Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(3):446-459. doi:10.7150/ijms.67094 This issue Cite
Review
Diagnostic value of circRNAs as effective biomarkers in human cardiovascular disease: an updated meta-analysis
1. Key Laboratory of Research in Maternal and Child Medicine and Birth Defects, Guangdong Medical University, Foshan, China.
2. Dongguan Key Laboratory of Environmental Medicine, School of Public Health, Guangdong Medical University, Dongguan, China.
3. Department of Surgery, the Third Affiliated Hospital of Guangdong Medical University (Longjiang Hospital of Shunde District), Foshan, China.
4. Key Laboratory of Stem Cell and Regenerative Tissue Engineering, Guangdong Medical University, Dongguan, China.
*These authors contributed equally to this work.
Received 2021-9-20; Accepted 2022-1-7; Published 2022-2-7
Abstract
Background: A growing body of literature has demonstrated that circular RNAs (circRNAs) are the potential biomarkers in human cardiovascular disease (CVD). Therefore, a meta-analysis based on current studies was accomplished to appraise the role of circRNAs in the diagnostic of CVD patients.
Methods: Studies before October 30, 2021, were searched using PubMed, EMBASE, the Web of Science, and Cochrane Library. The diagnostic odds ratio (DOR) with a confidence interval (CI) of 95% was used to investigate the associations between circRNAs and CVDs.
Results: A total of 27 eligible articles were selected, including 47 studies, with 6833 participants meeting the criteria standard constrain. The pooled overall sensitivity and specificity for circRNAs expression profile in differentiating CVD patients from controls (non-CVDs or healthy subjects) were 0.81 (95%CI 0.78-0.83) and 0.74 (95%CI 0.68-0.78), respectively; the overall positive likelihood ratio was 3.1 (95%CI 2.5-3.7); the negative likelihood ratio was 0.26 (95%CI 0.22-0.31); the overall diagnostic odds ratio corresponding to an area under the curve of 0.85 (95%CI 0.81-0.88) was 12 (95%CI 9-16). Subgroup analysis indicated that the serum rather than blood has higher diagnostic accuracy. Likewise, meta-regression analysis demonstrated that the specimen, detection method, sample size, and publication year were the main sources of heterogeneity. Sensitivity analysis and Deeks' funnel plot revealed that our results are relatively robust.
Conclusions: Our evidence-based analysis results suggested that circRNAs provide higher diagnostic accuracy in the prediction of CVDs. Thus, circRNAs might be potential biomarkers in CVDs.
Keywords: circRNAs, cardiovascular disease, diagnosis, biomarker, meta-analysis
Introduction
The concerns of epidemiology in the 20th century gradually shifted with the decrease in the mortality and disability rate and the increase in non-communicable diseases [1]. Among non-communicable diseases, cardiovascular disease (CVD) is the most severe public health problem, threatening the health of the general population [2]. CVDs are characterized by high morbidity, high disability rate, high mortality, high recurrence rate, and severe complications [3]. Therefore, CVDs have become one of the main causes of mortality around the world. With CVDs being such a huge burden, more and more genetic biomarkers of CVDs have been discovered, among which circRNAs have received considerable attention owing to their biological and clinical application in the diagnosis and treatment of CVDs [4,5].
CircRNA is a particular type of noncoding RNA with a closed circular structure, which is abundant in blood or tissue and acts as an indispensable member in a variety of biological processes [6-8]. CircRNA is widespread in human cells, with the expression abundance sometimes even exceeding 10 times of their linear transcripts [9]. It is formed by precursor mRNA (pre-mRNA) back-splicing, which is different from the classical shearing mode of general linear RNA [10]. Due to various cyclization forms, it can be divided into three categories: exonic circRNA, intronic circRNA, and exon-intron circRNA [11,12]. With the furthering research, several essential functions have been identified at the transcriptional and posttranscriptional levels, including acting as microRNA (miRNA) sponges, manipulating RNA binding proteins (RBPs), and controlling alternative splicing and parental gene expression [13]. Generally, current studies have confirmed that the role of miRNA sponge is the main mechanism of circular RNA in disease [14].
CircRNAs have covalently closed loop structures with neither 5'-3' polarities nor polyadenylated tails, which leads to its various characteristics: stability, specificity and conservatism. Based on these characteristics, circRNAs have advantages over other types of RNAs in the diagnosis of specific diseases. Simultaneously, increasing studies have demonstrated that circRNAs are related to the occurrence and development of various diseases. In the cardiovascular system, circRNAs are not only important for the development of the heart and blood vessels [15], but they also play a vital role in the pathophysiology of heart abnormalities such as coronary artery disease (CAD), stroke, heart failure (HF), essential hypertension (EH), idiopathic pulmonary arterial hypertension (IPAH), acute Stanford type A aortic dissection (AAAD) and so on [16-18]. Furthermore, under normal circumstances, the level of circRNAs in plasma and other body fluids is stable, suggesting that circRNAs have the potential to serve as diagnostic biomarkers for many diseases [19]. For instance, circ-STAB2 and circCHFR play a crucial role in up-regulating the related target genes by sponging miR-939 and miR-370, respectively, thus playing a crucial vital role in atherosclerosis and CVDs [20,21]. To date, a considerable number of biomarkers related to CVDs have been reported in the research while only a small part of circRNAs have been repeatedly verified and applied. Although it is doubtful whether circRNAs can be accurate and useful biomarkers for the diagnosis of CVDs, they have been extensively explored as biomarkers of cancer. Therefore, summarizing the connection between circRNAs and CVDs and providing new ideas are imperative. Herein, we summarize the latest research progress on circRNAs, focusing on exploring the accuracy of circRNAs-based biomarkers in the diagnosis of CVDs' outcomes, so as to demonstrate its role as a non-invasive diagnostic biomarker in this setting.
Materials and methods
Study selection and data collection
To get the high-quality research, systematic network document search was aimed at multiple website database, including PubMed, Embase, and the Web of science until October 30, 2021 , and the detailed search strategy were as follow: (a) 'circular RNA' or 'circRNA' and (b) 'Cardiovascular Disease' or 'Coronary Artery Disease' or 'Myocardial Infarction' or 'Heart Attack' or 'Coronary Heart Disease' or 'Ischemic Heart Disease' or 'Ischaemic Heart Disease' or 'Stroke' or 'Transient Ischemic Attack' or 'Transient Ischaemic Attack' or 'Vascular Accident' or 'Apoplexy' or 'High Blood Pressure' or 'Hypertension' or 'Atherosclerotic sclerosis' and (c) diagnosis or diagnostic or sensitivity or specificity or 'receiver operating characteristic curve. The relevant systematic reviews and references cited in the searched articles were also manually reviewed for additional studies of circRNAs in patients with CVDs. And all included studies were in English.
Selected and removed criteria
The selected criteria were as follows: (a) studies patients were diagnosed with cardiovascular events (including Cardiovascular Disease, Coronary Artery Disease, Myocardial Infarction, Heart Attack, Coronary Heart Disease, Ischemic Heart Disease, Stroke, Transient Ischemic Attack, Transient Ischaemic Attack, Vascular Accident, Apoplexy, High Blood Pressure, Hypertension and Atherosclerotic Sclerosis); (b) expression of circRNAs was measured in the blood (serum, plasma, peripheral whole blood and leukocyte in peripheral blood); (c) the study has clinical results related to circRNAs expression; (d) AUC, sensitivity, specificity or 95% CI between circRNAs expression could be picked up directly or calculated indirectly in the study; (e) case-control studies were included.
The exclusion criteria were: (a) repeated research publication; (b) content has nothing to do with CVDs or circRNAs; (c) non-human studies, letters, experiments, reviews, meta-analysis, etc.
Data extraction and quality evaluation
Two investigators independently extracted the information and data from all selected studies through cross-check. Any difference between investigators was discussed and resolved by a third investigator. The information and data were collected by a purpose-designed form: author, year of publication, country, the type of CVDs, the number of patients and controls, specimens, the basic information of circRNAs, and more. We collected the numbers of true-positive (TP), false-positive (FP), true-negative (TN), false-negative (FN), platform, fold changes (FC), cut-off criteria of the dysregulated circRNAs through original data. ∣FC∣ ≥ 1.5, P < 0.05 were selected as the cut-off threshold. The quality of studies was evaluated using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) checklist.
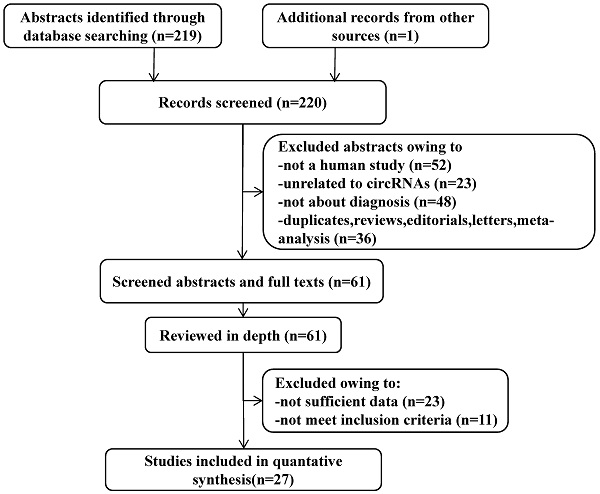
A flow diagram demonstrating the study selection process. The process of study selection including identification, screening, eligibility extraction, and inclusion steps were depicted in the flow diagram. Out of 220 records identified from three databases, 27 studies met the selection criteria.
However, there is still no unified naming standard for circRNAs, and different discoverers will use their own ways to name newly discovered circRNAs. Different circRNAs with different expressions will cause difficulties in the comparative analysis of circRNAs in CVDs. One needs to merge combine information to better unify circRNAs. Referring to the naming methods in the literature, we use the Gene symbol of the corresponding circRNAs to unify the naming format, which is convenient for finding potential biomarkers of CVDs. “Gene symbol” represents the name of the gene where the circRNAs is located. Therefore, we found the corresponding gene symbol by referring to the circRNAs database mentioned in the literature or circbase (http://www.circbase.org/). If the gene symbol still could not be found in the original literature data and circbase, the original circRNAs name will be retained (hsa_circ_0141720 and hsa-circRNA9102-5).
Statistical analysis
Stata 12.0, RevMan 5.0, and Meta-Disc 1.4 softwares were used for all statistical analyses. The sensitivity, specificity, positive, negative likelihood ratio (LR+ and LR-), and diagnostic odds ratio (DOR) of each circRNAs about the diagnostic value of CVD events were worked out from each study. The summary receiver operating characteristic (SROC) was curved to measure the overall performance of circRNAs in differentiating CVD patients from the controls by calculating the area under the curve (AUC). The heterogeneity among the qualified studies was indicated by the Cochran Q-test and Inconsistency Index (I2), in which I2 > 50 implied significant heterogeneity. If I2 <50% or P>0.5, then the random-effects model (Der Simonian and Laird) was applied. Or else, we selected the fixed-effects model (Mantel-Haenszel). Subgroup analysis, sensitivity analysis, and meta-regression should be carried out to explore the potential causes of heterogeneity. Additionally, publication bias was assessed by Deeks' funnel plots asymmetry test, and P < 0.05 indicated remarkable bias.
Results
Literatures search results and characteristics of the selected studies
After the initial search algorithm, a total of 219 articles were retrieved from PubMed, MEDLINE, and the Cochrane database (Figure 1), and 1 article was included by manually searching the publication bibliography and review articles. Among them, 36 articles were duplicates, reviews, letters, or meta-analysis, 52 articles were not human studies, 23 articles were unrelated to circRNAs and 48 articles were not about the diagnosis. After these articles were removed, 61 articles were further evaluated. The full-text review identified 23 articles without sufficient data and 11 articles that did not meet the inclusion criteria. Finally, 27 articles, including 47 eligible studies, were included in this meta-analysis (Table 1).
Characteristics of 21 articles included in the meta-analysis
| Study | Year | CVD | Patients (controls) | Source of control | Specimen | Method | Design type | CircRNA |
|---|---|---|---|---|---|---|---|---|
| Bazan [22] | 2017 | IS | 41(71) | Non-urgent | Serum | RT-PCR | Retrospective | circR-284(U) |
| Bu [23] | 2019 | CAD | 585(585) | Healthy | Blood | qRT-PCR | Retrospective | hsa_circ_0008507(U)hsa_circ_0001946(U) hsa_circ_0000284(D) |
| Chen [24] | 2020 | AIS | 80(30) | Healthy | Serum | qRT-PCR | Retrospective | hsa_circ_0141720(U) |
| Huang [25] | 2021 | CAD | 30(30) | Non-CAD | Blood | qRT-PCR | Retrospective | hsa_circ_0001946(U) |
| Jiang [26] | 2018 | PE | 35(35) | Non-PE | Blood | qRT-PCR | Prospective | hsa_circ_0004904(U)hsa_circ_0001855(U) |
| Jin [27] | 2021 | PAH | 21(21) | Healthy | Blood | qRT-PCR | Retrospective | hsa_circNFXL1_009(D) hsa_circMFN2_004(D) hsa_circ_ZNF302(D) hsa_circGSDMD_004(U) hsa_circWDR37_016(U) |
| Li [28] | 2021 | IS | 118(118) | Healthy | Blood | qRT-PCR | Retrospective | hsa_circ_0001599(U) |
| Liang [29] | 2020 | CAD | 330(209) | Non-CAD | Blood | qPCR | Retrospective | circZNF609(D) |
| Liu [30] | 2019 | EH | 89(89) | Healthy | Blood | qRT-PCR | Retrospective | hsa_circ_0126991(U) |
| Peng [31] | 2019 | IS | 160(160) | Non-IS | Blood | qPCR | Retrospective | circHECTD1(U) |
| Sun [32] | 2020 | HF | 30(30) | Non-HF | Plasma | qRT-PCR | Retrospective | hsa_circ_0062960(U) |
| Tian [33] | 2019 | AAAD | 30(30) | Non-AAAD | Serum | qRT-PCR | Retrospective | circMARK3(U) |
| Tian [34] | 2021 | MI | 47(18) | Healthy | Tissue | qRT-PCR | Retrospective | circSLC8A1(U) circNFIX(D) |
| Wang [35] | 2019 | CAD | 436(297) | Non-CAD | Blood | qRT-PCR | Retrospective | hsa_circ_0001879(U)hsa_circ_0004104(U) |
| Wu [36] | 2019 | KD | 56(56) | Healthy | Serum | qRT-PCR | Retrospective | circANRIL(D)hsa_circ_0123996(U) |
| Wu [37] | 2020 | CAD | 108(89) | Non-CAD | Plasma | qPCR | Retrospective | hsa_circ_0005540(U) |
| xiong [38] | 2021 | CAD | 109(70) | Non-CAD | Serum | qRT-PCR | Retrospective | circNPHP4(U) |
| Yang [39] | 2021 | MI | 30(60) | Non-MI | Blood | qRT-PCR | Retrospective | circRNA_104761(D) |
| Zhang [40] | 2018 | PoAF | 158(521) | Non-PoAF | Blood | qRT-PCR | Retrospective | hsa_circ_025016(U) |
| Zhang [41] | 2019 | IPAH | 82(82) | Healthy | Serum | qRT-PCR | Prospective | circ_0068481(U) |
| Zhao [42] | 2017 | CAD | 179(157) | Non-CAD | Blood | qRT-PCR | Retrospective | hsa_circ_0124644(U) |
| Zhao [43] | 2020 | IS | 75(90) | Non-IS | Blood | qRT-PCR | Retrospective | hsa_circ_0072309(D) |
| Zheng [44] | 2019 | EH | 89(89) | Healthy | Blood | qRT-PCR | Retrospective | hsa_circ_0014243(U) |
| Zheng [45] | 2020 | EH | 96(96) | Healthy | Blood | qRT-PCR | Retrospective | hsa-circRNA9102-5(U) |
| Zhu [46] | 2019 | IS | 170(170) | Non-IS | Blood | qPCR | Retrospective | circ-DLGAP4(D) |
| Zuo [47] | 2020 | AIS | 239(139) | Healthy | Blood | qRT-PCR | Retrospective | circFUNDC1+circPDS5B +circCDC14A(U) |
| Zuo [48] | 2020 | AIS | 26(42) | Non-AIS | Blood | qPCR | Retrospective | circFUNDC1(U) |
Abbreviations: CAD, coronary artery disease; AIS, acute ischemic stroke; IS, ischemic stroke; PE, preeclampsia; HF, heart failure; EH, essential hypertension; IPAH, idiopathic pulmonary arterial hypertension; AAAD, acute stanford type A aortic dissection; KD, kawasaki disease; PoAF, postoperative atrial fibrillation; CVD, cardiovascular disease; circRNA, circular RNA; qPCR, real-time polymerase chain reaction; qRT-PCR, RT-PCR/qPCR combined technique; RT-PCR, reverse transcription-polymerase chain reaction; D, downregulated; U, upregulated.
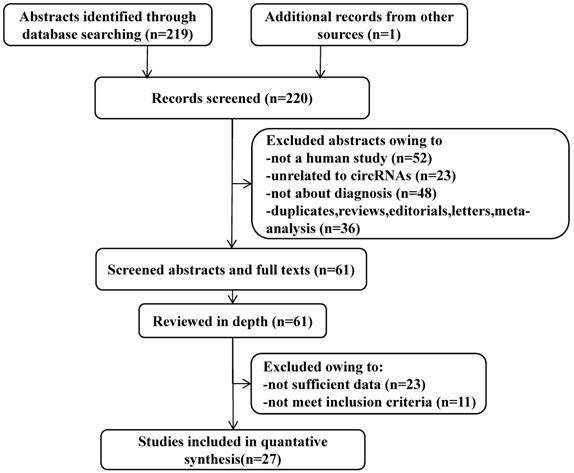
All of these studies were published from January 1, 2010, to October 30, 2021, containing 6833 participants (Table 2). Among them, only one was from the USA, the rest were from China. In this meta-analysis, 10 different types of CVDs were contained, namely, 8 studied strokes, 15 CADs, 3 EHs, 3 HF, 4 MI, 2 PE, 1 AAAD, 2 KD, 2 PoAF and 8 PAH. PCR (qPCR), RT-PCR, or qRT-PCR was used for quantitative detection of these circRNAs. A total of 39 circRNAs were investigated in these articles, of which 29 were up-regulated and 10 circRNAs were down-regulated. Participants' blood was the main specimen source. According to the result of the QUADAS-2 assessment, the included 27 articles were at a certain risk of bias regarding patient selection, index test, and flow and timing. Figure 2A shows the risk degree of bias risk and applicability of each study in detail, while Figure 2B uses a bar chart to visually show the ratio of bias risk and applicability risk of the overall included literatures (Figure 2A, 2B). As we can see from the figures, the main causes of these risks of bias and application concerns were the blinding of participants, a lack of information on exclusion criteria and not meeting the prior undetermined threshold.
Diagnostic performance
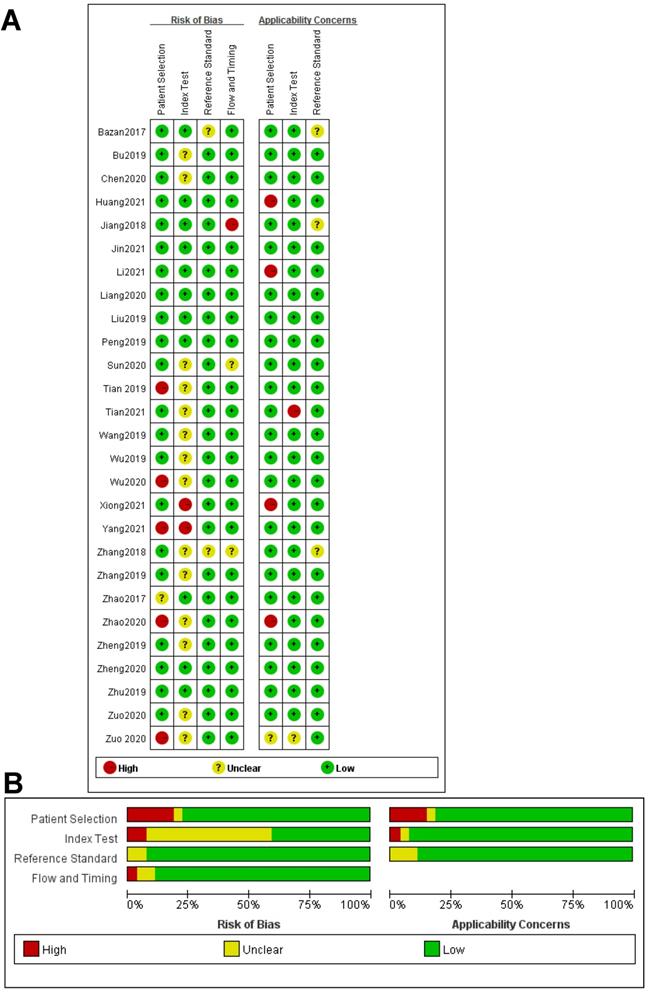
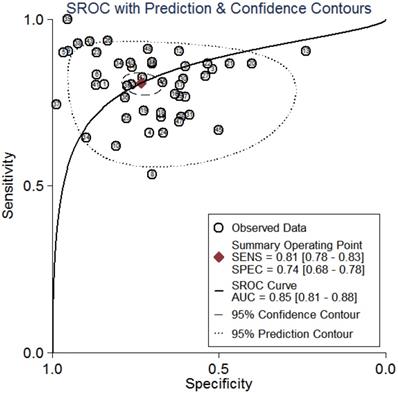
All the results of individual studies were summarized to understand the potential application of circRNAs in the diagnosis of CVDs. The Spearman correlation coefficient was 0.282 (P=0.052), indicating no threshold effect in the accuracy estimates of circRNAs. Forest plots of the sensitivity and specificity of circRNAs for diagnosing CVDs are displayed (Figure 3). The random-effects model was employed to estimate the combined effects in virtue of the significant heterogeneity (I²=78.42% and 86.97%). The indexes are as follows: sensitivity, 0.81 (95%CI 0.78-0.83); specificity, 0.74 (95%CI 0.68-0.78); the pooled LR+, LR- and DOR were 3.1 (95%CI 2.5-3.7), 0.26 (95%CI 0.22-0.31), and 12 (95%CI 9-16), respectively. Comprehensive diagnostic performance was evaluated by the summary receiver operating characteristic curve (SROC). The area under the SROC can accurately reflect the authenticity of the diagnostic test. The closer the area under the SROC is to 1, the better the authenticity of the diagnostic test is. The AUC was 0.85 (95%CI 0.81-0.88), which means that the overall circRNAs has good diagnostic accuracy (Figure 4). Furthermore, a series of analyses, including subgroup analysis, sensitivity analysis, meta-regression, and publication bias, were conducted to explore the potential sources of heterogeneity between studies.
Overall quality assessment of eligible studies by QUADAS-2 tool. A. Methodological quality summary (by study). B. Methodological quality graph (overall).
Characteristics of eligible studies included in the meta-analysis
| Author | Year | CircRNAs | CVD | Platform | Cut-off criteria | TP | FP | FN | TN | SEN% | SPE% | AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bazan | 2017 | circHIPK3 (1) | IS | RT-PCR | FC > 2, P < 0.05 | 33 | 11 | 8 | 60 | 80 | 85 | 0.91 |
| Bu¹ | 2019 | circGBA3 (2) | CAD | Microarray | FC > 2, P < 0.05 | 86 | 40 | 14 | 60 | 86 | 60 | 0.75 |
| Bu² | 2019 | circCDR1 (3) | CAD | Microarray | FC > 2, P < 0.05 | 85 | 48 | 15 | 52 | 85 | 52 | 0.71 |
| Bu³ | 2019 | circHIPK3 (4) | CAD | Microarray | FC > 2, P < 0.05 | 66 | 29 | 34 | 71 | 66 | 71 | 0.68 |
| Huang (2021) | 2021 | hsa_circ_0001946 (5) | CAD | qRT-PCR | FC > 2, P < 0.05 | 25 | 4 | 5 | 26 | 83.3 | 86.7 | 0.897 |
| Chen | 2020 | hsa_circ_0141720 (6) | AIS | Microarray | FC > 2, P < 0.05 | 72 | 1 | 8 | 29 | 89.7 | 95.6 | 0.911 |
| Jiang¹ | 2018 | circPOLE2 (7) | PE | Microarray | FC > 2, P < 0.05 | 23 | 12 | 7 | 18 | 76.67 | 60 | 0.728 |
| Jiang² | 2018 | circRNF38 (8) | PE | Microarray | FC > 2, P < 0.05 | 16 | 9 | 14 | 21 | 53.33 | 70 | 0.621 |
| Jin¹ | 2021 | hsa_circNFXL1_009 (9) | PAH | qRT-PCR | FC > 2, P < 0.05 | 19 | 1 | 2 | 20 | 90.48 | 95.24 | 0.941 |
| Jin² | 2021 | hsa_circMFN2_004 (10) | PAH | qRT-PCR | FC > 2, P < 0.05 | 13 | 4 | 8 | 17 | 61.9 | 80.95 | 0.747 |
| Jin³ | 2021 | hsa_circ_ZNF302 (11) | PAH | qRT-PCR | FC > 2, P < 0.05 | 18 | 5 | 3 | 16 | 85 | 76.19 | 0.847 |
| Jin4 | 2021 | hsa_circGSDMD_004 (12) | PAH | qRT-PCR | FC > 2, P < 0.05 | 19 | 8 | 2 | 13 | 90.04 | 60 | 0.731 |
| Jin5 | 2021 | hsa_circWDR37_016 (13) | PAH | qRT-PCR | FC > 2, P < 0.05 | 19 | 16 | 2 | 5 | 90 | 25 | 0.822 |
| Li | 2021 | hsa_circ_0001599 (14) | IS | qRT-PCR | FC > 2, P < 0.05 | 76 | 12 | 42 | 106 | 64.41 | 89.83 | 0.805 |
| Liang¹ | 2020 | circZNF609 (15) | CAD | RT-PCR | P < 0.0001 | 26 | 9 | 4 | 21 | 86.7 | 70 | 0.83 |
| Liang² | 2020 | circZNF609 (16) | CAD | RT-PCR | P < 0.0001 | 233 | 66 | 67 | 113 | 77.7 | 63 | 0.752 |
| Liang³ | 2020 | circZNF609 (17) | CAD | RT-PCR | P < 0.0001 | 265 | 80 | 65 | 129 | 80.4 | 61.5 | 0.761 |
| Liu | 2019 | circSep-11 (18) | EH | Microarray | FC > 2, P < 0.05 | 64 | 29 | 25 | 60 | 72.4 | 67.3 | 0.741 |
| Peng | 2019 | circ HECTD1 (19) | IS | qRT-PCR | P < 0.001 | 116 | 44 | 44 | 116 | 72.5 | 72.5 | 0.814 |
| Sun¹ | 2020 | circDEPDC5 (20) | HF | Microarray | FC > 2, P < 0.01 | 26 | 18 | 4 | 12 | 86.7 | 40 | 0.838 |
| Sun² | 2020 | circLTBP1 (21) | HF | Microarray | FC > 2, P < 0.01 | 26 | 16 | 4 | 14 | 86.2 | 46.7 | 0.759 |
| Sun³ | 2020 | circMARC2 (22) | HF | Microarray | FC > 2, P < 0.01 | 26 | 14 | 4 | 16 | 86.7 | 53.3 | 0.817 |
| Tian | 2019 | circMARK3 (23) | AAAD | Microarray | FC > 2, P < 0.05 | 27 | 4 | 3 | 26 | 90 | 86.7 | 0.9344 |
| Tian¹ | 2021 | circSLC8A1 (24) | MI | qRT-PCR | FC > 2, P < 0.05 | 31 | 6 | 16 | 12 | 66.7 | 67.1 | 0.706 |
| Tian² | 2021 | circNFIX (25) | MI | qRT-PCR | FC > 2, P < 0.05 | 33 | 4 | 14 | 14 | 71.1 | 77.8 | 0.868 |
| Wang¹ | 2019 | circNIPSNAP3A (26) | CAD | Microarray | FC ≥ 1.5, P < 0.05 | 342 | 133 | 70 | 157 | 83.1 | 54.3 | 0.703 |
| Wang² | 2019 | circSPARC (27) | CAD | Microarray | FC ≥ 1.5, P < 0.05 | 291 | 112 | 121 | 178 | 70.7 | 61.4 | 0.7 |
| Wang³ | 2019 | circNIPSNAP3A+circSPARC (28) 19hsa_circ_0004104 | CAD | Microarray | FC ≥ 1.5, P < 0.05 | 317 | 110 | 95 | 180 | 76.9 | 62 | 0.742 |
| Wu¹ | 2019 | circFBXW12 (29) | KD | qRT-PCR | P < 0.001 | 46 | 22 | 10 | 34 | 82.2 | 60 | 0.747 |
| Wu² | 2019 | circANRIL (30) | KD | qRT-PCR | P < 0.05 | 40 | 23 | 16 | 33 | 72.3 | 58.9 | 0.624 |
| Wu | 2020 | circMCTP1 (31) | CAD | Microarray | FC > 4, P < 0.05 | 87 | 21 | 21 | 68 | 81 | 76.5 | 0.853 |
| xiong | 2021 | circNPHP4 (32) | CAD | qRT-PCR | FC ≥ 2, P < 0.05 | 95 | 21 | 14 | 49 | 87.1 | 69.7 | 0.837 |
| Yang | 2021 | circRNA_104761 (33) | MI | qRT-PCR | FC ≥ 2, P < 0.05 | 26 | 12 | 4 | 48 | 86.7 | 80 | 0.89 |
| Zhang¹ | 2018 | circCACNA1C (34) | PoAF | Microarray | FC ≥ 2, P < 0.05 | 60 | 65 | 15 | 225 | 79.4 | 77.6 | 0.802 |
| Zhang² | 2018 | circCACNA1C (35) | PoAF | Microarray | FC ≥ 2, P < 0.05 | 50 | 48 | 18 | 168 | 73.52 | 77.83 | |
| Zhang¹ | 2019 | circST6GAL1 (36) | IPAH | qRT-PCR | P < 0.05 | 61 | 1 | 21 | 81 | 74.39 | 98.78 | 0.895 |
| Zhang² | 2019 | circST6GAL1 (37) | IPAH | qRT-PCR | P < 0.05 | 26 | 4 | 2 | 49 | 94.59 | 92.45 | 0.978 |
| Zhang³ | 2019 | circST6GAL1 (38) | IPAH | qRT-PCR | P < 0.05 | 16 | 3 | 0 | 63 | 1 | 95.45 | 0.993 |
| Zhao¹ | 2017 | circROBO2 (39) | CAD | Microarray | FC > 2, P < 0.01 | 119 | 27 | 18 | 88 | 86.7 | 76.7 | 0.872 |
| Zhao² | 2017 | circSRGAP1 (40) | CAD | Microarray | FC > 2, P < 0.01 | 110 | 15 | 27 | 100 | 80 | 86.7 | 0.82 |
| Zhao³ | 2017 | circROBO2+circSRGAP1 (41) hsa_circ_0098964 | CAD | Microarray | FC > 2, P < 0.01 | 113 | 31 | 24 | 84 | 82.5 | 73 | 0.811 |
| Zhao | 2020 | circLIFR (42) | IS | qRT-PCR | P < 0.0001 | 70 | 10 | 5 | 80 | 93.3 | 88.9 | 0.9505 |
| Zheng | 2019 | circCHTOP (43) | EH | qRT-PCR | P < 0.001 | 63 | 29 | 26 | 60 | 70.8 | 67.4 | 0.732 |
| Zheng | 2020 | hsa-circRNA9102-5 (44) | EH | Microarray | FC > 2, P < 0.01 | 64 | 43 | 32 | 53 | 66.7 | 55.2 | 0.62 |
| Zhu | 2019 | circ-DLGAP4 (45) | IS | qPCR | P < 0.001 | 138 | 56 | 32 | 114 | 81.2 | 67.1 | 0.816 |
| Zuo | 2020 | circFUNDC1 (46) | AIS | Microarray | FC > 4, P < 0.01 | 18 | 16 | 8 | 26 | 69.23 | 61.9 | 0.6612 |
| Zuo | 2020 | circFUNDC1+circPDS5B +circCDC14A (47) | AIS | Microarray | FC > 4, P < 0.01 | 215 | 39 | 21 | 97 | 91 | 0.715 | 0.897 |
Abbreviations: CAD, coronary artery disease; AIS, acute ischemic stroke; IS, ischemic stroke; PE, preeclampsia; HF, heart failure; EH, essential hypertension; IPAH, idiopathic pulmonary arterial hypertension; AAAD, acute stanford type A aortic dissection; KD, kawasaki disease; PoAF, postoperative atrial fibrillation; CVD, cardiovascular disease; circRNA, circular RNA; qPCR, real-time polymerase chain reaction; qRT-PCR, RT-PCR/qPCR combined technique; RT-PCR, reverse transcription-polymerase chain reaction; FC, fold change.
Subgroup analysis
Subgroup analyses based on the specimen, disease types, detection method, sample size, source of control, and published year were conducted. The main results of subgroup analyses were summarized (Table 3). Furthermore, we drew an SROC curve to evaluate the accuracy of the diagnosis (Supplementary Figure S1). From the perspective of disease types, the diagnostic value of CADs (DOR 8 (95%CI 6-11)) was lower than IS (DOR 20 (95%CI 8-50)). Compared with the blood sample, the specimen of serum has a higher overall diagnostic accuracy, indicating that circRNAs were serum-specific with extremely high diagnostic value. The subgroups analysis results of the detection method demonstrated that the diagnostic accuracy of qRT-PCR was not different from that of the qPCR test. In the stratified analysis based on the source of control, healthy subjects and non-CVDs subjects have similar recognition rates of identifying CVD patients with similar sensitivity, specificity, PLR, NLR, DOR and AUC. Studies using small sample size (<200) presented better diagnostic accuracy than those using large sample size (≥200), with increased DOR (14 vs. 9) and AUC (0.86 vs. 0.82). Similar results were observed in publication year (~2019: AUC = 0.84; 2019~: AUC = 0.84). Most groups still remained high heterogeneity, and only had a significant decrease in heterogeneity observed in qPCR and the year before 2019. In conclusion, significant differences occurred between CVD disease type, specimen, and sample size, while only slight differences were observed among the other subgroups.
Forest plots for studies on overall circRNAs used in the diagnosis of CVDs among 27 studies included in the meta-analysis. (A) Sensitivity of circRNAs in diagnosis of CVDs and (B) specificity of circRNAs in diagnosis of CVDs.
Assessment of diagnostic accuracy and heterogeneity in subgroup analysis
| Subgroups | N | SEN(95%CI) | SPE (95%CI) | LR+ (95%CI) | LR-(95%CI) | DOR (95%CI) | AUC (95%CI) |
|---|---|---|---|---|---|---|---|
| ALL | 47 | 0.81 (0.78-0.83),I²=78.42,P<0.001 | 0.74 (0.68-0.78),I²=86.97,P<0.001 | 3.1 (2.5-3.7) | 0.26 (0.22-0.31) | 12 (9-16) | 0.85 (0.81-0.88) |
| CVD | |||||||
| CAD | 15 | 0.75 (0.65-0.83),I²=77.28,P<0.001 | 0.67 (0.59-0.74),I²=83.49,P<0.001 | 2.4 (2.0-2.9) | 0.30 (0.25-0.35) | 8 (6-11) | 0.82 (0.78-0.85) |
| IS | 8 | 0.83 (0.74-0.89),I²=85.63,P<0.001 | 0.81 (0.71-0.88),I²=83.73,P<0.001 | 3.9 (2.5-6.2) | 0.19 (0.12-0.32) | 20 (8-50) | 0.89 (0.86-0.91) |
| Method | |||||||
| qRT-PCR | 39 | 0.81 (0.77-0.84),I²=80.35,P<0.001 | 0.74 (0.68-0.80),I²=89.89,P<0.001 | 3.0 (2.3-3.9) | 0.27 (0.22-0.34) | 11 (7-18) | 0.84 (0.81-0.87) |
| qPCR | 7 | 0.82 (0.77-0.86),I²=77.89,P<0.001 | 0.68 (0.64-0.72),I²=46.97,P<0.001 | 2.6 (2.2-3.0) | 0.27 (0.21-0.35) | 10 (7-14) | 0.78 (0.74-0.81) |
| Specimen | |||||||
| Blood | 27 | 0.79 (0.75-0.83),I²=83.68,P<0.001 | 0.71 (0.65-0.76),I²=83.05,P<0.001 | 2.4 (2.0-2.9) | 0.31 (0.25-0.39) | 8 (5-12) | 0.80 (0.76-0.83) |
| Serum | 9 | 0.85 (0.79-0.90),I²=72.66,P<0.001 | 0.88 (0.75-0.94),I²=92.34,P<0.001 | 8.0 (3.4-18.8) | 0.17 (0.11-0.27) | 47 (15-149) | 0.91 (0.89-0.94) |
| Others | 11 | 0.80 (0.77-0.83),I²=0.00,P<0.001 | 0.65 (0.56-0.73),I²=84.13,P<0.001 | 2.3 (1.8-2.9) | 0.31 (0.27-0.36) | 7 (5-10) | 0.81 (0.78-0.85) |
| Sample size | |||||||
| <200 | 30 | 0.82 (0.78-0.85),I²=73.51,P<0.001 | 0.76 (0.68-0.82),I²=89.02,P<0.001 | 3.4 (2.3-5.1) | 0.25 (0.18-0.34) | 14 (7-27) | 0.86 (0.82-0.88) |
| ≥200 | 17 | 0.79 (0.76-0.83),I²=81.82,P<0.001 | 0.70 (0.65-0.75),I²=85.81,P<0.001 | 2.5 (2.2-3.0) | 0.29 (0.24-0.34) | 9 (7-12) | 0.82 (0.78-0.85) |
| Source of control | |||||||
| Healthy | 22 | 0.79 (0.74-0.83),I²=85.03,P<0.001 | 0.72 (0.63-0.79),I²=86.91,P<0.001 | 2.5 (1.8-3.6) | 0.31 (0.23-0.41) | 8 (5-15) | 0.82 (0.78-0.85) |
| Non-CVD | 25 | 0.81 (0.78-0.84),I²=66.06,P<0.001 | 0.73 (0.68-0.78),I²=84.31,P<0.001 | 3.2 (2.5-4.2) | 0.24 (0.20-0.31) | 13 (8-21) | 0.86 (0.82-0.88) |
| Year | |||||||
| ~2019 | 8 | 0.79 (0.73-0.84),I²=64.24,P<0.001 | 0.77 (0.73-0.81),I²=54.87,P<0.001 | 3.5 (2.8-4.3) | 0.27 (0.21-0.36) | 13 (8-20) | 0.84 (0.81-0.87) |
| 2019~ | 39 | 0.81 (0.78-0.84),I²=82.63,P<0.001 | 0.73 (0.67-0.78),I²=88.18,P<0.001 | 2.8 (2.2-3.7) | 0.26 (0.21-0.33) | 11 (7-17) | 0.84 (0.81-0.87) |
Abbreviations: CVDs, cardiovascular diseases; CAD, coronary artery disease; IS, ischemic stroke; SEN, sensitivity; SPE, specificity; CI, confidence interval; LR+, positive likelihood ratio; LR-, negative likelihood ratio; DOR, diagnostic odds ratio; AUC, area under the curve.
Summary receiver operator characteristic curves (SROC) of circRNAs for the diagnosis of CVDs in overall population.
Sensitivity analysis, meta-regression analysis, and publication bias
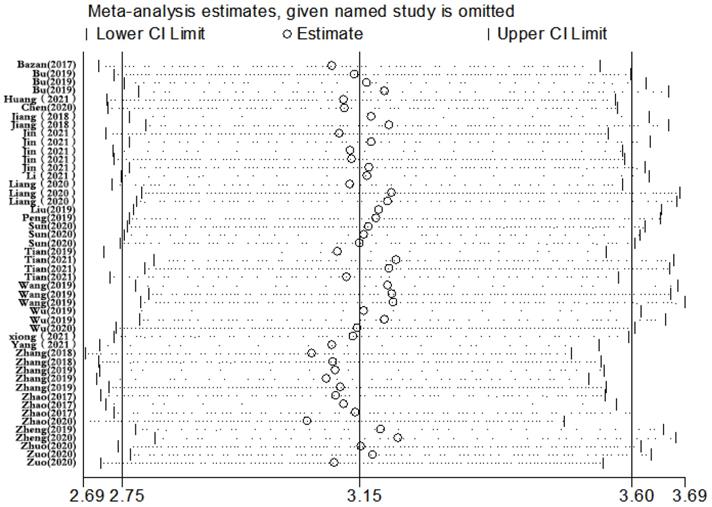
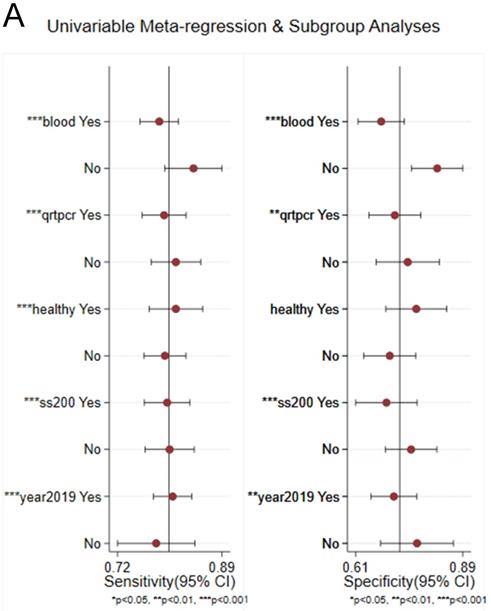
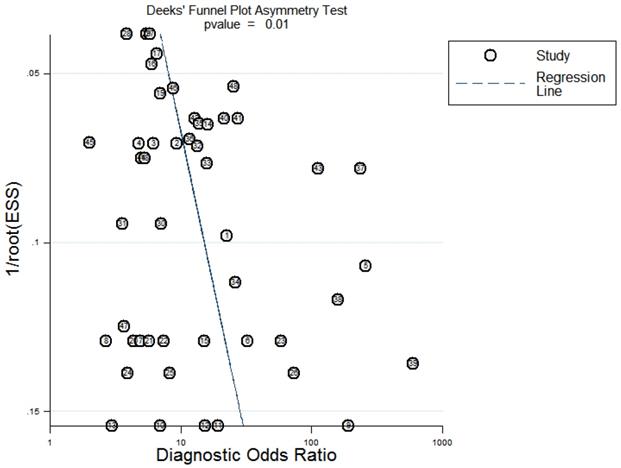
Sensitivity analysis was conducted simultaneously. One study was deleted at a time, and other studies were analyzed to estimate whether a single study would significantly affect the results. The results demonstrated that the pooled DOR in the result of the meta-analysis of CVDs were not significantly affected, indicating that this analysis confirmed the stability of our results (Figure 5). Subsequently, meta-regression analysis was performed on the bias of specimen, detection method, the source of control, sample size, and publication year (Figure 6). As illustrated in the figure, sensitivity was affected by all of these factors, whereas specificity was affected by specimen, detection method, sample size and publication year. Deeks' funnel plot asymmetry test was employed to assess publication bias, and showed a statistically significant value (P=0.01), reflecting data asymmetry and the possibility of publication bias (Figure 7).
Clinical utility of index test
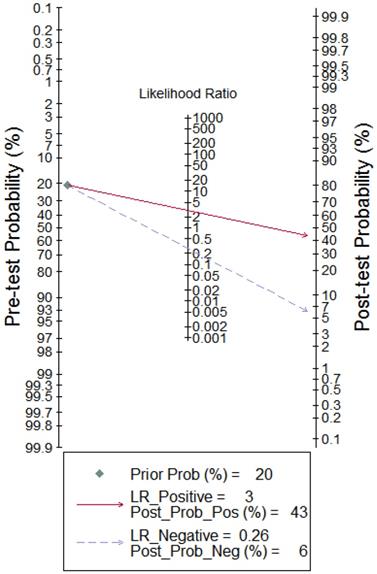
To better illustrate the clinical value of circRNA diagnosis in CVDs, Fagan's nomogram was adopted to calculate post-test probabilities. With a pre-test probability setting at 20%, the post-test probability increased to 43% with a positive LR of 3, while the post-test probability decreased to 6% with a negative LR of 0.26 (Figure 8). These results indicate that circRNAs can be good diagnostic biomarkers for CVDs.
Sensitivity analysis of the result of the meta-analysis for CVDs.
Univariable meta-regression for sensitivity and specificity of circRNAs for diagnosis of CVDs.
Deeks' funnel plot evaluating the potential publication bias of the included studies.
Fagan's nomogram evaluating the overall value of circRNAs for diagnosis of CVDs. The pre-test probability was set at 20%.
Discussion
With high morbidity and mortality rate, CVDs are the predominant cause of death around the world [49]. Therefore, improving the efficiency of early diagnosis is crucial for the improvement in CVDs prognosis. CircRNAs have outstanding characteristics such as high selective abundance [50], high stability [51], conservation [52,53] and specific expression [19,54] in human body fluids and tumor tissues, making it a suitable biomarker for disease diagnosis. There is an abundance of literature on the roles of circRNA in the clinical diagnosis of various CVDs. The association between circRNAs and clinical outcomes needs to be further explored.
Our meta-analysis, comprising 27 articles that described 47 studies, with a total of 6833 participants (3449 cases and 3384 controls), can be considered the most comprehensive meta-analysis to evaluate the diagnostic value of circRNAs in CVDs. Within high diagnostic value, circRNAs represent a promising diagnostic biomarker for CVDs. According to our results, the pooled DOR of circRNAs was 12, demonstrating a powerful discriminating capacity of circRNAs for CVDs diagnosis. Among the circRNAs we analyzed, circST6GAL1 might serve as a biomarker of CVDs for the clinical outcome, because its serum expression in patients with CVDs was not only significantly higher than that in healthy volunteers, but also significantly correlated with the death [41]. Simultaneously, the combined circRNAs will provide a better alternative, as more than two circRNAs would have better diagnostic value with significant sensitivity and specificity. The quality of our study was evaluated by the QUADAS-2 tool [55], with four domains of patients' selection, index tests, risk of bias, and flow and timing. The results of the quality assessment vary from low to high and may affect the stability of the combined results. The included studies were at high risk due to a lack of information on patient selection, exclusion criteria, or pre-specified thresholds, etc. Meanwhile, most of the literature used retrospective analysis, affecting the stability of the meta-analysis results. Therefore, it is urgent to make clear the specific situation of patient selection and adopt prospective research, so as to improve the results of these findings.
The pooled results revealed that there was a large degree of heterogeneity among the overall study. Then, the various causes of heterogeneity were clarified in our study. Spearman correlation coefficient was 0.282 (P = 0.052), suggesting that threshold effect was not the cause of heterogeneity. The results of the subgroup analyses study may provide some useful information for the high heterogeneity. As illustrated from the overall results, most of the subgroup analyses exhibited significant sensitivity and specificity heterogeneity (Table 3), except among studies detected by qPCR in specificity and published before 2019. Second, the subgroup analyses performed based on CVD revealed that the diagnostic value of circRNAs in patients with IS was higher compared with CAD. Due to the small sample size for other types of CVDs, we can't reach conclusions statistically. Both healthy subjects and non-CVDs controls can achieve the same diagnostic accuracy, demonstrating that circRNAs could be used for both population screening and as biomarkers to distinguish patients from non-CVDs controls. It can be seen from the results of subgroup analysis of the specimen, the serum is a better specimen in detecting circRNAs for CVDs. Therefore, the serum should be more widely used in clinical detection, such as blood biochemical tests and protein marker assays. Meanwhile, we made an interesting observation that combining multiple circRNAs or combining circRNAs with miRNAs yielded better diagnostic performance compared to single circRNAs in studied by Wang et al. [35] and Zheng et al. [45].
Additionally, a subsequent meta-regression analysis was performed. The results suggested that detection methods, the source of controls, sample size, and the publication year were not the origin of heterogeneity. Among them, the specimen is most likely to be the biggest factor affecting the heterogeneity. As with all meta-analysis, our study had potential publication bias with a statistically significant value, implying that better quality researches need to be included. Sensitivity analysis was also applied to analyze heterogeneity. However, no outlier studies were found, reflecting that our results were fairly reliable. Confounding factors such as age and gender have been statistically analyzed in the literature, and the risk factors have been excluded.
After searching, we found that a meta-analysis has been published on the diagnostic value of circRNAs in CVDs [56]. However, our study exhibited superiority in the following two areas compared to the previous work. First, our sample size was larger than that and the research data has been updated. The data we collected is due in October 2021, while the last study was due in September 2018, indicating that our study is more robust and reliable. Moreover, our study observed a higher diagnostic odds ratio (DOR:12, (95%CI: 9-16) than the previous study (DOR:2.94, (95%CI: 2.35-3.85)). Next, various subgroup analyses such as disease types, detection method, sample size, source of control, and published year were conducted to provide more information on and to give a more comprehensive insight into the diagnostic role of circRNAs in CVDs.
At the end of the analysis, the potential target and related pathways of circRNAs in the database were collected (Table 4), revealing its internal mechanism of promoting the occurrence and development of CVDs. Part of the circRNAs we mentioned are involved in the metabolic pathway and the PI3K-Akt signaling pathway, which may be involved in the activation of metabolic regulation, hypertrophy, and survival pathways, essential for the cardiovascular system cells [57]. By over-expressing hsa_circ_0004104 in THP-1-derived macrophages, it was revealed that atherosclerotic susceptibility genes, such as IDO1, MMP8, and CD40, were up-regulated, and anti-atherosclerotic genes, such as ApoA I, RNASE1, were down-regulated, indicating that hsa_circ_0004104 plays a crucial role in the pathogenesis of atherosclerosis [35]. Some of the circRNAs, such as circ_0072309, can induce apoptosis and inhibit cell proliferation and migration by acting on endothelial cells or smooth muscle cells, leading to the formation of atherosclerosis. Other circRNAs can induce inflammation and cause abnormalities in endothelial function by promoting the abnormal expression of pro-inflammatory factors, chemokines, and adhesion molecules. Although related pathways between circRNAs and CVDs have been listed, the detailed relationship between them remains to be explored.
In real life, many patients suffering from CVDs are not diagnosed until the middle or late stage of the CVDs, delaying the best time for earlier treatment. Therefore, early diagnosis of CVDs is of great clinical significance. As non-invasive and pain-free biomarkers for detection, circRNAs may regulate cellular functions by competing with “sponge” miRNAs. Rather than the classical detection method, only serum or plasma, which has broad application prospects due to the characteristics of non-invasive, high acceptability, and high stability, is required for detection. Statistical analysis of the data demonstrated that combining circRNAs with existing clinical diagnostic methods, such as coronary angiography (CAG), routine electrocardiogram (ECG), and Holter monitoring, will achieve a higher diagnostic sensitivity and specificity compared to a single indicator. However, the research on the clinical application of circRNAs as biomarkers is still in fancy. Thus, turning this idea into reality is full of difficulties and challenges. First, the use of circRNAs as biomarkers has not been validated by enough experiments. Second, the specificity of circRNAs as biomarkers for CVDs still needs to be improved. For example, among the circRNAs collected in this meta-analysis, circZNF609 is used as a new biomarker for the diagnosis of CAD [29], Nevertheless, circZNF609 can also serve as a diagnostic biomarker for Hirschsprung disease (HSCR) [58]. The expression levels of circRNAs are also downregulated in these diseases, generating a challenge for the diagnosis of a certain disease. Third, risk factors affecting the expression level of circRNAs, such as gender, age, environment,and diabetes, need to be further explored. Lastly, although a considerable number of circRNAs have been reported as biomarkers for the diagnosis of CVDs, only a few of them could be replicated. Thus, there is still a long way to go before circRNAs can be used as standard biomarkers of CVDs under a series of technical barriers and financial difficulties.
Research on the theoretical mechanism should be considered to realize the clinical application of circRNAs. Further studies are needed to confirm the safety and efficiency of circRNAs in diagnosis. Clinical trials of circRNAs as biomarkers should be conducted as soon as possible. Since further studies and rigorous experiments should be implemented, putting circRNA as biomarkers in clinical diagnosis still has a long way to go.
Summary of circRNAs with their potential targets and related pathways
| Author | CircRNA | Disease | Control | Expression | Potential target(s) | Related pathways |
|---|---|---|---|---|---|---|
| Bazan2017 | circR-284 | IS | Non-IS | U | miR-221 | ↓miR-221 |
| Bu2019 | hsa_circ_0001946 | CHD | Healthy | U | PARP1 | ↓cell proliferation, migration and invasion; ↓has-miR-7-5p, ↑PARP1, ↑cell apoptosis; ↓EGFR mRNA, ↓downstream protein kinases |
| hsa_circ_0008507 | CHD | Healthy | U | / | ↑adhesion of peripheral blood leukocytes to blood vessels | |
| hsa_circ_0000284 | CHD | Healthy | D | / | ↓endothelial cell proliferation, migration and tube formation, ↓growth, ↓plaque rupture of vascular endothelial cells, ↓adhesion of peripheral blood leukocytes to blood vessels | |
| Chen2020 | hsa_circ_0141720 | AIS | Healthy | U | / | ↑hs-CRP, ↑IL-6, ↑immune cells |
| Jiang2018 | hsa_circ_0004904 | PE | Non-PE | U | PAPP-A | ↓MREs, ↑PAPP-A, IGF axis |
| hsa_circ_0001855 | PE | Non-PE | U | PAPP-A | ↓MREs, ↑PAPP-A, IGF axis | |
| Liang2020 | circZNF609 | CAD | Non-CAD | D | AKT1, Smad7 | ↓cardiac inflammation, ↓atherosclerosis chronic inflammation, ↑anti-inflammation genes, ↓pro-inflammatory cytokines, ↑anti-inflammatory cytokines; ↓miR-138-5p, ↑AKT1, ↓IL-6; ↓miR-150-5p, ↓miR-615-5p |
| Liu2019 | hsa_circ_01269919 | EH | Healthy | U | miR-10a-5p | ↑inflammation, ↓miR-10a-5 |
| Peng2019 | circRNA HECTD1 | AIS | Non-AIS | U | TRAF3 | ↓autophagy; ↑pro-inflammatory cytokines, ↑inflammation, ↑immune responses; ↓TRAF3, ↓miR-142, ↓astrocyte activation; ↓miR-133b, ↑infarct |
| Sun2020 | hsa_circ_0062960 | HF | Non-HF | U | / | platelet activity |
| hsa_circ_0053919 | HF | Non-HF | U | / | platelet activity | |
| hsa_circ_0112085 | HF | Non-HF | U | / | platelet activity | |
| Tian2019 | circMARK | AAAD | Non-AAAD | U | Fgr | ↓apoptosis, ↑inflammation, ↓p53, ↑Fgr, ↑HASMC |
| Wang2019 | hsa_circ_0001879 | CAD | Healthy | U | / | PI3K/Akt pathway |
| hsa_circ_0004104 | CAD | Healthy | U | MAPK, TGF-β | PI3K/Akt pathway, ↑cardiac fibrosis; ↑IDO1, ↑MMP8, ↑CD40, ↓ApoA I, ↓RNASE1 | |
| Wu2019 | hsa_circ_0123996 | KD | Healthy | U | / | / |
| circANRIL | KD | Healthy | D | PES1 | ↑antiatherogenic cell functions; ↑apoptosis, ↓proliferation, ↓excessive the proliferation of VSMC; ↓protein translation rate, ↓cell growth | |
| Wu2020 | hsa_circ_0005540 | CAD | Non-CAD | U | miR-221, miR-145 | regulate endothelial cells, migration, capillary tube formation |
| Zhang2018 | hsa_circRNA_025016 | PoAF | No-PoAF | U | / | regulate melanogenesis, insulin and thyroid hormone secretion |
| Zhang2019 | circ_0068481 | IPAH | Healthy | U | / | / |
| Zhao2017 | hsa_circ_0124644 | CAD | Non-CAD | U | / | cell apoptosis |
| hsa_circ_0098964 | CAD | Non-CAD | U | / | / | |
| Zhao2020 | circ_0072309 | AIS | Non-AIS | D | miR-100, hsa-miR-519e-5p, hsa-miR-516b-5p, miR-492 | ↑apoptosis, ↓cell proliferation, migration; ↑necrosis, ↑apoptosis of SMC, PI3K/Akt pathway; ↑miR-100, ↑mTOR signal pathway; ↑hsa-miR-519e-5p, ↑hsa-miR-516b-5p,↑miR-492 |
| Zheng2019 | hsa_circ_0014243 | EH | Healthy | U | hsa-miR-10a-5p | ↓hsa-miR-10a-5p |
| Zheng2019 | hsa-circRNA9102-5 | EH | Healthy | U | hsa-miR-150-5p | ↑endothelial dysfunction, ↓angiogenesis, ↓hsa-miR-150-5p |
| Zhu2019 | circ-DLGAP4 | AIS | Non-AIS | D | miR-143 | ↓cardiomyocytes apoptosis, ↓inflammation; ↓miR-143 |
| Zuo2020 | circFUNDC1 | AIS | Non-AIS | U | FUNDC1 | mitophagy |
| Zuo2020 | AIS | Non-AIS | U | / | / |
Abbreviations: PARP1, poly (ADP-ribose) polymerase 1; EGFR: epidermal growth factor receptor; hs-CRP, high-sensitivity C relative protein; IL-6, interleukin 6; MREs, miRNA recognition elements; PAPP-A, pregnancy-associated plasma protein A; AKT1, a member of the three serine/threonine protein kinase family; Smad7, a negative regulator of TGF-β signaling; TRAF3, the tumor necrosis factor receptor-associated factor 3; Fgr, tyrosine-protein kinase; HASMC: human aortic smooth muscle cells; MAPK: mitogen-activated protein kinase; IDO1/MMP8/CD40, atherosclerosis-susceptible genes; PES1: pescadillo homologue 1; VSMC: vascular smooth muscle cell; SMC: smooth muscle cells; ApoA I/RNASE1, anti-atherosclerosis genes; D, downregulated; U, upregulated.
Although we strive to achieve a comprehensive and accurate analysis, several limitations remained, the main one being the significant heterogeneity among the included studies. Excessive heterogeneity cannot be completely eliminated, though subgroup analysis and meta-regression analysis have been conducted to explore the sources of heterogeneity. Notably, that specimen diversity and baseline differences between samples will affect the stability of our research results. Meanwhile, all kinds of CVDs are not the same, and unified analysis will lead to greater heterogeneity. The diagnostic value of circRNAs and specific types of CVDs should be further clarified. Secondly, only one study was conducted in American, and the rest studies were conducted in China, which will cause other ethnicities to be left out, leading to population selection bias. In addition, we have included studies with the small sample size, which may lead to publication bias. Small sample studies tend to report a higher effect value, though the research quality of small samples is often inferior to that of the large sample [59]. Finally, the credibility of causality between circRNAs and CVDs needs to be increased by more cell culture or animal model studies. Moreover, these candidate circRNAs biomarkers from cells or animal models must be verified in patients to illustrate their clinical value. Therefore, more high-quality, well-designed studies are required to confirm our findings.
Conclusion
Despite the limitations of the existing medical literature, research has verified that circRNAs can be useful biomarkers with advantages both in early screening and in distinguishing patients from non-CVD controls for its good diagnostic efficiency (AUC = 0.85). However, the feasibility, reliability, and repeatability of circRNA as next-generation CVD biomarkers should be fully verified by more studies.
Supplementary Material
Supplementary figure.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81874260), the Natural Science Foundation of Guangdong Province (2019A1515011567) and the Young Innovative Talents Projects of Universities in Guangdong (2018KQNCX097, 4SG19003Gj).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Global regional, national disability-adjusted life-years (DALYs) for 315 diseases injuries, healthy life expectancy (HALE) 1990-2015. a systematic analysis for the Global Burden of Disease Study 2015. Lancet (London, England). 2016;388(10053):1603-1658
2. LaRocca TJ, Martens CR, Seals DR. Nutrition and other lifestyle influences on arterial aging. Ageing research reviews. 2017;39:106-119
3. Mozaffarian D, Benjamin EJ, Go AS. et al. Executive Summary: Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133(4):447-454
4. Viereck J, Thum T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circulation research. 2017;120(2):381-399
5. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiological reviews. 2016;96(4):1297-1325
6. Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333-338
7. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nature biotechnology. 2014;32(5):453-461
8. Meng S, Zhou H, Feng Z. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Molecular cancer. 2017;16(1):94
9. Panda AC, Grammatikakis I, Kim KM. et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic acids research. 2017;45(7):4021-4035
10. Liu J, Li D, Luo H, Zhu X. Circular RNAs: The star molecules in cancer. Molecular aspects of medicine. 2019;70:141-152
11. Schindewolf C, Braun S, Domdey H. In vitro generation of a circular exon from a linear pre-mRNA transcript. Nucleic acids research. 1996;24(7):1260-1266
12. Conn SJ, Pillman KA, Toubia J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125-1134
13. Abdelmohsen K, Panda AC, Munk R. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA biology. 2017;14(3):361-369
14. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nature reviews Genetics. 2019;20(11):675-691
15. Tang Y, Bao J, Hu J, Liu L, Xu DY. Circular RNA in cardiovascular disease: Expression, mechanisms and clinical prospects. Journal of cellular and molecular medicine. 2021;25(4):1817-1824
16. Li M, Ding W, Sun T. et al. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. The FEBS journal. 2018;285(2):220-232
17. Cao Q, Guo Z, Du S, Ling H, Song C. Circular RNAs in the pathogenesis of atherosclerosis. Life sciences. 2020;255:117837
18. Devaux Y, Creemers EE, Boon RA. et al. Circular RNAs in heart failure. European journal of heart failure. 2017;19(6):701-709
19. Bahn JH, Zhang Q, Li F. et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clinical chemistry. 2015;61(1):221-230
20. Mao YY, Wang JQ, Guo XX, Bi Y, Wang CX. Circ-SATB2 upregulates STIM1 expression and regulates vascular smooth muscle cell proliferation and differentiation through miR-939. Biochemical and biophysical research communications. 2018;505(1):119-125
21. Yang L, Yang F, Zhao H, Wang M, Zhang Y. Circular RNA circCHFR Facilitates the Proliferation and Migration of Vascular Smooth Muscle via miR-370/FOXO1/Cyclin D1 Pathway. Molecular therapy Nucleic acids. 2019;16:434-441
22. Bazan HA, Hatfield SA, Brug A, Brooks AJ, Lightell DJ Jr, Woods TC. Carotid Plaque Rupture Is Accompanied by an Increase in the Ratio of Serum circR-284 to miR-221 Levels. Circulation Cardiovascular genetics. 2017 10(4)
23. Sun Y, Chen R, Lin S. et al. Association of circular RNAs and environmental risk factors with coronary heart disease. BMC cardiovascular disorders. 2019;19(1):223
24. Chen Y, Wang B, Liu W, Xu P, Song L. Diagnostic Value of Serum hsa_circ_0141720 in Patients with Acute Ischemic Stroke. Clinical laboratory. 2020 66(8)
25. Huang S, Zeng Z, Sun Y. et al. Association study of hsa_circ_0001946, hsa-miR-7-5p and PARP1 in coronary atherosclerotic heart disease. International journal of cardiology. 2021;328:1-7
26. Jiang M, Lash GE, Zhao X, Long Y, Guo C, Yang H. CircRNA-0004904, CircRNA-0001855, and PAPP-A: Potential Novel Biomarkers for the Prediction of Preeclampsia. Cell Physiol Biochem. 2018;46(6):2576-2586
27. Jin X, Xu Y, Guo M. et al. hsa_circNFXL1_009 modulates apoptosis, proliferation, migration, and potassium channel activation in pulmonary hypertension. Molecular therapy Nucleic acids. 2021;23:1007-1019
28. Li S, Hu W, Deng F. et al. Identification of Circular RNA hsa_circ_0001599 as a Novel Biomarker for Large-Artery Atherosclerotic Stroke. DNA and cell biology. 2021;40(3):457-468
29. Liang B, Li M, Deng Q. et al. CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease. Annals of translational medicine. 2020;8(12):741
30. Liu L, Gu T, Bao X, Zheng S, Zhao J, Zhang L. Microarray Profiling of Circular RNA Identifies hsa_circ_0126991 as a Potential Risk Factor for Essential Hypertension. Cytogenet Genome Res. 2019;157(4):203-212
31. Peng X, Jing P, Chen J, Xu L. The role of circular RNA HECTD1 expression in disease risk, disease severity, inflammation, and recurrence of acute ischemic stroke. Journal of clinical laboratory analysis. 2019;33(7):e22954
32. Sun Y, Jiang X, Lv Y. et al. Circular RNA Expression Profiles in Plasma from Patients with Heart Failure Related to Platelet Activity. Biomolecules. 2020 10(2)
33. Tian C, Tang X, Zhu X. et al. Expression profiles of circRNAs and the potential diagnostic value of serum circMARK3 in human acute Stanford type A aortic dissection. PloS one. 2019;14(6):e0219013
34. Tian M, Xue J, Dai C, Jiang E, Zhu B, Pang H. CircSLC8A1 and circNFIX can be used as auxiliary diagnostic markers for sudden cardiac death caused by acute ischemic heart disease. Scientific reports. 2021;11(1):4695
35. Wang L, Shen C, Wang Y. et al. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis. 2019;286:88-96
36. Wu J, Zhou Q, Niu Y. et al. Aberrant expression of serum circANRIL and hsa_circ_0123996 in children with Kawasaki disease. Journal of clinical laboratory analysis. 2019;33(5):e22874
37. Wu WP, Pan YH, Cai MY. et al. Plasma-Derived Exosomal Circular RNA hsa_circ_0005540 as a Novel Diagnostic Biomarker for Coronary Artery Disease. Disease markers. 2020;2020:3178642
38. Xiong F, Mao R, Zhang L. et al. CircNPHP4 in monocyte-derived small extracellular vesicles controls heterogeneous adhesion in coronary heart atherosclerotic disease. Cell death & disease. 2021;12(10):948
39. Yang W, Sun L, Cao X. et al. Detection of circRNA Biomarker for Acute Myocardial Infarction Based on System Biological Analysis of RNA Expression. Frontiers in genetics. 2021;12:686116
40. Zhang J, Xu Y, Xu S. et al. Plasma Circular RNAs, Hsa_circRNA_025016, Predict Postoperative Atrial Fibrillation After Isolated Off-Pump Coronary Artery Bypass Grafting. Journal of the American Heart Association. 2018 7(2)
41. Zhang Y, Chen Y, Yao H. et al. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ. 2019;9(4):2045894019888416
42. Zhao Z, Li X, Gao C. et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Scientific reports. 2017;7:39918
43. Zhao Y, Li J, Li J, Xu L, Lian W. The decreased circular RNA hsa_circ_0072309 promotes cell apoptosis of ischemic stroke by sponging miR-100. European review for medical and pharmacological sciences. 2020;24(8):4420-4429
44. Zheng S, Gu T, Bao X. et al. Circular RNA hsa_circ_0014243 may serve as a diagnostic biomarker for essential hypertension. Exp Ther Med. 2019;17(3):1728-1736
45. Zheng S, He X, Sun J, Li Q, Zhang T, Zhang L. The up-regulated hsa-circRNA9102-5 may be a risk factor for essential hypertension. Journal of clinical laboratory analysis. 2020;34(8):e23339
46. Zhu X, Ding J, Wang B, Wang J, Xu M. Circular RNA DLGAP4 is down-regulated and negatively correlates with severity, inflammatory cytokine expression and pro-inflammatory gene miR-143 expression in acute ischemic stroke patients. International journal of clinical and experimental pathology. 2019;12(3):941-948
47. Zuo L, Zhang L, Zu J. et al. Circulating Circular RNAs as Biomarkers for the Diagnosis and Prediction of Outcomes in Acute Ischemic Stroke. Stroke. 2020;51(1):319-323
48. Zuo L, Li C, Zu J, Yao H, Yan F. Circular RNA FUNDC1 improves prediction of stroke associated infection in acute ischemic stroke patients with high risk. Biosci Rep. 2020 40(6)
49. Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nature medicine. 2011;17(11):1410-1422
50. Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends in genetics: TIG. 2016;32(5):309-316
51. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cellular and molecular life sciences: CMLS. 2020;77(9):1661-1680
52. Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. CircRNAs and cancer: Biomarkers and master regulators. Seminars in cancer biology. 2019;58:90-99
53. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife. 2015;4:e07540
54. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development (Cambridge, England). 2016;143(11):1838-1847
55. Whiting PF, Rutjes AW, Westwood ME. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of internal medicine. 2011;155(8):529-536
56. Li JJ, Wang W, Wang XQ, He Y, Wang SS, Yan YX. A novel strategy of identifying circRNA biomarkers in cardiovascular disease by meta-analysis. Journal of cellular physiology. 2019;234(12):21601-21612
57. Ghigo A, Laffargue M, Li M, Hirsch E. PI3K and Calcium Signaling in Cardiovascular Disease. Circulation research. 2017;121(3):282-292
58. Peng L, Chen G, Zhu Z. et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2017;8(1):808-818
59. Nüesch E, Trelle S, Reichenbach S. et al. Small study effects in meta-analyses of osteoarthritis trials: meta-epidemiological study. BMJ (Clinical research ed). 2010;341:c3515
Author contact
![]() Corresponding authors: Tangbin Zou, Email: zoutbcom; Dongguan Key Laboratory of Environmental Medicine, School of Public Health, Guangdong Medical University, Dongguan, China. Hongfu Wu, Email: hongfuwcom; Key Laboratory of Stem Cell and Regenerative Tissue Engineering, Guangdong Medical University, Dongguan 523808, China.
Corresponding authors: Tangbin Zou, Email: zoutbcom; Dongguan Key Laboratory of Environmental Medicine, School of Public Health, Guangdong Medical University, Dongguan, China. Hongfu Wu, Email: hongfuwcom; Key Laboratory of Stem Cell and Regenerative Tissue Engineering, Guangdong Medical University, Dongguan 523808, China.

 Global reach, higher impact
Global reach, higher impact