX24 suppressed the growth at 72 and 96 h (p = 0.0076 and p = 0.0042 respectively) and expression of E271K-DDX24 suppressed the growth at 72 and 96 h (p = 0.0218 and p = 0.0019 respectively) compared with WT-DDX24 (Figure 2E). Moreover, there were no significant differences in PC3 cells (Figure 2F). Likewise, expression of K11E-DDX24 suppressed the growth of HeLa cells at 72 and 96 h (p = 0.0022 and p < 0.0001 respectively) and expression of E271K-DDX24 suppressed the growth at 72 and 96 h (p < 0.0001) compared with WT-DDX24 (Figure 2G).
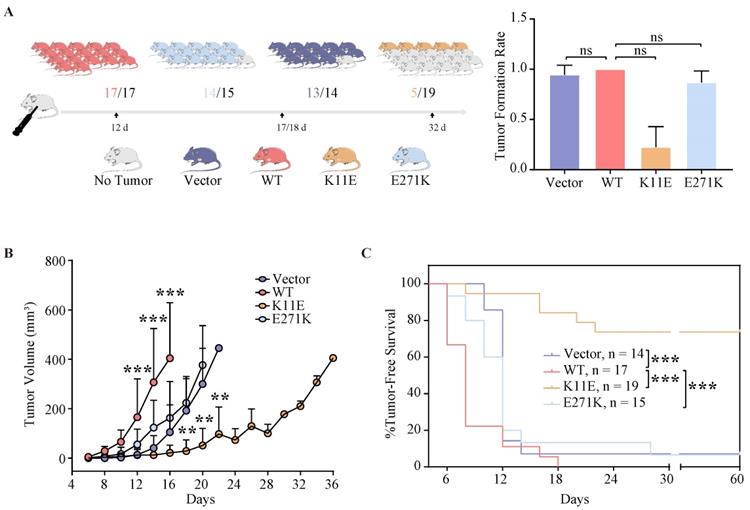
The tumor growth rate of mice inoculated with CHO-K11E-DDX24 was markedly slowed down
All mice (17 of 17) that were inoculated with CHO-WT-DDX24 cells developed tumors. 93.3% mice (14 of 15) inoculated with CHO-E271K-DDX24 cells developed tumors, which was nearly equal to CHO-Vector (92.86%, 13 of 14). However, mice inoculated with CHO-K11E-DDX24 cells showed a lower tumor formation rate (only 26.3% mice, 5 of 19) (Figure 3A). The median day of tumor volume reaching 200 mm3 in mice inoculated with CHO-WT-DDX24 was 12. While the median day of tumor volume reaching 200 mm3 in mice inoculated with CHO-E271K-DDX24 was 17, which was nearly close to CHO-Vector group (18 days). However, the tumor in mice inoculated with CHO-K11E-DDX24 spent 32 days to reaching 200 mm3 (Figure 3B). The tumor growth rate of mice inoculated with CHO-K11E-DDX24 was slowed down significantly.
Expression of DDX24 affected cell proliferation. (A) Cell proliferation assay in constructed CHO cell lines. P values were calculated with Dunnett multiple comparisons test. ***, p < 0.001. WT vs. K11E: p 72 h < 0.0001, p 96 h < 0.0001; WT vs. E271K: p 72 h < 0.0001, p 96 h = 0.006. (B) Colony formation assay in constructed CHO cell lines. P values were calculated with Dunnett multiple comparisons test. ***, p < 0.001. (C) The colonies were pictured under the Chemiluminescence Reaction System and counted by Image J. Cell Proliferation assay (D, E, F, G) in specific tumor cell lines. P values were calculated with Tukey multiple comparisons test. *, p < 0.05; **, p < 0.01; ***, p < 0.001. (D) WT vs. K11E: p = 0.0194; WT vs. E271K: p = 0.0131. (E) WT vs. K11E: p 72 h = 0.0076, p 96 h = 0.0042; WT vs. E271K: p 72 h = 0.0218, p 96 h = 0.0019. (G) WT vs. K11E: p 72 h = 0.0022, p 96 h < 0.0001; WT vs. E271K: p < 0.0001.
Wild-type DDX24 enhanced tumorigenicity in vivo, while mutant DDX24 inhibited tumor-growth. (A) The numbers under the gray line segment represented the median day of tumorigenesis and above the gray line segment represented the number of mice with tumor/total number of mice in four independent replicated experiments. Tumor formation rates showed no statistics significance. (B) Tumor volume was measured every 2 days after cell transplantation. CHO-Vector group was used as negative control. (C) Kaplan-Meier tumor-free survival curves of the mice were shown. P values in (B) and (C) were calculated with Log-rank (Mantel-Cox) test. **, p < 0.01; ***, p < 0.001.
The survival time of mice inoculated with CHO-K11E-DDX24 was prolonged
The median survival time of mice was 6 ~ 8 days in CHO-WT-DDX24 group, while the CHO-E271K-DDX24 or CHO-Vector groups were 10 ~ 12 days. However, the median survival time of mice inoculated with CHO-K11E-DDX24 was exceed 32 days (Figure 3C). In general, the survival time of mice inoculated with CHO-K11E-DDX24 was significantly prolonged.
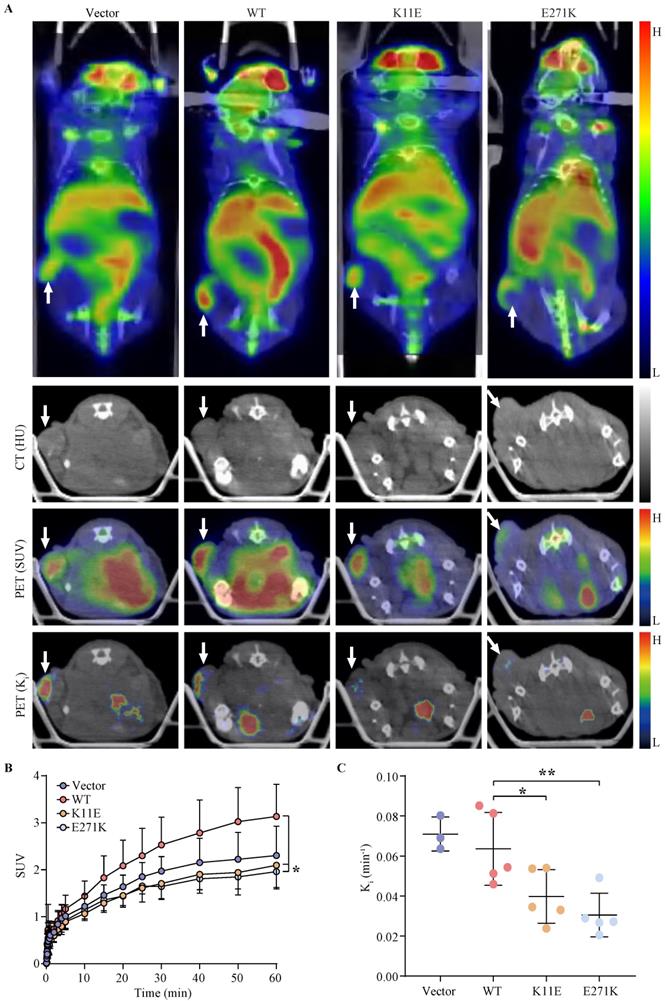
The SUV and Ki of tumors formed by CHO-K11E-DDX24 or CHO-E271K-DDX24 were reduced
Tumor bearing mice were scheduled for dynamic 18F-FDG PET/CT scan when the tumor grew up to 200 mm3. The TAC of tumors was calculated. Representative PET/CT images were shown in Figure 4A. Higher 18F-FDG uptakes of WT-DDX24 group could be observed with corresponding SUV values at 3.13 ± 0.68 (Figure 4B), while the SUV values of K11E (2.10 ± 0.50) and E271K (1.96 ± 0.34) groups were obviously lower (p = 0.0410 and p = 0.0192 respectively). Similarly, the values of Ki from WT-DDX24 group (0.064 ± 0.018 min-1) were higher than that of K11E (0.040 ± 0.013 min-1) (p < 0.05) and E271K (0.030 ± 0.011 min-1) groups (p = 0.0484 and p = 0.0099 respectively) (Figure 4C, 4D), which indicated that K11E or E271K mutation of DDX24 resulted in suppressing the tumor cell proliferation.
The number of nucleoli in tumor tissue formed by CHO-K11E-DDX24 or CHO-E271K-DDX24 was decreased
DDX24 was highly expressed in tumor tissue formed by CHO-WT-DDX24, CHO-K11E-DDX24 or CHO-E271K-DDX24, except CHO-Vector (Figure S6). At the same magnification, the tumor tissues formed by CHO-WT-DDX24 showed higher density of nucleolus, while the mutant tumor exhibited less density of nucleolus.
Loss-of-function K11E or E271K mutation showed decreased glucose utilization rate. Mice PET/CT scan was scheduled when tumor volume reached 200 mm³. (A) Representative transaxial PET/CT images showed similar and intermediate signal in the constructed CHO cells formed tumors (white arrow). The level of signal was represented by a color range from black (low) to red (high). SUV (B) and Ki (C) showed significant difference among the constructed tumors. P values in (B) were calculated with Tukey multiple comparisons test; *, p < 0.05; WT vs. K11E, p = 0.0410; WT vs. E271K, p = 0.0192. P values in (C) were calculated with Holm-Sidak multiple comparisons test. *, p < 0.05; **, p < 0.01; WT vs. K11E, p = 0.0484; WT vs. E271K, p = 0.0099.
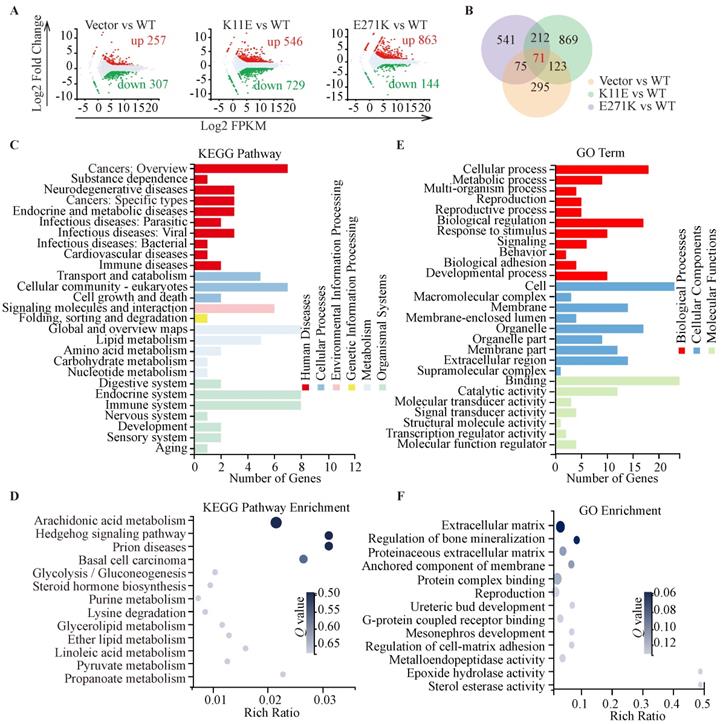
Sequencing-based transcriptomic analyses. (A) Red dots represented the up-regulated DEGs, while green dots represented the down-regulated DEGs, and gray dots represented non-DEGs. (B) The Venn diagram showed that there were 71 overlapped DEGs in three groups. KEGG pathway classification and enrichment (C, D) and GO feature classifications and enrichment (E, F).
CHO-K11E-DDX24 or CHO-E271K-DDX24 caused increased expression of tumor necrosis factors (TNF) and chemokines, along with alteration in immune-related signal pathways
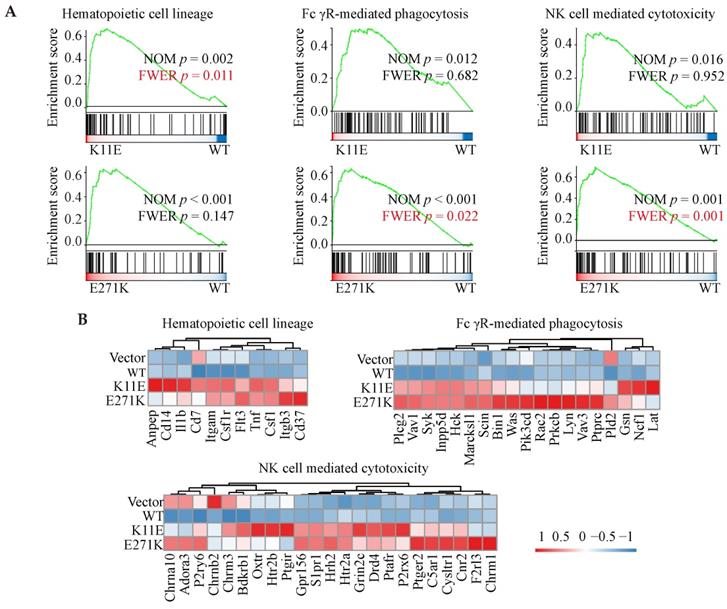
In total, we identified 564, 1275, and 1007 DEGs in the three comparisons (Vector vs. WT, K11E vs. WT, and E271K vs. WT, respectively). Contrasted with the WT group, there were 257 genes up-regulated and 307 genes down-regulated in the Vector group, 546 genes up-regulated and 729 genes down-regulated in the K11E group, as well as 863 genes up-regulated and 144 genes down-regulated in the E271K group (Figure 5A). To guarantee the contrast, we selected 71 overlapped DEGs from our datasets (Figure 5B). The KEGG pathway enrichment (Figure 5C, 5D) and Go term enrichment (Figure 5E, 5F) based on the 71 DEGs indicated there was no significant difference between WT group and other group. However, GSEA enrichment showed that there were 25 pathways between K11E group and WT group and there were 68 pathways between E271K group and WT group. There were 15 overlapping pathways between the two contrasts (Table S2), where 8 pathways were related with immune regulation [hematopoietic cell lineage, Fc-γR-mediated phagocytosis, natural killer (NK) cell mediated cytotoxicity, leukocyte transendothelial migration, T helper 17 (Th17) cell differentiation, chemokine signaling pathway, C-type lectin receptor signaling pathway, and T cell receptor signaling pathway]. In addition, there were 3 pathways (hematopoietic cell lineage, Fc-γR-mediated phagocytosis and NK cell mediated cytotoxicity) (Figure 6A) still of significant difference after corrected by Bonferonni test. The heatmap analyses of gene expression in these 3 pathways (Figure 6B) showed that the expression of TNF and chemokines increased in mutant groups.
Discussion
DDX24 was identified as a drug target in cancer therapeutics by screening of a shRNA library, which was agreed with the report by Li F in gastric cancer [19]. Moreover, Yamauchi T, Shi D and their colleagues showed that the depletion of DDX24 might block p53 functions [11, 12], which indicated DDX24 seemed to be an oncogene. In previous study, the point mutations of DDX24, K11E and E271K, were related to vascular malformations disease, MOVLD. But whether these mutations work as “loss of function” or “gain of function” remains unknown. We found that the above two mutation sites were highly conserved in different species. It was generally believed mutations in highly conserved regions would lead to inability to survive or functional subversion. Many members of DDX family were reported to show decreased alteration in enzyme activity and function due to mutations in highly conserved regions. For example, DDX5 (Dbp5/DDX19B) shuttled between the nucleus and the cytoplasm to mediate the export of messenger ribonucleoprotein (mRNP). L12A at the N-terminal caused defection of mRNP export, which led to the failing growth of corresponding strain [20]. Moreover, the point mutation in conserved residues of the ATPase active sites of DbpA in Escherichia coli (E. coli) reduced its ATPase activity and helicase activity [21]. In other DExD/H helicases, such as Prp16, Prp22, Prp43 and eIF-4A, K53 or R331 mutations can lead to be lethal [22-26]. We tried to purify recombinant wild-type DDX24 or mutant DDX24 proteins from E. coli or CHO cell lines to detect their activity of RNA helicase or ATPase, but all failed due to the instability of DDX24 itself in vitro. Analogous to other DDX, whether K11E or E271K DDX24 would lose its enzyme activity is still worthy to study further.
GSEA enrichment analyses based on RNA-seq. (A) Three immune-related pathways with p value corrected by Bonferonni test. (B) The heatmap analyses of gene in above three pathways.
Recent advances highlighted the nucleolus, as an important membrane-less organelle, usually formed via phase separation [27-29]. Our data showed DDX24 was co-localized with Npm1, a marker in the granular compartment [30] and a key driver of nucleolar phase separation [31, 32], which indicated that DDX24 distributed in nucleolus and participated in the composition of nucleolus. However, the specific and detailed compositions of nucleolus are not clear yet; previous study showed that the nucleolus consisted of three sub-structures including the fiber center, the dense fiber component and the granular components [33]. A range of studies have characterized that the nucleolus played a significant role in genomic stability, stress response, DNA repair and recombination, transcription regulation, telomere maintenance, ribosome biogenesis and other essential cellular processes, which were closely related to cancer development [34, 35]. In addition, nucleolar size was one of important parameters to estimate cell growth and proliferative intensity; meanwhile it has been used in some cancers as a predictive and prognostic biomarker for chemotherapy and clinical outcomes [36-38]. Our IF and IHC staining revealed that compared with cells expressing wild-type DDX24, the number of droplets in K11E-DDX24 or E271K-DDX24 cells was significantly reduced, which implied that these mutations altered the quantity of nucleolus. Indeed, the heteroligomer state of native E. coli recombination human DDX24 has been studied in our institute by Dr. Shoudeng Chen, who has observed dimer or tetramer of DDX24 in solution (data not yet published). We also searched the literature, and found recent study regarding the eukaryotic expression of DDX24 with heterooligomer [39]. Our existing study provided evidence that DDX24 formed heteroligomer with the mutated DDX24 proteins, therefore “dominant negatives” may be the underline mechanism that these mutations in cell and in tumor bearing mouse changed the oncogenicity of DDX24. As Lasry I reported, the alterational location of target protein in cell loss of zinc transport activity of the WT ZnT-2 due to homodimerization acted as dominant negatives [40]. Nevertheless, the specific mechanism behind this alteration still remains unknown and is needed to do more explore.
Proliferation assays in human tumor cells showed that the expression of K11E or E271K DDX24 may inhibit relate cell proliferation in tumors with high expression of DDX24 or bad prognosis of DDX24, but not in tumors with low expression of DDX24 or none-risk prognostic of DDX24. Although we did not knockdown or knockout the endogenous expression of DDX24 in above human tumor cells, MHCC-97L, KYSE410 and HeLa cells transfected with mutant DDX24 still exhibited proliferation suppression, which implied these mutations have lost these roles in cellular proliferation. Moreover, proliferation assays in CHO cells along with further in vivo tumor implantation studies also suggested “loss of function of cell proliferation” for these two mutations.
To further investigate the in vivo oncogenicity of DDX24 mutations, 18F-FDG PET/CT was applied to investigate the imaging quantification of DDX24 expressed tumors. SUV was widely used for PET imaging quantification [41]. However, many factors such as the tumor heterogeneity, uptake kinetics and post injection time can influence the feedback of SUV [42, 43], which appeared less specific for tumor proliferation. Thus, the Ki value, a quantitative measure of the influx rate constant of 18F-FDG derived from the dynamic PET scans was calculated [17]. In this study, tumors of each group were size-matched. From this quantitative analysis, both SUV and Ki value of WT-DDX24 group were higher than that of the vector controls, which convincingly implied that WT-DDX24 accelerated the glucose utilization rate and enhanced the tumor proliferation rate. The majority of mutant groups (K11E and E271K) showed less glucose utilization than that of WT groups, suggested “loss of function” of in vivo oncogenicity for these two mutations of DDX24.
Moreover, RNA-seq from the tumors formed by mutant DDX24 showed that at least 8 immune-related pathways (hematopoietic cell lineage, Fc-γR-mediated phagocytosis, NK cell mediated cytotoxicity, leukocyte transendothelial migration, Th17 cell differentiation, chemokine signaling pathway, C-type lectin receptor signaling pathway, and T cell receptor signaling pathway) were altered in comparison between wild-type group and mutant groups. In addition, several immune-correlated factors such as TNF and chemokines in mutant tissues were highly expressed. Even though nude mice do not have conventional CD4+ and CD8+ T cells or enough effective B cells to have the signaling altered, nude mice possessed precursor cells for the T compartment of the lymphoid system as previous Loor F reported [44]. Unconventional T cells including natural killer T cells, mucosal-associated invariant T cells and γδ T cell were thymus-independent and made up ~10% of circulating T cells [45]. Given nude mice were not defective in the responder cells of NK cell lineage [46], we speculated that K11E-DDX24 or E271K-DDX24 may enhance non-specific immune killing to inhibit cell proliferation and tumor growth.
In summary, our present work verified K11E or E271K mutation led to “loss-of-function” of DDX24 in cell proliferation and tumor bearing mice by using a CHO cell model. The tumor suppression induced by K11E or E271K mutation of DDX24 is expected to provide a novel idea or direction for tumors prevention and treatment, especially for those tumors in which DDX24 plays an oncogenic role in proliferation.
Abbreviations
DDX: DEAD-box RNA helicase; MOVLD: multi-organ venous lymphatic malformation syndrome; CHO: Chinese hamster ovary cell; IF: immunofluorescence; vs.: verses; LIHC: liver hepatocellular carcinoma; ESCA: esophageal carcinoma; CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma; PRAD: prostate adenocarcinoma; UCEC: uterine corpus endometrial carcinoma; IHC: immunohistochemistry; PET: positron emission tomography; SUV: standardized uptake value; RNA-Seq: RNA-sequencing; DEG: differentially expressed genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene Ontology; GSEA: Gene Set Enrichment Analyses; NK cell: natural killer cell; Th17 cell: T helper 17 cell; E. coli: Escherichia coli.
Supplementary Material
Supplementary materials and methods, figures and tables.
Acknowledgements
This work was supported by the National Key R&D Program of China [2018YFC0910600], the National Natural Science Foundation of China [No.81871382], Key Realm R&D Program of Guangdong Province [2018B030337001] and Starting Fund from Sun Yat-sen University Fifth Affiliated Hospital.
Author Contributions
Hongjun Jin and Hong Shan conceived and designed the study. Xinglin Li and Xiaoyun Chen conducted the main experiments and drafted the initial manuscript. Jiebing Gao, Jianzhong Xian and Zhijun Li performed the main data analysis. Ming Yang, Lei Bi and Shuai Yang approved the final version. All authors read and approved the manuscript.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary.
Ethical standards
All animal experiments were performed according to the institutional ethical guidelines for animal experiments of the Fifth Affiliated Hospital of Sun Yat-sen University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Banroques J, Cordin O, Doere M. et al. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol Cell Biol. 2008;28(10):3359-3371
2. Sarkar M, Ghosh M. DEAD box RNA helicases: crucial regulators of gene expression and oncogenesis. Front Biosci (Landmark Ed). 2016;21:225-250
3. Linder P. Dead-box proteins: a family affair-active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34(15):4168-4180
4. Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505-516
5. Tang J, Chen H, Wong C. et al. DEAD-box helicase 27 promotes colorectal cancer growth and metastasis and predicts poor survival in CRC patients. Oncogene. 2018;37(22):3006-3021
6. Kim D, Camacho C, Nagari A. et al. Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. Mol Cell. 2019;75(6):1270-1285 e1214
7. He H, Song D, Sinha I. et al. Endogenous interaction profiling identifies DDX5 as an oncogenic coactivator of transcription factor Fra-1. Oncogene. 2019;38(28):5725-5738
8. Ma Z, Moore R, Xu X. et al. DDX24 negatively regulates cytosolic RNA-mediated innate immune signaling. PLoS Pathog. 2013;9(10):e1003721
9. Peng X, Sun T, Yao P. et al. Differential expression of innate immunity regulation genes in chronic HIV-1 infected adults. Cytokine. 2020;126:154871
10. Oliver D, Ji H, Liu P. et al. Identification of novel cancer therapeutic targets using a designed and pooled shRNA library screen. Sci Rep. 2017;7:43023
11. Yamauchi T, Nishiyama M, Moroishi T. et al. MDM2 mediates nonproteolytic polyubiquitylation of the DEAD-Box RNA helicase DDX24. Mol Cell Biol. 2014;34(17):3321-3340
12. Shi D, Dai C, Qin J. et al. Negative regulation of the p300-p53 interplay by DDX24. Oncogene. 2016;35(4):528-536
13. Pang P, Hu X, Zhou B. et al. DDX24 Mutations Associated With Malformations of Major Vessels to the Viscera. Hepatology. 2019;69(2):803-816
14. Assis R, Kondrashov A. Conserved proteins are fragile. Mol Biol Evol. 2014;31(2):419-424
15. Omer S, Harlow T, Gogarten J. Does Sequence Conservation Provide Evidence for Biological Function? Trends Microbiol. 2017;25(1):11-18
16. Fischer S, Handrick R, Otte K. The art of CHO cell engineering: A comprehensive retrospect and future perspectives. Biotechnol Adv. 2015;33(8):1878-1896
17. Yang M, Lin Z, Xu Z. et al. Influx rate constant of (18)F-FDG increases in metastatic lymph nodes of non-small cell lung cancer patients. Eur J Nucl Med Mol Imaging. 2020;47(5):1198-1208
18. Wang L, Feng Z, Wang X. et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136-138
19. Li F, Li J, Yu J. et al. Identification of ARGLU1 as a potential therapeutic target for gastric cancer based on genome-wide functional screening data. EBioMedicine. 2021;69:103436
20. Lari A, Rajan A, Sandhu R. et al. A nuclear role for the DEAD-box protein Dbp5 in tRNA export. Elife. 2019 8
21. Elles L, Uhlenbeck O. Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucleic Acids Res. 2008;36(1):41-50
22. Schmid S, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991;11(7):3463-3471
23. Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem. 2002;277(20):17743-17750
24. Hotz H, Schwer B. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics. 1998;149(2):807-815
25. Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11(7):2643-2654
26. Schneider S, Hotz H, Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J Biol Chem. 2002;277(18):15452-15458
27. Lafontaine D, Riback J, Bascetin R. et al. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22(3):165-182
28. Hyman A, Weber C, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39-58
29. Riback JA, Zhu L, Ferrolino M. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581(7807):209-214
30. Colombo E, Alcalay M, Pelicci P. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene. 2011;30(23):2595-2609
31. Mitrea D, Cika J, Guy C. et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife. 2016 5
32. Feric M, Vaidya N, Harmon T. et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016;165(7):1686-1697
33. Iarovaia O, Minina E, Sheval E. et al. Nucleolus: A Central Hub for Nuclear Functions. Trends Cell Biol. 2019;29(8):647-659
34. Lindström M, Jurada D, Bursac S. et al. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene. 2018;37(18):2351-2366
35. Larsen D, Stucki M. Nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2016;44(2):538-544
36. Carotenuto P, Pecoraro A, Palma G. et al. Therapeutic Approaches Targeting Nucleolus in Cancer. Cells. 2019 8(9)
37. Derenzini M, Trerè D, Pession A. et al. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152(5):1291-1297
38. Derenzini M, Nardi F, Farabegoli F. et al. Distribution of silver-stained interphase nucleolar organizer regions as a parameter to distinguish neoplastic from nonneoplastic reactive cells in human effusions. Acta Cytol. 1989;33(4):491-498
39. Zhang X, Spiegel J, Martínez-Cuesta S. et al. Chemical profiling of DNA G-quadruplex-interacting proteins in live cells. Nat Chem. 2021;13(7):626-633
40. Lasry I, Seo Y, Ityel H. et al. A dominant negative heterozygous G87R mutation in the zinc transporter, ZnT-2 (SLC30A2), results in transient neonatal zinc deficiency. J Biol Chem. 2012;287(35):29348-29361
41. Huang S. Anatomy of SUV. Standardized uptake value. Nucl Med Biol. 2000;27(7):643-646
42. Kurtipek E, Cayci M, Duzgun N. et al. (18)F-FDG PET/CT mean SUV and metabolic tumor volume for mean survival time in non-small cell lung cancer. Clin Nucl Med. 2015;40(6):459-463
43. Soret M, Bacharach S, Buvat I. Partial-volume effect in PET tumor imaging. J Nucl Med. 2007;48(6):932-945
44. Loor F, Kindred B. Differentiation of T-cell precursors in nude mice demonstrated by immunofluorescence of T-cell membrane markers. J Exp Med. 1973;138(5):1044-1055
45. Pellicci D, Koay H, Berzins S. Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge. Nat Rev Immunol. 2020;20(12):756-770
46. Hinuma S, Naruo K, Shiho O. et al. Characteristics of mouse lymphoid cells proliferating in vitro by recombinant human interleukin 2 (TGP-3): comparison between normal and nude mice. Microbiol Immunol. 1989;33(9):761-773
Author contact
![]() Corresponding authors: Hong Shan, E-mail: shanhongsysu.edu.cn; Hongjun Jin, E-mail: jinhj3sysu.edu.cn.
Corresponding authors: Hong Shan, E-mail: shanhongsysu.edu.cn; Hongjun Jin, E-mail: jinhj3sysu.edu.cn.

 Global reach, higher impact
Global reach, higher impact