Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(14):3261-3270. doi:10.7150/ijms.61552 This issue Cite
Research Paper
Alendronate-induced Perturbation of the Bone Proteome and Microenvironmental Pathophysiology
1. Departments of Surgery and Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, USA
2. Department of Medicine, University of California Los Angeles, CA, USA
3. Smidt Heart Institute, Department of Cardiology, Cedars-Sinai Medical Center, Los Angeles, CA, USA
4. Division of Orthodontics, College of Dentistry, The Ohio State University, Columbus, OH, USA
5. Division of Oral Surgery, College of Dentistry, The Ohio State University, Columbus, OH, USA
6. Department of Cell Biology, Microbiology, and Molecular Biology, University of South Florida, Tampa, FL, USA
Received 2021-4-14; Accepted 2021-7-11; Published 2021-7-23
Abstract
Objectives: Bisphosphonates (BPs) are powerful inhibitors of osteoclastogenesis and are used to prevent osteoporotic bone loss and reduce the risk of osteoporotic fracture in patients suffering from postmenopausal osteoporosis. Patients with breast cancer or gynecological malignancies being treated with BPs or those receiving bone-targeted therapy for metastatic prostate cancer are at increased risk of bisphosphonate-related osteonecrosis of the jaw (BRONJ). Although BPs markedly ameliorate osteoporosis, their adverse effects largely limit the clinical application of these drugs. This study focused on providing a deeper understanding of one of the most popular BPs, the alendronate (ALN)-induced perturbation of the bone proteome and microenvironmental pathophysiology.
Methods: To understand the molecular mechanisms underlying ALN-induced side-effects, an unbiased and global proteomics approach combined with big data bioinformatics was applied. This was followed by biochemical and functional analyses to determine the clinicopathological mechanisms affected by ALN.
Results: The findings from this proteomics study suggest that the RIPK3/Wnt/GSK3/β-catenin signaling pathway is significantly perturbed upon ALN treatment, resulting in abnormal angiogenesis, inflammation, anabolism, remodeling, and mineralization in bone cells in an in vitro cell culture system.
Conclusion: Our investigation into potential key signaling mechanisms in response to ALN provides a rational basis for suppressing BP-induced adverse effect and presents various therapeutic strategies.
Keywords: Osteonecrosis of the jaw, bisphosphonate, GSK signaling, clinical cone beam computed tomography, bone mineral density, proteomics, biomarker
Introduction
Bone tissue undergoes continuous cycles of bone resorption by osteoclasts and bone formation by osteoblasts, which were orchestrated by osteocytes[1]. Bone tissue is also highly vascularized providing O2, nutrients, and precursor cells for bone remodeling and serving as routes for blood and immune cells into bone tissue. Regulatory interactions between cells of these hematopoietic, immune, and skeletal (bone) systems closely regulate bone remodeling and repair processes via secreted factors such as VEGF, M-CSF, RANKL, Wnt3a, and Osteoprotegerin, etc. and their cell surface receptors.
Several key signal pathway has been shown to play pivotal roles in bone remodeling/repair processes, enhancing osteoblast differentiation and angiogenesis and modulating immune cell functions[2]. Specifically, Wnt pathway activation via GSK3 inactivation leads to osteoblast differentiation and stimulates bone anabolism while GSK3 gain-of-function promotes osteogenesis of adipose-derived stromal cells, making GSK3 as a possible therapeutic target for bone diseases [3-5]. Mice expressing constitutively active GSK3β (GSK3β S9A) mutant, exhibited a marked increase in osteogenesis, whereas ones with catalytically inactive GSK3β (GSK3β K85A) showed decreased osteogenic differentiation by regulating β-catenin[5]. Wnt/GSK3/β-catenin pathway also plays important roles in angiogenesis and vasculogenesis, supporting wound healing and regeneration of oral mucosa and jaw tissue [6]. Wnt signaling activation by Wnt1, VEGF, or CHIR99021 (GSK3β inhibitor) enhanced, while its inactivation by JW67 (targeting APC/GSK3/β-catenin complex) or β-catenin kinase dead form suppressed, vascular differentiation of mesenchymal stem cells (MSCs) derived from dental pulp [7]. GSK3β regulates β-catenin level in endothelial cells. Expression of β-catenin in HUVEC cells increases VEGF-A and -C level and induces capillary formation [8].
Bisphosphonates (BPs) have been suggested to modulate the proliferation and differentiation rates of osteoblasts and trigger survival signaling leading to bone homeostasis and antiresorptive effect [9-11]. First approved by the FDA in 1995, alendronate (ALN) is currently one of the most used BPs in the medical field[12]. ALN has been used successfully for the treatment of osteoporosis [13]. Several pieces of evidence indicate that there is a strong association between ALN and lower risk of bone metastases in postmenopausal women with early breast cancer [14, 15]. Cancer patients undergoing BPs-based treatments are at a 10-fold greater risk of developing bisphosphonate-related osteonecrosis of the jaw (BRONJ) [16], which is suggested to be a result of osteoclast inhibition and apoptosis[17]. Due to the prevalent usage of BPs in many bone-related diseases, more understanding on underlying mechanisms of adverse effect caused by BPs is crucial in providing better care and improving patient quality of life [18]. In oncology patients, incidence of BRONJ has been estimated to be as high as 18.6%[19], and risk of developing BRONJ increases with longer duration or higher dosages of BPs-based therapy[20].
This study sought to understand the pathogenesis of BP-associated adverse effects by looking into proteome perturbation and potential molecular biomarkers and mechanisms using an in vitro cell culture system.
Materials and Methods
Reagents and cell culture
Several cell lines, including MG-63, SCC-9, SCC-15, and HUVEC cells, were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). Culture condition, antibodies and reagents used for this study are available in Supplementary Materials.
Quantitative proteomics
Sample preparation methods for this study are available in Supplementary Materials. For protein quantification and statistical analysis, mapDIA was used. Data was analyzed based on the established workflows previously described [21, 22]. Briefly, peptides were identified using the openSWATH workflow [23], searched against the pan human library [24] with decoy sequences appended for false discovery rate calculation using the pyprophet algorithm [25]. Peptides with no greater than 5% identified false discovery rate (FDR) across all samples were compiled into the final experimental results using the TRIC alignment algorithm [26]. Following removal of non-proteotypic peptides (e.g., sequences matching more than one gene product from the Pan Human Library), the final aligned results were analyzed using mapDIA to select only high-quality performing fragments for quantification and to compile fragment level data into peptide and protein level abundance estimates [27]. The mapDIA software was also used to perform pairwise comparisons between ALN and control groups, including adjustment for multiple testing effects to produce a comparison FDR, which filtered proteins with significant or non-significant differential abundance in response to ALN treatment. The MS proteomics data has been deposited to the PRIDE repository with the dataset identifier, PXD024585.
Identification of differentially expressed proteins (DEPs)
Proteins with more than 3 nonredundant peptides in each sample were selected. Further selection of proteins detected in at least 2 samples in the same group was performed for statistical testing. A median difference test and Welch's t-test were performed separately, and the resulting two p-values were combined to compute adjusted p-values using Stouffer's method. The DEPs were identified based on an adjusted p-values<0.05 and absolute log2 fold-change (FC) ≧0.58.
VEGF ELISA assay
To determine vascular endothelial growth factor (VEGF-A) levels of conditioned medium from MG-63 cells incubated with ALN, supernatants from cell cultures were analyzed using the Human VEGF Quantikine ELISA Kit (R&D Systems, Minneapolis, Mass).
Cytokine array
Cell lysates and conditioned media from RAW 264.7 macrophages were collected and analyzed using a cytokine array, per standard provided protocols (R&D Systems, Minneapolis, MN, USA). ImageJ was used to measure the signal intensities.
Mineralization assay using Alizarin Red-S staining
The formation of calcium phosphate was quantified in MG-63 bone cells via Alizarin Red-S mineralization assay. Optical density was detected at an absorbance of 562 nm.
Statistical analysis
Most of the experiments were repeated at least six (6) times with independent treatments, while all the cases were repeated at least three times. Each of the experiments did not show significantly different results across replications. Statistical analyses were conducted using GraphPad Prism, version 7.03 (GraphPad Software Inc., La Jolla, CA). Mean values from technical replicates were used for statistical analyses, and all data were presented as the mean ± standard deviation (SD). A one-way analysis of variance (ANOVA) or Student's t-test was conducted to compare the groups of data. Differences were considered statistically significant when P < 0.05.
Results
Comprehensive analysis with large unbiased global proteomic assays suggested perturbed proteins in response to BP in bone cells
Mass spectrometry (MS) has several important attributes that make it amenable to providing reproducible and accurate assays for proteins and metabolites. It provides a scalable number of analytes quantified in a single assay and absolute quantification, which leads to a standardized path from assay development to validation of new candidate biomarkers applicable in any clinical chemistry laboratory. To understand the molecular mechanisms underlying specific diseases, an unbiased and global omics approach combined with big data analysis using bioinformatics is critical.
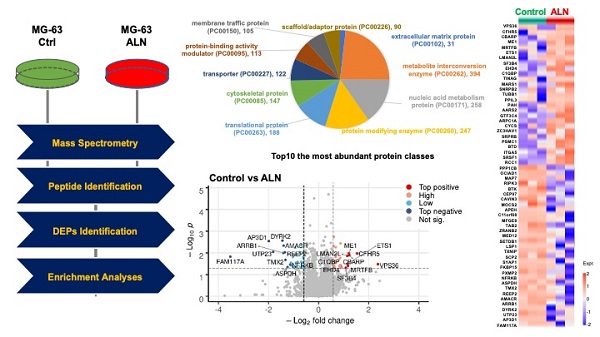
As described in the Materials and Methods, a proteomics approach was implemented (Fig 1A). The top 10 most abundant protein classes are shown in Fig 1B. Global proteomics analysis identified a highly confident and comprehensive list of perturbed proteins in MG-63 bone cells treated with ALN. Protein quantification and statistical analysis using mapDIA identified perturbed proteins in MG-63 cells treated with 10 μM ALN. A total of 2,865 proteins with UniProtKB IDs were identified. Further analysis with the PANTHER Protein Classification Tool revealed that the most abundant top 10 proteins classes included extracellular matrix, metabolite interconversion, nucleic acid metabolism, protein modification, translational regulation, cytoskeletal, transporter, protein-binding activity modulator, membrane traffic, and scaffold/adaptor[28]. To identify DEPs, the integrated hypothesis testing method was applied. Briefly, the median difference test and Welch's t-test was performed on high confidence proteins, which in the case of this experiment, were proteins detected with more than 3 non-redundant peptides encompassing at least 2 samples in the same group. The median test p-value and Welch's t-test p-value were then combined to adjust for multiple testing errors. Finally, 27 up- and 31 downregulated DEPs were selected for based on adjusted p-values < 0.05 and log2 FC ≧0.58. Significant expression was assessed using a volcano plot (Fig 1C and Fig 1D) and heatmap (Fig 1E). The DEPs are listed in Table 1.
Angiogenesis alteration in response to ALN treatment
When verifying proteins associated with angiogenesis-related Gene Ontology Biological Processes (GOBPs), several proteins were identified, including ETS proto-oncogene 1 (ETS1) (log2 FC, 1.1566), integrin subunit alpha 5 (ITGA5) (log2 FC, 0.6102), and milk fat globule-EGF factor 8 (MFGE8) (log2 FC, -0.7468) (Table 1). To further investigate these findings, the effects of ALN on several well-known angiogenic factors were investigated. Secretion of VEGF-A, a potent angiogenic factor, was examined in bone cells after stimulation with ALN. Consistent with similarly designed work from previous trials [29], treatment of MG-63 cells with ALN led to a statistically significant but modest decrease (approximately 30%) of VEGF secretion into the conditioned medium compared to control (Fig 2A). Furthermore, HUVEC stimulation in the collected culture medium also exhibited modest but meaningful suppression of proliferation (Fig 2B). Collectively, the reduction of VEGF secretion and HUVEC proliferation by ALN strongly implies angiogenic signals to vessel cells from bone cells. This finding suggests the potential microenvironment-level regulation of bone remodeling in ONJ. For proteomics profiling, necrotic and apoptotic conditions were avoided to fully investigate the effects of ALN on bone cells. Additional analysis confirmed that there was no induced cell death with ALN treatment in MG-63 cells. Cell viability and proliferation rates, which were determined using MTT (Fig 2C) and crystal violet staining assays (Fig 2D), showed no cytotoxicity.
List of differentially expressed proteins (DEPs) with corresponding statistics.
| Uniprot ID | Gene Symbol | Full Name | Log2 FC (ALN/ Ctrl) | Median P-Value | T Test P- Value | Adj. P |
|---|---|---|---|---|---|---|
| Q86VN1 | VPS36 | Vacuolar protein-sorting-associated protein 36 | 2.3697 | 0.0191 | 0.3002 | 0.0332 |
| Q9BXR6 | CFHR5 | Complement factor H-related protein 5 | 1.5412 | 0.0306 | 0.0824 | 0.0106 |
| Q8N350 | CBARP | Voltage-dependent calcium channel beta subunit-associated regulatory protein | 1.23 | 0.0776 | 0.0415 | 0.0129 |
| P48163 | ME1 | NADP-dependent malic enzyme | 1.2145 | 0.1359 | 0.0149 | 0.0103 |
| Q9ULH7 | MRTFB | Myocardin-related transcription factor B | 1.1752 | 0.1301 | 0.0877 | 0.0397 |
| P14921 | ETS1 | Protein C-ets-1 | 1.1566 | 0.1653 | 0.0497 | 0.0319 |
| Q9H0V9 | LMAN2L | VIP36-like protein | 1.1446 | 0.013 | 0.1852 | 0.0136 |
| Q15427 | SF3B4 | Splicing factor 3B subunit 4 | 1.0902 | 0.3234 | 0.0296 | 0.0487 |
| Q9H223 | EHD4 | EH domain-containing protein 4 | 0.8995 | 0.113 | 0.1213 | 0.0463 |
| Q07021 | C1QBP | Complement component 1 Q subcomponent-binding protein, mitochondrial | 0.8848 | 0.1071 | 0.1049 | 0.0388 |
| Q9UJW2 | TINAG | Tubulointerstitial nephritis antigen | 0.8833 | 0.0976 | 0.1428 | 0.0474 |
| P56192 | MARS1 | Methionine--tRNA ligase, cytoplasmic | 0.8772 | 0.0039 | 0.1283 | 0.0037 |
| P08579 | SNRPB2 | U2 small nuclear ribonucleoprotein B | 0.8721 | 0.0322 | 0.2438 | 0.0361 |
| Q9H4B7 | TUBB1 | Tubulin beta-1 chain | 0.8644 | 0.0467 | 0.0944 | 0.0172 |
| Q9H2H8 | PPIL3 | Peptidyl-prolyl cis-trans isomerase-like 3 (PPIase) | 0.8257 | 0.1129 | 0.1055 | 0.0408 |
| P00439 | PAH | Phenylalanine-4-hydroxylase (PAH) | 0.8024 | 0.0475 | 0.1114 | 0.0206 |
| Q5JTZ9 | AARS2 | Alanine--tRNA ligase, mitochondrial | 0.7071 | 0.0877 | 0.0325 | 0.0118 |
| Q9UKN8 | GTF3C4 | General transcription factor 3C polypeptide 4 | 0.6861 | 0.0038 | 0.1761 | 0.0055 |
| Q92747 | ARPC1A | Actin-related protein 2/3 complex subunit 1A (SOP2-like protein) | 0.6813 | 0.1877 | 0.0156 | 0.0158 |
| P99999 | CYCS | Cytochrome c | 0.664 | 0.0456 | 0.1572 | 0.0283 |
| Q7Z2W4 | ZC3HAV1 | Zinc finger CCCH-type antiviral protein 1 | 0.6468 | 0.1397 | 0.0465 | 0.0254 |
| Q9Y5M8 | SRPRB | Signal recognition particle receptor subunit beta | 0.645 | 0.0983 | 0.1459 | 0.0486 |
| P62191 | PSMC1 | 26S proteasome regulatory subunit 4 | 0.6447 | 0.1717 | 0.0328 | 0.0243 |
| P43251 | BTD | Biotinidase (Biotinase) | 0.6273 | 0.1495 | 0.0734 | 0.0392 |
| P08648 | ITGA5 | Integrin alpha-5 | 0.6102 | 0.0891 | 0.1285 | 0.0398 |
| Q07955 | SRSF1 | Serine/arginine-rich splicing factor 1 | 0.6084 | 0.0123 | 0.0315 | 0.0018 |
| P18754 | RCC1 | Regulator of chromosome condensation | 0.5984 | 0.1047 | 0.008 | 0.0048 |
| P62140 | PPP1CB | Serine/threonine-protein phosphatase PP1-beta catalytic subunit | -0.6001 | 0.1545 | 0.0111 | 0.0097 |
| Q9NX40 | OCIAD1 | OCIA domain-containing protein 1 | -0.601 | 0.0912 | 0.1301 | 0.041 |
| Q14244 | MAP7 | Microtubule-associated protein 7 | -0.6115 | 0.0198 | 0.1806 | 0.0178 |
| Q9Y572 | RIPK3 | Receptor-interacting serine/threonine-protein kinase 3 | -0.6157 | 0.0717 | 0.0554 | 0.0153 |
| Q06187 | BTK | Bruton tyrosine kinase | -0.6434 | 0.2767 | 0.0165 | 0.027 |
| Q8IW35 | CEP97 | Centrosomal protein of 97 kDa | -0.6669 | 0.0989 | 0.1153 | 0.0394 |
| Q969G5 | CAVIN3 | Caveolae-associated protein 3 | -0.6891 | 0.0308 | 0.0337 | 0.0045 |
| O96033 | MOCS2 | Molybdopterin synthase sulfur carrier subunit | -0.6914 | 0.0731 | 0.0987 | 0.0262 |
| P13798 | APEH | Acyl-peptide hydrolase | -0.6929 | 0.0508 | 0.239 | 0.0485 |
| Q9H3H3 | C11orf68 | UPF0696 protein C11orf68 | -0.7029 | 0.0819 | 0.1651 | 0.0471 |
| Q08431 | MFGE8 | Milk fat globule-EGF factor 8 | -0.7468 | 0.1464 | 0.0932 | 0.0466 |
| Q9NYJ8 | TAB2 | TGF-beta-activated kinase 1 | -0.7572 | 0.1619 | 0.0095 | 0.0092 |
| O95218 | ZRANB2 | Zinc finger Ran-binding domain-containing protein 2 | -0.8387 | 0.1273 | 0.054 | 0.0261 |
| Q93074 | MED12 | Mediator of RNA polymerase II transcription subunit 12 | -0.91 | 0.0239 | 0.2422 | 0.0291 |
| Q15047 | SETDB1 | Histone-lysine N-methyltransferase SETDB1 | -0.9105 | 0.2702 | 0.0382 | 0.0459 |
| P33241 | LSP1 | Lymphocyte-specific protein 1 | -0.9375 | 0.0021 | 0.242 | 0.0058 |
| Q9H3M7 | TXNIP | Thioredoxin-interacting protein | -0.9387 | 0.0348 | 0.086 | 0.0123 |
| P22307 | SCP2 | Sterol carrier protein X | -1.0061 | 0.1877 | 0.0682 | 0.0465 |
| Q96A49 | SYAP1 | Synapse-associated protein 1 | -1.0203 | 0.0638 | 0.0634 | 0.0155 |
| Q5T1M5 | FKBP15 | FK506-binding protein 15 | -1.0953 | 0.0796 | 0.135 | 0.0379 |
| Q9NR77 | PXMP2 | Peroxisomal membrane protein 2 | -1.1041 | 0.397 | 0.0075 | 0.0284 |
| Q6P4R8 | NFRKB | Nuclear factor related to kappa-B-binding protein | -1.131 | 0.0431 | 0.1973 | 0.0347 |
| A6ND91 | ASPDH | Aspartate dehydrogenase domain-containing protein | -1.2357 | 0.0503 | 0.2253 | 0.0451 |
| Q9Y320 | TMX2 | Thioredoxin-related transmembrane protein 2 | -1.3223 | 0.1468 | 0.0342 | 0.0211 |
| Q9BRK0 | REEP2 | Receptor expression-enhancing protein 2 | -1.3431 | 0.0303 | 0.0738 | 0.0094 |
| Q9UHK6 | AMACR | Alpha-methylacyl-CoA racemase | -1.3986 | 0.0022 | 0.2013 | 0.0046 |
| P49407 | ARRB1 | Beta-arrestin-1 (Arrestin beta-1) | -1.4064 | 0.1261 | 0.016 | 0.01 |
| Q92630 | DYRK2 | Dual specificity tyrosine-phosphorylation-regulated kinase 2 | -1.4443 | 0.0139 | 0.0418 | 0.0027 |
| Q9BRU9 | UTP23 | rRNA-processing protein UTP23 homolog | -1.8162 | 0.0833 | 0.0247 | 0.009 |
| O14617 | AP3D1 | AP-3 complex subunit delta-1 | -1.9901 | 0.0471 | 0.0127 | 0.0029 |
| Q9C073 | FAM117A | Protein FAM117A (C/EBP-induced protein) | -3.5266 | 0.0051 | 0.308 | 0.015 |
Proteomics profiling revealing alendronate (ALN)-induced protein alteration in the global proteome of MG-63 bone cells. (A) Experimental mass spectrometry (MS) workflow for this study. (B) Top 10 most abundant protein classes. (C) Volcano plot shows DEPs. (D) Heatmap depicts the differential expression patterns of proteins in response to ALN. Red and blue dots represent upregulated and downregulated proteins, respectively. Per row z-score of protein intensity is calculated. Each dot represents one protein. Proteins used are identical with those in the volcano plot. Experiments were done in triplicate.
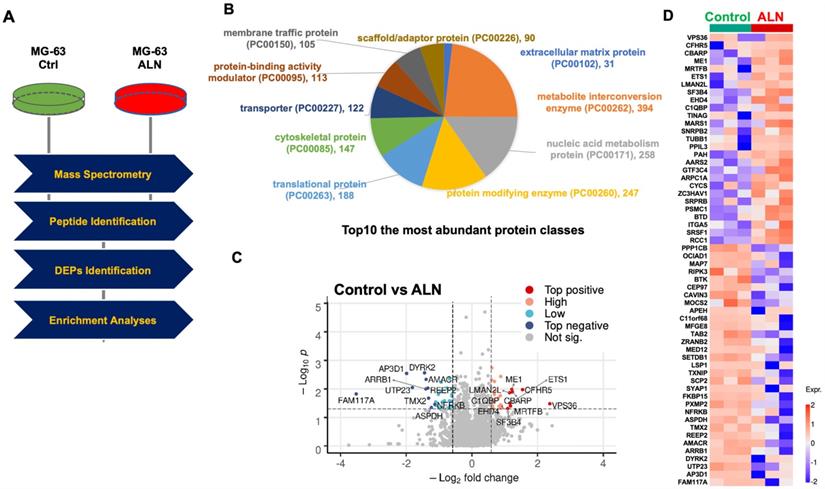
Angiogenic pathways may be upregulated by ALN treatment. (A) Secretion of VEGF in MG-63 bone cells treated with ALN. Effect of ALN treatment on the secreted VEGF levels into conditioned medium by MG-63 cells. Values (mean and standard deviation (SD)) are expressed as fold-changes compared to untreated cells (Ctrl, control). (B) Proliferation of HUVEC in the collected media of MG-63 cells. **p < 0.001, compared to control (Student's t-test). (C-D) No apoptosis was observed within the treatment period of 6 h. (C). Cell viability of MG-63 cells. MTT assay revealed no viability changes by ALN treatment. (D) Crystal violet staining assay showed no cell mass changes in response to varying concentration of ALN for 6 days. Experiments were done 6 times. Representative images were shown.
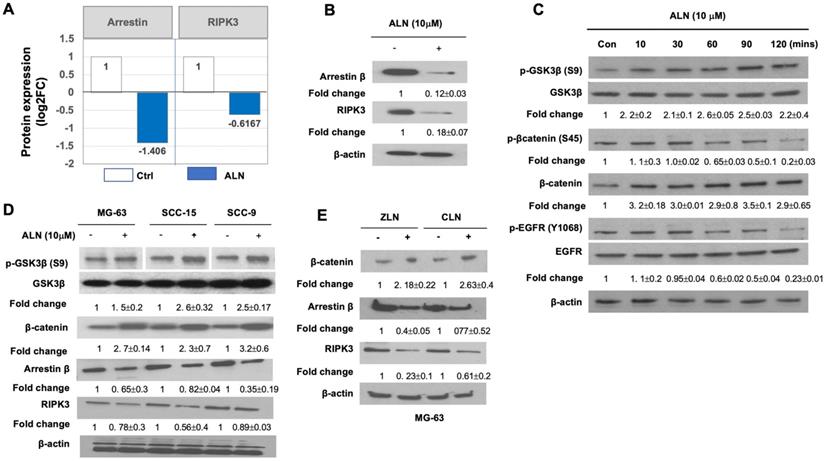
The RIPK3/arrestin/GSK3β/ β-catenin/VEGF pathway is altered by ALN treatment. (A-B). Quantification results showed that arrestin β and RIPK3 are significantly suppressed with ALN treatment. (A) Data from proteomics profiling. DEP levels obtained from proteomics analysis are shown in Table 1. (B) Western blot analysis to measure the expression levels of arrestin β and RIPK3 proteins in the presence or absence of ALN. β-actin was used as the loading control. (C) ALN-induced phosphorylation of GSK3β (S9) and β-catenin (S45) led to stabilization of β-catenin in MG-63 cells. (D) Comparison of phosphorylation of GSK3β and expression of β-catenin, arrestin β, and RIPK3 in MG-63, SCC-15, and SCC-9 cells after treatment with ALN. (E) Effects of several BPs (ZLN and CLN) on β-catenin, arrestin β, and RIPK3 in MG-63 cells. After stimulation with 10 μM of ALN, ZLN, or CLN at various times, cells were harvested for protein extraction and western blot analysis. Representative western blot images were selected after experiments were repeated 6 times.
Receptor-interacting protein kinase 3 (RIPK3), a necroptosis factor, is altered in the ALN-treated proteome
Among the DEPs regulated by ANL treatment, proteins involved in angiogenesis, inflammation, and necrosis were of particular interest due to their relevance in ONJ. Proteomics profiling revealed downregulation of RIPK3 in MG63 cells treated with ANL (Fig 3A). RIPK3 has recently been reported as a mediator of necroptosis, programmed non-apoptotic cell death, and necroinflammation in response to immune signaling and cytokines, such as TNF-α [30]. The inhibition of RIPK3 activity suppressed Enterococcus faecalis infection-induced cell death in MG-63 cells[31]. RIPK3 expression is inhibited by hypoxia, which contributes to angiogenesis [32]. Loss of RIPK3 leads to the activation of the Wnt/β-catenin signaling pathway in the ripk3-/- colon cancer mouse model, and enhances inflammation, immune cell infiltration, and angiogenesis [33].
Western blot analysis was able to validate that the protein expression levels of arrestin β1 (ARRB1) was significantly diminished by ALN treatment (Fig 3B), which was consistent with proteomics analysis. Given that ARRB1 is reported as a necessary component for Wnt/β-catenin signaling and as a regulator of GSK-3β activation/inactivation [34], the effects of ALN and ARRB1 on the Wnt/GSK3/β-catenin signaling cascades were another point of interest. Proteomics profiling and biochemical analysis revealed the downregulation of RIPK3 and ARRB1 by ALN treatment, which suggests that the effects of ALN on MG-63 cells are likely to be mediated by the Wnt/GSK3/β-catenin signaling pathway.
The glycogen synthase kinase 3 (GSK3) network is an ALN regulatory signaling pathway
To understand the activation of signaling cascades in response to BP treatment in bone cells, the phosphorylation of important signaling proteins in MG-63 cells treated with ALN was assessed. The involvement of Wnt/GSK3/β-catenin signaling aberration was first determined, and the downstream secreted effectors of the Wnt pathway were evaluated as a part of the ALN signaling pathway.
Based on previous findings in literature, the Wnt/GSK3/β-catenin pathway has been shown to play a pivotal role in bone remodeling/repair processes, enhancement of osteoblast differentiation, angiogenesis, and modulation of immune cell functions[2]. This evaluation further suggests that the Wnt/GSK3/β-catenin pathway may play a key role in the biological effects of response to ALN treatment in MG-63 cells.
After treatment with ALN at varying incubation times (0, 20, 30, 60, 90, and 120 min), the phospohorylation status of a series of crucial signaling molecules was evaluated using western blot analysis. The phospohorylation of GSK-3β (S9) increased with ALN treatment (Fig 3C). GSK-3, a serine/threonine protein kinase that phosphorylates and inactivates glycogen synthase, is a key downstream regulator of the PI3K/Akt pathway. GSK-3 signaling is inactivated by phosphorylation of Ser9 in GSK-3β. Since the phospohorylation of GSK-3β (S9) increased, this suggests that ALN treatment inactivates GSK-3 signaling in MG-63 cells.
As an important downstream effector of the Wnt signaling pathway, β-catenin is phosphorylated at S45 by a complex of axin and casein kinase I (CKI), which initiates the β-catenin phosphorylation-degradation cascade [35]. While the phospohorylation of GSK-3β (S9) increased with ALN treatment, phosphorylation of β-catenin (S45) and EGFR (Y1068) decreased (Fig 3C). The decreased phosphorylation of β-catenin may increase protein stability and protein expression (Fig 3B). Increased phosphorylation of GSK-3β (S9) was consistently observed in other cells, including SCC-9 and SCC-15, with ALN, zoledronic acid (ZLN), or clodronate (CLN) treatment (Fig 3D and Fig 3E). These results suggest that ANL suppresses ARRB1, inactivates GSK-3β, and stabilizes β-catenin. The RIPK3/arrestin/Wnt/GSK/β-catenin network may be a potential molecular regulatory network whose activation is altered upon ALN therapy.
Cytokine production and secretion in RAW 264.7 macrophages may be enhanced by ALN treatment
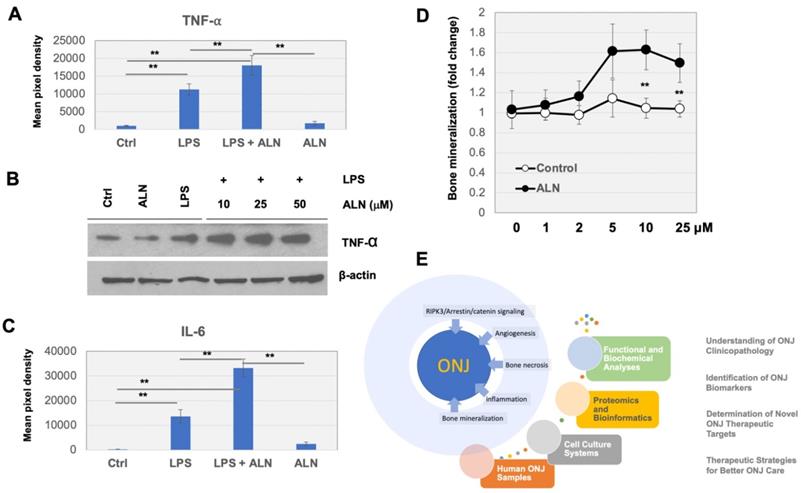
To test the effects of ALN on the immune system, a commercially available cytokine array was used to screen for potentially stimulated cytokines. RAW 264.7 macrophages were incubated with ALN both with and without the presence of lipopolysaccharides (LPS) (100 ng/ml) for 24 h. As shown in figure 4A, the production of tumor necrosis factor alpha (TNF-α) was stimulated by LPS and the levels of TNF-α were significantly increased with ALN. Western blot analysis also supported these findings (Fig 4B). The secretion of IL-6 also greatly increased with ALN (Fig 4C). However, there were no dramatic additional effects across other cytokines.
Pro-inflammatory cytokines are produced and secreted in response to ALN treatment in RAW 264.7 macrophage cells. (A-B) Cytokine array was conducted as described in Materials and Methods. Production of TNF-α (A) and secretion of IL-6 (B) increased with ALN treatment. (C) Western blot analysis for further validation. ***p < 0.001 and **p < 0.001, compared to control (Student's t-test). Representative images are shown. (D) ALN treatment impaired homeostasis in bone mineralization. Quantification of mineral deposition by Alizarin Red-S staining shown as a graph. Data represent average±SD (n= 6). Statistical analysis was compared between ALN and vehicle only (ctrl) (p-value<0.05).
Abnormalities in calcium phosphate formation in bone cells and bone mineral density (BMD) distribution in ONJ-associated osteonecrosis
ALN is regularly used to help osteoporosis patients with bone mineralization loss. To test the effects of ANL on the quantification of mineral deposition, Alizarin Red-S staining assays were used to further assess mineralization levels after treatment. MG-63 cells were incubated with ALN or vehicle control (0, 1, 5, 10, and 25 μM) for 2 days. Incubation of cells with ALN led to a marked increase in mineralization (to ~1.6 fold) compared to controls (Fig 4D).
Discussion
Our proteomics profiling revealed the downregulation of RIPK3 in response to ALN treatment in MG-63 bone cells. RIPK3 has been reported to play a fundamental role in inhibiting inflammation and mediating necroptosis and necroinflammation through the RIPK3-MLKL (mixed lineage kinase domain-like protein) pathway [30]. Inhibitors of RIPK3 and MLKL suppressed cell death from Enterococcus faecalis infection in MG-63 cells[31]. Although not encompassed in the current study, the role, and mechanisms of RIPK3 and its downstream signaling cascades in ALN-induced bone biology are under further investigation by our group. In addition, this study showed that the presence of ALN enhanced production or secretion of inflammatory cytokines in LPS-activated macrophage cells. A previous study found that ZLN, a potent BP, stimulated and increased inflammatory osteoclastic mediators [36]. Furthermore, ZLN was found to suppress proliferation and migration of vascular endothelial cells [37]. Expression of VEGF receptor 2 in vascular endothelial cells was also reported in response to treatment with ZLN[38]. In our experimental system, we observed modest decreases in VEGF secretion in response to ALN treatment.
The experimental data further suggested the potential role of the Wnt/GSK3/β-catenin signaling pathway in the BP-perturbated proteome and its effects on bone homeostasis. This study demonstrated that the Wnt/GSK3/β-catenin signaling pathways may play a fundamental role in bone metabolism, homeostasis, and remodeling. Multifaceted roles of GSK3 under each cellular context have been reported. In cytotoxic T lymphocytes (CTL), GSK3 inhibition blocks programmed cell death protein-1 (PD-1) transcription; thereby, enhancing CTL functioning [39]. GSK3 is a serine/threonine kinase that regulates Wnt/β-catenin, PI3K/PTEN/AKT, RAS/RAF/MAPK, hedgehog, Notch, and other signaling pathways and has been implicated in multiple diseases [40, 41]. Phosphorylation of GSK-3α/β at multiple serine and threonine sites inactivates the kinase, while Tyr279/216 phosphorylation (pY) activates the kinase. GSK3 is reported to have both tumor promoting (glioblastoma, pancreatic, ovarian, and blood cancers) and tumor suppressive (breast and skin cancers) roles[42]. GSK3 stabilizes anti-apoptotic Bcl2, Bcl2L12A, c-Myb, Mcl-1, and VEGF, promoting tumors. On the other hand, GSK3 phosphorylates and destabilizes β-catenin leading to the downregulation of c-Myc and cyclin D1. GSK3 also phosphorylates T286 on cyclin D1, leading to its nuclear export and degradation[43]. Consistent with this study, previous findings have suggested an important role for the Wnt/GSK-3 signaling pathway in osteogenesis; inhibition of Wnt/GSK-3 activity induced osteoblast differentiation and significantly increased BMD in an ovariectomized rat model [44].
Experimental observation from this study suggests that a systematic overview of changes in the microenvironmental landscape is important for understanding ALN-induced pathophysiology in bone cells (Fig 4E). Treatment with ALN also leads to alterations in bone mineralization, which may further impair bone biology. In ONJ patients, our previous studies quantifying bone density and mineralization found that cone-beam computed tomography (CBCT) and micro-computed tomography image-based histomorphometric evaluation may be an efficient method to check bone health[45]. Abnormal BMD distribution in ONJ-associated osteonecrosis was observed by clinical CBCT imaging[46]. It would be worthwhile to determine if the patterns and severity of abnormal mineralization densities within jaw-bone biopsy samples can be implemented in ONJ patient care.
Collectively, the main innovative deliverables from this study are expected to lead to a better understanding of the mechanisms underlying ALN-induced pathological effects on bone and immune cells. The findings in this paper are promising but have several limitations; (1) the effects of BPs on osteoblast function are throughout the skeleton, and (2) ALN targets osteoclasts, not osteoblasts. In conjunction with standard diagnostic procedures, the more mechanistic data related to the adverse effects of ALN can also act as an applicable supplement for clinical judgment.
Supplementary Material
Supplementary methods.
Acknowledgements
This research was supported by the Samuel Oschin Comprehensive Cancer Institute (SOCCI) at Cedars-Sinai Medical Center through 2019 Lucy S. Gonda Award. We appreciate technical support from the Cedars-Sinai Proteomics and Metabolomics Core.
Kim J: Contributed to conception or design, drafted the manuscript, and critically revised the manuscript.
Yeon A: Contributed to analysis, drafted the manuscript, and critically revised the manuscript.
Parker SJ: Contributed to analysis, drafted the manuscript, and critically revised the manuscript.
Shahid M: Contributed to analysis, and critically revised the manuscript.
Thiombane A: Contributed to analysis, and critically revised the manuscript.
Cho E: Contributed to analysis, and critically revised the manuscript.
You S: Contributed to analysis, drafted the manuscript, and critically revised the manuscript.
Emam H: Contributed to analysis, drafted the manuscript, and critically revised the manuscript.
Kim D-G: Contributed to conception or design, drafted the manuscript, and critically revised the manuscript.
Kim M: contributed to conception or design, drafted the manuscript, and critically revised the manuscript.
Funding
This research was funded by National Institutes of Health (1U01DK103260, 1R01DK100974, U24 DK097154, NIH NCATS UCLA CTSI UL1TR000124), Department of Defense (W81XWH-15-1-0415 and W81XWH-19-1-0109), Centers for Disease Controls and Prevention (1U01DP006079), and the U.S.-Egypt Science and Technology Development Fund by the National Academies of Sciences, Engineering, and Medicine (all to J.K.). This article is derived from the Subject Data funded in whole or part by National Academies of Sciences, Engineering, and Medicine (NAS) and The United States Agency for International Development (USAID). Any opinions, findings, conclusions, or recommendations expressed in this article are those of the authors alone, and do not necessarily reflect the views of USAID or NAS.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Florencio-Silva R, Sasso GR, Sasso-Cerri E, Simoes MJ, Cerri PS. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. Biomed Res Int. 2015;2015:421746
2. Issack PS, Helfet DL, Lane JM. Role of Wnt signaling in bone remodeling and repair. HSS J. 2008;4:66-70
3. Bertacchini J, Magaro MS, Poti F, Palumbo C. Osteocytes Specific GSK3 Inhibition Affects In Vitro Osteogenic Differentiation. Biomedicines. 2018 6
4. Schoeman MA, Moester MJ, Oostlander AE, Kaijzel EL, Valstar ER, Nelissen RG. et al. Inhibition of GSK3beta Stimulates BMP Signaling and Decreases SOST Expression Which Results in Enhanced Osteoblast Differentiation. J Cell Biochem. 2015;116:2938-46
5. Huh JE, Ko R, Jung HJ, Lee SY. Glycogen synthase kinase 3beta promotes osteogenic differentiation of murine adipose-derived stromal cells. PloS one. 2013;8:e54551
6. Pabst AM, Kruger M, Blatt S, Ziebart T, Rahimi-Nedjat R, Goetze E. et al. Angiogenesis in the Development of Medication-Related Osteonecrosis of the Jaws: An Overview. Dent J (Basel). 2016 5
7. Zhang Z, Nor F, Oh M, Cucco C, Shi S, Nor JE. Wnt/beta-Catenin Signaling Determines the Vasculogenic Fate of Postnatal Mesenchymal Stem Cells. Stem Cells. 2016;34:1576-87
8. Skurk C, Maatz H, Rocnik E, Bialik A, Force T, Walsh K. Glycogen-Synthase Kinase3beta/beta-catenin axis promotes angiogenesis through activation of vascular endothelial growth factor signaling in endothelial cells. Circ Res. 2005;96:308-18
9. Bellido T, Plotkin LI. Novel actions of bisphosphonates in bone: preservation of osteoblast and osteocyte viability. Bone. 2011;49:50-5
10. Plotkin LI, Manolagas SC, Bellido T. Dissociation of the pro-apoptotic effects of bisphosphonates on osteoclasts from their anti-apoptotic effects on osteoblasts/osteocytes with novel analogs. Bone. 2006;39:443-52
11. Plotkin LI, Weinstein RS, Parfitt AM, Roberson PK, Manolagas SC, Bellido T. Prevention of osteocyte and osteoblast apoptosis by bisphosphonates and calcitonin. J Clin Invest. 1999;104:1363-74
12. Wilkins Parker LR, Preuss CV. Alendronate. StatPearls. Treasure Island (FL). 2021
13. Weinstein RS, Roberson PK, Manolagas SC. Giant osteoclast formation and long-term oral bisphosphonate therapy. N Engl J Med. 2009;360:53-62
14. Rouach V, Goldshtein I, Wolf I, Catane R, Chodick G, Iton A. et al. Exposure to alendronate is associated with a lower risk of bone metastases in osteoporotic women with early breast cancer. J Bone Oncol. 2018;12:91-5
15. Jackson C, Freeman ALJ, Szlamka Z, Spiegelhalter DJ. The adverse effects of bisphosphonates in breast cancer: A systematic review and network meta-analysis. PloS one. 2021;16:e0246441
16. Dodson TB. The Frequency of Medication-related Osteonecrosis of the Jaw and its Associated Risk Factors. Oral Maxillofac Surg Clin North Am. 2015;27:509-16
17. George EL, Lin YL, Saunders MM. Bisphosphonate-related osteonecrosis of the jaw: a mechanobiology perspective. Bone Rep. 2018;8:104-9
18. Zymperdikas VF, Yavropoulou MP, Kaklamanos EG, Papadopoulos MA. Bisphosphonates as Supplement to Dental Treatment: A Network Meta-Analysis. J Dent Res. 2021;100:341-51
19. Khan A, Morrison A, Cheung A, Hashem W, Compston J. Osteonecrosis of the jaw (ONJ): diagnosis and management in 2015. Osteoporos Int. 2016;27:853-9
20. Fung P, Bedogni G, Bedogni A, Petrie A, Porter S, Campisi G. et al. Time to onset of bisphosphonate-related osteonecrosis of the jaws: a multicentre retrospective cohort study. Oral Dis. 2017;23:477-83
21. Parker SJ, Venkatraman V, Van Eyk JE. Effect of peptide assay library size and composition in targeted data-independent acquisition-MS analyses. Proteomics. 2016;16:2221-37
22. Parker SJ, Stotland A, MacFarlane E, Wilson N, Orosco A, Venkatraman V. et al. Proteomics reveals Rictor as a noncanonical TGF-beta signaling target during aneurysm progression in Marfan mice. Am J Physiol Heart Circ Physiol. 2018;315:H1112-H26
23. Rost HL, Rosenberger G, Navarro P, Gillet L, Miladinovic SM, Schubert OT. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219-23
24. Rosenberger G, Koh CC, Guo T, Rost HL, Kouvonen P, Collins BC. et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci Data. 2014;1:140031
25. Teleman J, Rost HL, Rosenberger G, Schmitt U, Malmstrom L, Malmstrom J. et al. DIANA-algorithmic improvements for analysis of data-independent acquisition MS data. Bioinformatics. 2015;31:555-62
26. Rost HL, Liu Y, D'Agostino G, Zanella M, Navarro P, Rosenberger G. et al. TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat Methods. 2016;13:777-83
27. Teo G, Kim S, Tsou CC, Collins B, Gingras AC, Nesvizhskii AI. et al. mapDIA: Preprocessing and statistical analysis of quantitative proteomics data from data independent acquisition mass spectrometry. J Proteomics. 2015;129:108-20
28. Mi H, Ebert D, Muruganujan A, Mills C, Albou LP, Mushayamaha T. et al. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021;49:D394-D403
29. Ishtiaq S, Edwards S, Sankaralingam A, Evans BA, Elford C, Frost ML. et al. The effect of nitrogen containing bisphosphonates, zoledronate and alendronate, on the production of pro-angiogenic factors by osteoblastic cells. Cytokine. 2015;71:154-60
30. Chen H, Fang Y, Wu J, Chen H, Zou Z, Zhang X. et al. RIPK3-MLKL-mediated necroinflammation contributes to AKI progression to CKD. Cell Death Dis. 2018;9:878
31. Dai X, Deng Z, Liang Y, Chen L, Jiang W, Zhao W. Enterococcus faecalis induces necroptosis in human osteoblastic MG63 cells through the RIPK3 / MLKL signalling pathway. Int Endod J. 2020;53:1204-15
32. Moriwaki K, Bertin J, Gough PJ, Orlowski GM, Chan FK. Differential roles of RIPK1 and RIPK3 in TNF-induced necroptosis and chemotherapeutic agent-induced cell death. Cell Death Dis. 2015;6:e1636
33. Bozec D, Iuga AC, Roda G, Dahan S, Yeretssian G. Critical function of the necroptosis adaptor RIPK3 in protecting from intestinal tumorigenesis. Oncotarget. 2016;7:46384-400
34. Li H, Sun X, LeSage G, Zhang Y, Liang Z, Chen J. et al. beta-arrestin 2 regulates Toll-like receptor 4-mediated apoptotic signalling through glycogen synthase kinase-3beta. Immunology. 2010;130:556-63
35. Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M. et al. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066-76
36. Tamari T, Elimelech R, Cohen G, Cohen T, Doppelt O, Eskander-Hashoul L. et al. Endothelial Progenitor Cells inhibit jaw osteonecrosis in a rat model: A major adverse effect of bisphosphonate therapy. Sci Rep. 2019;9:18896
37. Lang M, Zhou Z, Shi L, Niu J, Xu S, Lin W. et al. Influence of zoledronic acid on proliferation, migration, and apoptosis of vascular endothelial cells. Br J Oral Maxillofac Surg. 2016;54:889-93
38. Basi DL, Lee SW, Helfman S, Mariash A, Lunos SA. Accumulation of VEGFR2 in zoledronic acid-treated endothelial cells. Mol Med Rep. 2010;3:399-403
39. Taylor A, Harker JA, Chanthong K, Stevenson PG, Zuniga EI, Rudd CE. Glycogen Synthase Kinase 3 Inactivation Drives T-bet-Mediated Downregulation of Co-receptor PD-1 to Enhance CD8(+) Cytolytic T Cell Responses. Immunity. 2016;44:274-86
40. McCubrey JA, Steelman LS, Bertrand FE, Davis NM, Abrams SL, Montalto G. et al. Multifaceted roles of GSK-3 and Wnt/beta-catenin in hematopoiesis and leukemogenesis: opportunities for therapeutic intervention. Leukemia. 2014;28:15-33
41. McCubrey JA, Steelman LS, Bertrand FE, Davis NM, Sokolosky M, Abrams SL. et al. GSK-3 as potential target for therapeutic intervention in cancer. Oncotarget. 2014;5:2881-911
42. Beurel E, Jope RS. The paradoxical pro- and anti-apoptotic actions of GSK3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog Neurobiol. 2006;79:173-89
43. Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24
44. Kulkarni NH, Onyia JE, Zeng Q, Tian X, Liu M, Halladay DL. et al. Orally bioavailable GSK-3alpha/beta dual inhibitor increases markers of cellular differentiation in vitro and bone mass in vivo. J Bone Miner Res. 2006;21:910-20
45. Kim DG. Can dental cone beam computed tomography assess bone mineral density? Journal of bone metabolism. 2014;21:117-26
46. Nye SN CE, Emam H, Kim D-G. Heterogeneity of Bone Mineral Density in Osteonecrosis of the Jaw. 2018 AADR/CADR Annual Meeting (Fort Lauderdale, Florida). 2018
Author contact
![]() Corresponding author: Jayoung Kim, PhD. Departments of Surgery and Biomedical Sciences, Cedars-Sinai Medical Center, Davis 5071, 8700 Beverly Blvd., Los Angeles, CA 90048. Tel: +1-310-423-7168; Fax: +1-310-967-3809; E-mail: Jayoung.kimedu
Corresponding author: Jayoung Kim, PhD. Departments of Surgery and Biomedical Sciences, Cedars-Sinai Medical Center, Davis 5071, 8700 Beverly Blvd., Los Angeles, CA 90048. Tel: +1-310-423-7168; Fax: +1-310-967-3809; E-mail: Jayoung.kimedu

 Global reach, higher impact
Global reach, higher impact