Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(8):1877-1885. doi:10.7150/ijms.52177 This issue Cite
Research Paper
Hydroxyurea regulates the development and survival of B16 Melanoma Cells by upregulating MiR-7013-3p
Department of Laboratory Animals, Jilin Provincial Key Laboratory of Animal Model, Jilin University, Changchun 130062, Jilin, P.R. China.
Received 2020-8-19; Accepted 2020-12-18; Published 2021-3-3
Abstract
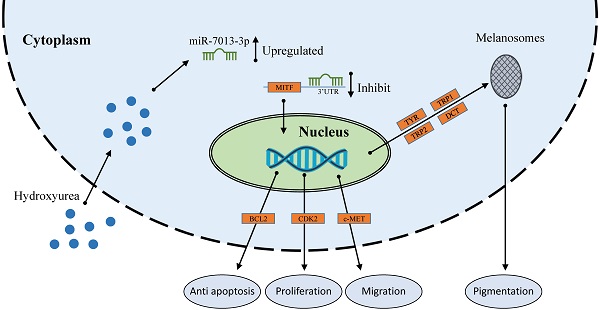
miRNAs are a family of short, noncoding RNAs that are involved in many processes in melanoma cells. MITF acts as a master regulator of melanocyte function, development and survival by modulating various genes. Hydroxyurea (HU) is used to treat melanoma, and miRNA expression is altered after HU treatment in B16 melanoma cells. In this study, we screened for miRNAs that were upregulated after HU treatment and that targeted the MITF gene. We found that miR-7013-3p exhibited increased expression after HU treatment and could bind to MITF. miR-7013-3p inhibited melanin production, proliferation, and migration and promoted apoptosis in B16 melanoma cells. The results may provide more information on the roles of miR-7013-3p in B16 melanoma cells.
Keywords: miR-7013-3p, B16 melanoma cells, MITF, hydroxyurea (HU), pigmentation
Introduction
Melanin prevents the skin, eyes and brain from being damaged by ultraviolet radiation [1-3]. In addition, melanin in the inner ear affects sound conduction [4]. However, melanin is aberrantly regulated in human skin disorders such as vitiligo and melisma [5, 6]. The pigment is produced by melanocytes through melanosomes and is transferred within melanocyte dendrites to adjacent keratinocytes [1]. Melanoma, which stems from melanocytes, is a malignant cancer of the skin and has a tendency to metastasize to distant locations [7-9]. This kind of skin cancer develops through abnormal hyperplasia of melanocytes and is characterized by rapid proliferation, resistance to apoptosis, unlimited replication, and significant increases in melanin content over time [10, 11]. Although the incidences of many cancer types are decreasing, the melanoma incidence continues to increase; in addition, the incidence and mortality of melanoma are higher than those of other cancers of the skin [12]. More than 1 million Americans currently suffer from melanoma according to current data from the American Academy of Dermatology (AAD) [13]. All of this information suggests that more effective treatments for melanoma are urgently needed.
Microphthalmia-associated transcription factor (MITF) acts as a master regulator of melanocyte function, development and survival by modulating various differentiation-related, cell cycle progression-related and antiapoptotic genes and can act as an oncogene in melanoma [14-17]. MITF can also activate the transcription of pigmentation-related genes, including tyrosinase (TYR), TYR-related protein 1 (Tyrp1), TYR-related protein 2 (Tyrp2), dopachrome tautomerase (DCT), and others [16, 18]. MITF directly regulates the gene expression of the antiapoptotic factor B-cell lymphoma 2 (BCL2). Constitutive overexpression of BCL2 partially attenuates MITF deletion-induced apoptosis of primary melanoma cells and melanoma [13, 19, 20]. MITF can regulate cell cycle progression by modulating cyclin-dependent kinase-2 (CDK2), which is important for melanoma clonogenic growth. Furthermore, MITF can regulate cell proliferation mediated by CDK2. In normal skin, cellular mesenchymal-epithelial transition factor (c-Met) is present on epithelial cells and melanocytes [21, 22]. The c-Met receptor tyrosine kinase is a multifaceted regulator of growth, motility, and invasion in a number of lineages in vivo. c-Met is also one of the MITF target genes [22, 23]. Overexpression of c-Met correlates with the invasive growth phase of melanoma cells [24], and knockdown of MITF can inhibit the invasion of melanoma cells by affecting the expression of c-met [19, 25, 26]. These findings indicate that MITF is a potential target gene for melanoma therapy [15, 20, 27-29].
Hydroxyurea (HU) is a metabolic inhibitor of ribonucleotide reductase and an anticancer drug that inhibits the synthesis of DNA [30, 31]. It works by scavenging free radicals of tyrosine to inhibit the production of deoxyribonucleotide, thereby reducing the production of ribonucleotide reductase [31, 32]. HU has been previously reported as a treatment for melanoma [33, 34]. In addition, miRNAs are a family of short, noncoding RNAs that can be cleaved by the DICER enzyme to form mature miRNAs after forming precursor RNA molecules in the nucleus. HU treatment can cause miRNA expression changes in cells [35, 36]. However, there have been no relevant experiments to verify the expression levels of miRNAs in melanoma cells treated with HU.
We identified miRNAs that may bind to MITF with TargetScan software, which predicted that four miRNAs may regulate MITF gene expression. We then chose the miRNAs upregulated by HU treatment to study the roles of miRNAs in the treatment of melanoma. In this experiment, we expected to find influences of miRNAs on MITF gene expression and transcription under HU stimulation. In addition, we sought to determine whether miRNAs affect melanogenesis, proliferation, apoptosis, and migration in B16 cells by regulating MITF gene expression.
Methods
Materials
HU with over 98% purity was obtained from Sigma and dissolved in dimethyl sulfoxide (DMSO) to make a stock solution.
Cell culture
Murine B16 melanoma cells were obtained from the National Infrastructure of Cell Line Resource (China) and cultured with high-glucose DMEM containing 5% heat-inactivated FBS, 100 units/ml penicillin, and 100 µg/mL streptomycin in a humidified atmosphere with 5% CO2 at 37 °C.
RNA isolation and quantitative RT-PCR (qRT-PCR) detection
Total RNA was extracted by using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Next, cDNA was obtained with a FastQuant RT kit, and raw data were obtained by qRT-PCR with SuperReal PreMix (SYBR Green). GAPDH was used as an internal control for the mRNA qRT-PCR analysis, while the small nuclear U6 was used as an internal control for miRNA quantitation. The mRNA and miRNA primers used in these assays are listed in Table 1.
Primers used for RT-qPCR
| Primer name | Sequence (5'-3') |
|---|---|
| GAPDH F | TCATCCCTGCATCCACTGGT |
| GAPDH R | TGTCCCAAGTCACTGTCACAC |
| U6 RT | CGCTTCACGAATTTGCGTGTCAT |
| MITF-F | AGGACCTTGAAAACCGACAG |
| MITF-R | GTGGATGGGATAAGGGAAAG |
| DCT-F | TCTCCAGAAGTTTGACAGCCC |
| DCT-R | AGAGTCCAGTGTTCCGTCTG |
| TYRP1-F | TCGAAGCCTTCACAACCTGG |
| TYRP1-R | TCGTCAAAGACCGCATCAGT |
| TYRP2-F | TCATCTGAGCACCCCTGTCT |
| TYRP2-R | CGTGGAAACTGAGCCCAAAC |
| TYR-F | AGCCTGTGCCTCCTCTAA |
| TYR-R | AGGAACCTCTGCCTGAAA |
| miR-7013-3p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGAAG |
| miR-7013-3p-f | ACACTCCAGCTGGGCCACACTTACTGTTGCCTCT |
| c-MET-F | GCATGTCAGCATCGCTCAA |
| c-MET-R | TGCAGGCCCAGCTGTTTC |
| ur | CTCAAGTGTCGTGGAGTCGGCAA |
| BCL2-F | CTGTGGATGACTGAGTACCT |
| BCL2-R | AGCCAGGAGAAATCAAACAG |
| miR-124-3p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGCATTC |
| miR-25-3p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAGACCG |
| miR-32-5p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGCAACTT |
| miR-124-3p-F | ACACTCCAGCTGGGTTAAGGCACGCGGTGA |
| miR-25-3p-F | ACACTCCAGCTGGGCATTGCACTTGTCTCG |
| miR-32-5p-F | ACACTCCAGCTGGGTATTGCACATTACTAA |
| CDK2-R | GGGTCCATCAAGCTGGCAGA |
| CDK2-F | CCACAGGGTCACCACCTCAT |
Melanin content measurement
Melanin content was measured as previously described [16]. Briefly, 24 h after transfection, B16 melanoma cells were collected, washed with phosphate-buffered saline (PBS) three times, dissolved in 1 mL of 1 mM NaOH and incubated at 80 °C for 30 min. The total melanin content was measured at 475 nm and normalized to the total cell number. All experiments were performed in triplicate.
Detection of apoptosis by flow cytometry
Flow cytometry was used to detect B16 melanoma cell apoptosis in order to evaluate the effect of miR-7013-3p on cells. To analyze apoptosis, we performed an assay in strict accordance with the apoptosis assay kit instructions (Tianjin Sun Gene Biotech Co., Ltd, Tianjin, China). B16 cells were transfected with a mimic negative control (NC) and miR-7013-3p mimic for 24 h, the cells and culture medium were harvested, and the cells were digested with trypsin without EDTA. The cells were then centrifuged at 1000 r/min for 5 min and washed with ice-cold PBS; we repeated this step 3 times. Then, 5 µL of FITC solution and 5 µL of propidium iodide (PI) were added to each centrifuge tube. The cells were incubated for 15 min at room temperature in the dark. Finally, we analyzed apoptosis via flow cytometry within 2 h.
Cell cycle assay
Cells were incubated with PI for 15 min in the dark, and then the cell cycle was detected by flow cytometry.
miRNA mimic transfection and RNA interference
All miRNA mimics were purchased from Guangzhou RiboBio Biotech Co., Ltd. The siRNAs were synthetized by GenePharma (Shanghai, China). B16 cells were seeded at a density of 4×105 cells per well in a 6-well plate. Transfection was performed with a Lipofectamine 2000 Transfection Kit (Thermo Fisher Scientific, USA) according to the manufacturer's protocol. After transfection, the cells were incubated for 24 h for gene expression analysis. The siRNA sequences are shown in Table S1.
Wound-healing migration assay
A wound-healing assay was used to evaluate cell migration. When B16 melanoma cells grew to 80-90% confluence, 10 µL pipette tips were used to scratch a cell-free trace in the middle of the 6-well plate. After 24 h of incubation, the cell scratches were observed by optical microscopy. Finally, the average width of the scratches was calculated using Image-Pro Plus to determine the cell migration changes.
Proliferation assay
B16 melanoma cells were transfected with the mimic NC and miR-7013-3p mimic for 24 h. Then, cells were collected for the proliferation experiment and seeded in an E-plate at a density of 5000 cells/well. Proliferation was monitored every 15 min for a minimum of 18 h by recording the cell impedance produced as the cells attached and detached from the gold electrodes in the CIM and E-plates. Real-Time Cell Analysis (RTCA) software was used to generate a survival curve and estimate the cell survival or cell index (CI). The CI correlated directly with the cell number. The data are expressed in bar graphs as the CI % relative to the control.
Western blot analysis
Forty-eight hours after transfection, protein lysis buffer was used to lyse B16 melanoma cells. Next, the proteins were transferred to PVDF membranes for 1 h. After protein transfer, the PVDF membranes were blocked in Tris-buffered saline with Tween-20 (TBST) containing 5% bovine serum albumin (BSA) for 90 min and then incubated with rabbit polyclonal MITF IgG (ImmunoWay Biotechnology Company, Newark, DE, USA), rabbit polyclonal TYR IgG (Abcam, Cambridge, MA, USA) and rabbit polyclonal GAPDH (Cell Signaling Technology, USA) primary antibodies at 4 °C overnight before being washed 3 times with TBST for 15 min per wash. After being washed with TBST, the membrane were incubated with secondary antibodies for 55 min, after which the membranes were washed with TBST 3 times for 15 min each. The final protein bands were visualized with a chemiluminescent substrate, and protein quantification was performed with the ImageJ program.
Statistical analysis
All statistical analyses were performed using SPSS 19.0 software (IBM, SPSS, Chicago, IL, USA). Unpaired t tests were used for two-group comparisons, and all data are expressed as the mean ± SEM from three independent experiments. Comparisons among three or more groups was performed by independent-sample tests. Statistical significance was indicated by p values less than 0.05 (*p <0.05).
Results
Identification of differentially expressed miRNAs that bind to MITF after HU treatment
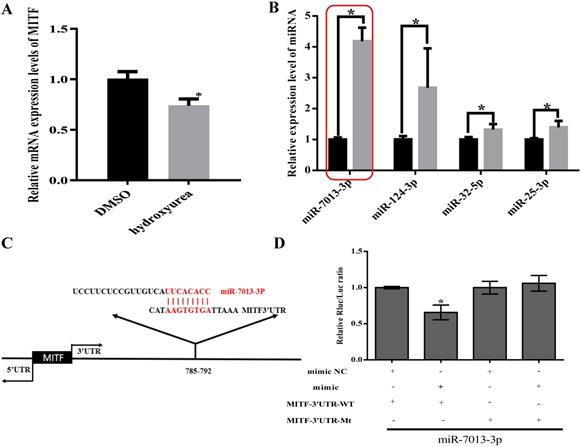
We used TargetScan to predict miRNAs that bind to MITF and measured MITF gene and miRNA expression levels after stimulating B16 cells with HU (400 μM) for 24 h [36, 37]. We found that MITF gene expression decreased, while miR-25-3p, miR-32-5p, miR-124-3p and miR-7013-3p expression increased (Fig. 1A-1B). We chose miR-7013-3p for further examination of the effect of this miRNA on B16 melanoma cells after treatment with HU. Information on the base complementary region between miR-7013-3p and the MITF 3'UTR was acquired via the TargetScan program (http://www.targetscan.org/) (Fig. 1C). Then, to further confirm that miR-7013-3p targets the MITF 3'UTR, we successfully mutated the target complementary sequence AAGTGTGA to TTCACACT (Table S1) and constructed an MITF 3'UTR wild-type (WT) plasmid and an Mitf-3'UTR mutated (MT) plasmid. Finally, 293T cells were cotransfected with the constructed plasmids and miR-7013-3p mimic. As we expected, cotransfection of the pmiR-Mitf-3'UTR WT plasmid and miR-7013-3p mimic into 293T cells reduced the luciferase activity (by 34%). However, cotransfection of cells with the Mitf-3'UTR MT plasmid and miR-7013-3p mimic did not reduce luciferase activity (Fig. 1D). Therefore, it can be concluded that miR-7013-3p can regulate MITF expression by directly targeting the MITF gene.
Identification of differentially expressed miRNAs after HU treatment that bind to MITF. After stimulation of B16 cells with HU (400 µM) for 24 h, MITF gene expression levels (A) and miRNA expression levels were altered (B). (C) Predictive maps of complementary targeting sites for miR-7013-3p and the MITF mRNA 3'UTR. (D) Luciferase activity after cotransfection with the MITF-3'UTR WT plasmid and miR-7013-3p mimic. The data summarize three independent experiments and are given as the mean±SD. *P<0.05.
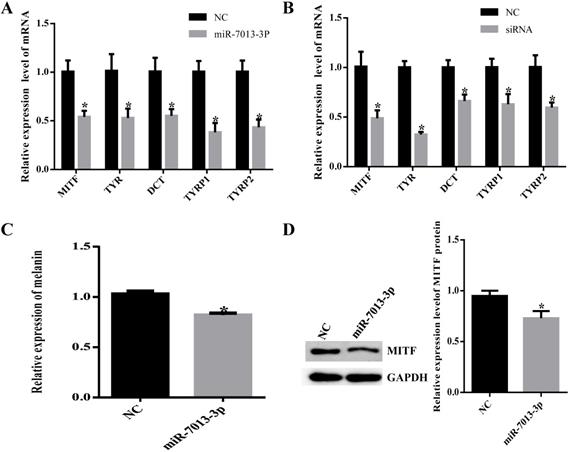
MiR-7013-3p inhibited pigmentation. (A) Relative MITF and pigmentation-related gene expression after transfection with the miR-7013-3p mimic. (B) Relative MITF and pigmentation-related gene expression after transfection with MITF siRNA. (C) Melanin levels in B16 melanoma cells transfected with the miR-7013-3p mimic and mimic NC. (D, E) Relative protein expression levels of MITF after transfection with the miRNA-7013-p mimic and mimic NC. The data summarize three independent experiments and are given as the mean±SD. *P<0.05.
MiR-7013-3p inhibited pigmentation
To detect the efficiency of transfection, we transfected B16 cells with mimic NC with fluorescent markers and subsequently detected red fluorescence in the cells, indicating that the mimic NC was successfully transfected into B16 cells. (Table S2). We examined MITF and pigmentation-related gene (TYR, DCT, TYRP1, TYRP2) mRNA levels after transfection of B16 cells with the miR-7013-3p mimic for 24 h to further verify that miR-7013-3p affects MITF and pigmentation-related gene expression and regulates melanin synthesis (Fig. 2A). MITF siRNA was also transfected as a positive control into B16 cells (Fig. 2B). As expected, MITF levels and pigmentation-related genes significantly decreased after transfection (P < 0.05). In addition, the expression levels of melanin after transfection with the miR-7013-3p mimic were lower (P < 0.05) than those after transfection with the mimic NC (Fig. 2C). Moreover, Western blot analysis showed that the protein expression levels of MITF in B16 melanoma cells transfected with the miR-7013-3p mimic were lower than those in cells transfected with the mimic NC (Fig. 2D). Taken together, the results suggest that overexpression of miR-7013-3p inhibits both the mRNA and protein expression of MITF.
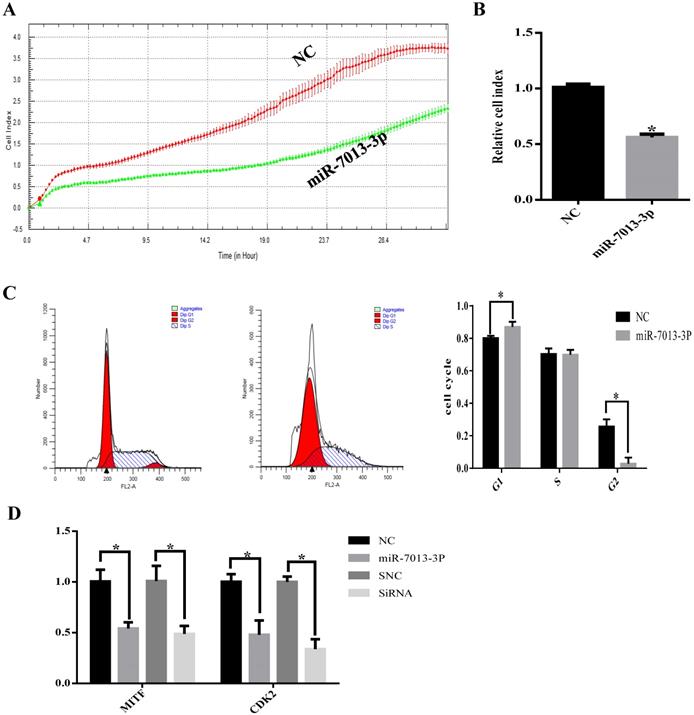
MiR-7013-3p inhibited proliferation in B16 melanoma cells
The RTCA results showed that transfection with the miR-7013-3p mimic inhibited B16 melanoma cell proliferation (Fig. 3A). To determine the possible intrinsic mechanism by which miR-7013-3p overexpression inhibits B16 melanoma cell proliferation, we detected the cell cycle phase distribution by flow cytometry with PI staining. We found that the compared with the control treatment, overexpression of miR-7013-3p in B16 melanoma cells induced G0/G1 phase accumulation (Fig. 3B). The qRT-PCR results showed that CDK2 mRNA expression levels decreased significantly after transfection with the miR-7013-3p mimic (MITF siRNA was used as a positive control) (Fig. 3C). The results also showed that overexpression of miR-7013-3p significantly reduced CDK2 gene expression and inhibited proliferation in B16 melanoma cells.
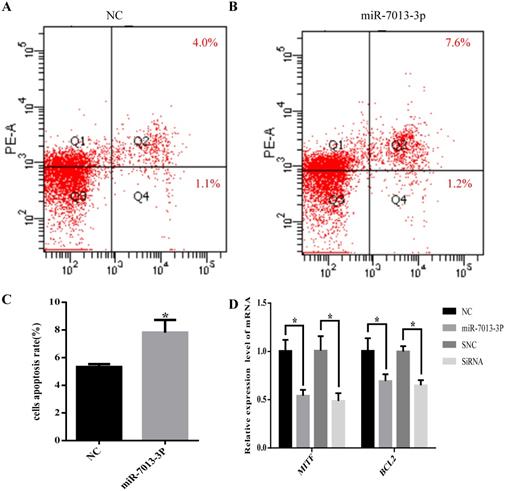
MiR-7013-3p promoted apoptosis in B16 melanoma cells
The flow cytometry results showed that the apoptotic rate of B16 melanoma cells was significantly increased after overexpression of miR-7013-3p (P <0.05, Fig. 4A-C). In addition, the qRT-PCR results showed that the BCL2 mRNA expression level was reduced significantly after transfection with the miR-7013-3p mimic; MITF siRNA was used as a positive control (Fig. 4D).
MiR-7013-3p inhibited proliferation in B16 melanoma cells. (A) The RTCA system was used to measure B16 melanoma cell proliferation. (B) Relative ratio of cell proliferation after transfection with the miR-7013-3p mimic compared with the mimic NC. (C) Flow cytometry was used to measure the cell cycle distribution. (D) qRT-PCR was used to analyze relative CDK2 gene expression after transfection with the miR-7013-3p mimic and MITF siRNA. The data summarize three independent experiments and are given as the mean±SD. *P<0.05.
miR-7013-3p promoted apoptosis in B16 melanoma cells. (A-C) Flow cytometry was used to measure apoptosis. The apoptosis rate of the cells of the miR-7013-3p overexpression group (Fig. 4B) was higher than that in the NC group (Fig. 4A). (D) The qRT-PCR results show the relative expression of BCL2 genes after transfection with the miR-7013-3p mimic and MITF siRNA. The data summarize three independent experiments and are given as the mean±SD. *P<0.05.
MiR-7013-3p inhibited cell migration and invasion. (A-E) The migration of B16 melanoma cells was measured by a wound-healing assay. (F) The qRT-PCR results show the relative expression of BCL2 genes after transfection with the miR-7013-3p mimic and MITF siRNA. The data summarize three independent experiments and are given as the mean±SD. *P<0.05.
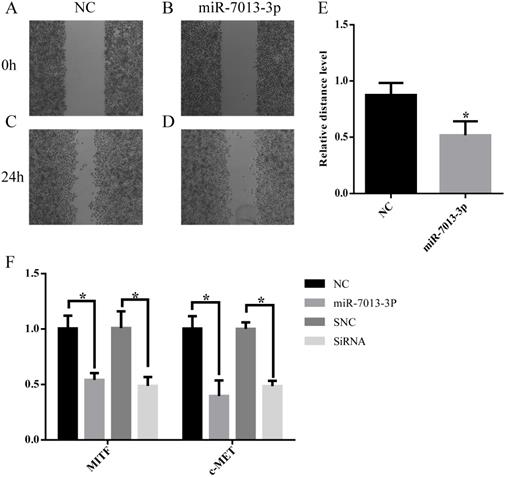
MiR-7013-3p inhibited cell migration
Wound-healing migration assays were used to evaluate the migration of miR-7013-3p mimic-transfected B16 melanoma cells. As shown in Fig. 5A-5E, miR-7013-3p mimic-transfected cells showed significantly reduced migration compared with mimic NC-transfected cells. The qRT-PCR results showed that the c-MET mRNA expression level was reduced significantly after transfection with the miR-7013-3p mimic; MITF siRNA was used as a positive control (Fig. 5F).
Discussion
Malignant melanoma is the most aggressive form of cutaneous carcinoma, with high metastatic potential and high resistance to conventional chemotherapy drugs [38, 39]. The incidence of malignant melanoma has been increasing worldwide; although melanoma was a rare cancer a century ago, the average lifetime risk of melanoma is now 1 in 50 in many Western populations [39].
MITF is considered to be the master regulator of melanocytes and an oncogene in melanoma. It is essential for the proliferation and survival of melanoma cells [40]. MITF stimulates melanin production by activating the transcription of pigmentation genes, including TYR, TYRP1, TYRP2 and DCT. MITF also regulates some genes that are important for melanoma survival (e.g., BCL2), proliferation (e.g., CDK2) and metastatic potential (e.g., c-MET). As an antiapoptotic factor, BCL2 can increase the antiapoptotic ability of melanoma cells. Knockdown of BCL2 can cause melanoma cells to exhibit increased apoptosis rates upon external stimulation and can improve the efficiency of drug action [41-44]. CDK2 is a member of the protein kinase family. It plays important roles in regulating various events of the eukaryotic cell division cycle. Accumulating evidence suggests that overexpression of CDK2 may lead to abnormal cell cycle regulation, which may be directly related to overproliferation of cancer cells [45-48]. Hepatocyte growth factor (HGF), a scatter factor and tumor cytotoxic factor, is a large multidomain heterodimeric protein that belongs to the HGF cytokine family and is the exclusive ligand of c-MET [49, 50]. Preclinical studies have shown that enhancement of HGF/c-MET activity can increase the proliferation of melanoma cells [51], increase the invasive ability of melanoma cells [52, 53] and protect melanoma cells from apoptosis [22]. These results indicate that MITF can be used as a target gene for melanoma treatment [15, 54, 55].
[Legend]
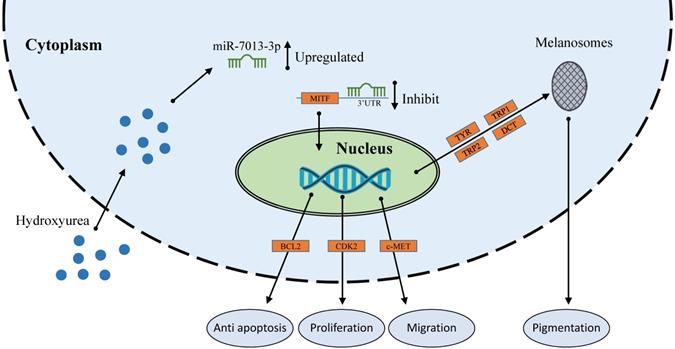
HU therapy is used for sickle cell anemia [56], transfusion-dependent β-thalassemia [57], melanoma [34] and other conditions. miRNAs that were differentially expressed under HU treatment are also involved in these treatments [35, 36, 58]. These differentially expressed miRNAs regulate the expression of some genes to affect cell apoptosis, proliferation, migration and invasion. Extensive reports indicating the functions of various miRNAs in melanoma cells have been published [59]. miRNAs play important roles in melanoma cells; for example, miR-137 and miR-182 can inhibit the invasion of melanoma cells through downregulation of MITF and other oncogenic target genes [25, 26]. However, the function of miR-7013-3p has rarely been reported. In this study, we found that miR-7013-3p expression was upregulated after HU treatment. HU reduced MITF gene expression in B16 melanoma cells, and miR-7013-3p assisted HU in reducing MITF gene expression levels. MITF acts as a transcriptional regulator to mediate the pigmentation, proliferation, apoptosis and migration of B16 melanoma cells. Notably, MITF suppression has been found to improve the sensitivity of melanoma cells to drugs [27]. In this study, miR-7013-3p was found to inhibit the function, proliferation and migration of B16 melanoma cells and to promote apoptosis of these cells by regulating the MITF gene directly. The findings of this study will help improve our understanding of the regulatory functions of miRNAs in melanoma, enrich current knowledge regarding the mechanism underlying miRNA-assisted drug function and provide a reference for later research on melanoma cells.
Conclusion
In conclusion, our study demonstrates that HU treatment alters miRNA expression and that such miRNA changes are involved in the effects of HU on cells. For example, upregulation of miR-7013-3p inhibits melanin production, proliferation, and migration and promotes apoptosis in B16 melanoma cells. The findings add to the growing body of evidence that miRNAs are important mediators of the therapeutic effects of HU on melanoma.
Supplementary Material
Supplementary file legends and figures.
Supplementary file.
Acknowledgements
This study was supported by the Science and Technology Project of Jilin Province (20191004002TC, 20190201166JC).
Competing Interests
The authors have declared that no competing interest exists.
References
1. D'Orazio J, Jarrett S, Amaro-Ortiz A, Scott T. UV radiation and the skin. Int J Mol Sci. 2013;14(6):12222-48 Epub 2013/06/12. doi: 10.3390/ijms140612222. PubMed PMID: 23749111; PubMed Central PMCID: PMCPMC3709783
2. Mohania D, Chandel S, Kumar P, Verma V, Digvijay K, Tripathi D. et al. Ultraviolet Radiations: Skin Defense-Damage Mechanism. Adv Exp Med Biol. 2017;996:71-87 Epub 2017/11/11. doi: 10.1007/978-3-319-56017-5_7. PubMed PMID: 29124692
3. Zucca FA, Segura-Aguilar J, Ferrari E, Munoz P, Paris I, Sulzer D. et al. Interactions of iron, dopamine and neuromelanin pathways in brain aging and Parkinson's disease. Prog Neurobiol. 2017;155:96-119 Epub 2015/10/13. doi: 10.1016/j.pneurobio.2015.09.012. PubMed PMID: 26455458; PubMed Central PMCID: PMCPMC4826627
4. Ganesan AK, Ho H, Bodemann B, Petersen S, Aruri J, Koshy S. et al. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 2008;4(12):e1000298. Epub 2008/12/06. doi: 10.1371/journal.pgen.1000298. PubMed PMID: 19057677; PubMed Central PMCID: PMCPMC2585813
5. Yamaguchi Y, Hearing VJ. Melanocytes and their diseases. Cold Spring Harb Perspect Med. 2014 4(5). Epub 2014/05/03. doi: 10.1101/cshperspect.a017046. PubMed PMID: 24789876; PubMed Central PMCID: PMCPMC3996377
6. Nicolaidou E, Katsambas AD. Pigmentation disorders: hyperpigmentation and hypopigmentation. Clin Dermatol. 2014;32(1):66-72 Epub 2013/12/10. doi: 10.1016/j.clindermatol.2013.05.026. PubMed PMID: 24314378
7. Liu K, Jin J, Rong K, Zhuo L, Li P. MicroRNA675 inhibits cell proliferation and invasion in melanoma by directly targeting metadherin. Mol Med Rep. 2018;17(2):3372-9 Epub 2017/12/20. doi: 10.3892/mmr.2017.8264. PubMed PMID: 29257296
8. Wang D, Xu W, Chen X, Han J, Yu L, Gao C. et al. Icariin induces cell differentiation and cell cycle arrest in mouse melanoma B16 cells via Erk1/2-p38-JNK-dependent pathway. Oncotarget. 2017;8(59):99504-13 Epub 2017/12/17. doi: 10.18632/oncotarget.20118. PubMed PMID: 29245919; PubMed Central PMCID: PMCPMC5725110
9. Mitsiogianni M, Amery T, Franco R, Zoumpourlis V, Pappa A, Panayiotidis MI. From chemo-prevention to epigenetic regulation: The role of isothiocyanates in skin cancer prevention. Pharmacol Ther. 2018;190:187-201 Epub 2018/06/12. doi: 10.1016/j.pharmthera.2018.06.001. PubMed PMID: 29890115
10. Li X, Wang R, Zhang J, Yang S, Ji K, Du B. et al. Cyclin-dependent kinase 5 regulates proliferation, migration, tyrosinase activity, and melanin production in B16-F10 melanoma cells via the essential regulator p-CREB. In vitro Cell Dev Biol Anim. 2019;55(6):416-25 Epub 2019/05/10. doi: 10.1007/s11626-019-00343-6. PubMed PMID: 31069610
11. Hsan KM, Chen CC, Shyur LF. Current research and development of chemotherapeutic agents for melanoma. Cancers (Basel). 2010;2(2):397-419 Epub 2010/01/01. doi: 10.3390/cancers2020397. PubMed PMID: 24281076; PubMed Central PMCID: PMCPMC3835084
12. Liu W, Liu X, Pan Z, Wang D, Li M, Chen X. et al. Ailanthone Induces Cell Cycle Arrest and Apoptosis in Melanoma B16 and A375 Cells. Biomolecules. 2019 9(7). Epub 2019/07/25. doi: 10.3390/biom9070275. PubMed PMID: 31336757; PubMed Central PMCID: PMCPMC6681521
13. Basu R, Kulkarni P, Qian Y, Walsh C, Arora P, Davis E. et al. Growth Hormone Upregulates Melanocyte-Inducing Transcription Factor Expression and Activity via JAK2-STAT5 and SRC Signaling in GH Receptor-Positive Human Melanoma. Cancers (Basel). 2019 11(9). Epub 2019/09/25. doi: 10.3390/cancers11091352. PubMed PMID: 31547367; PubMed Central PMCID: PMCPMC6769493
14. Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12(9):406-14 Epub 2006/08/11. doi: 10.1016/j.molmed.2006.07.008. PubMed PMID: 16899407
15. Hartman ML, Czyz M. MITF in melanoma: mechanisms behind its expression and activity. Cell Mol Life Sci. 2015;72(7):1249-60 Epub 2014/12/01. doi: 10.1007/s00018-014-1791-0. PubMed PMID: 25433395; PubMed Central PMCID: PMCPMC4363485
16. Chen T, Zhao B, Liu Y, Wang R, Yang Y, Yang L. et al. MITF-M regulates melanogenesis in mouse melanocytes. J Dermatol Sci. 2018;90(3):253-62 Epub 2018/03/03. doi: 10.1016/j.jdermsci.2018.02.008. PubMed PMID: 29496358
17. Flesher JL, Paterson-Coleman EK, Vasudeva P, Ruiz-Vega R, Marshall M, Pearlman E. et al. Delineating the role of MITF isoforms in pigmentation and tissue homeostasis. Pigment Cell Melanoma Res. 2019. Epub 2019/09/29. doi: 10.1111/pcmr.12828. PubMed. PMID: 31562697
18. Seo EY, Jin SP, Sohn KC, Park CH, Lee DH, Chung JH. UCHL1 Regulates Melanogenesis through Controlling MITF Stability in Human Melanocytes. J Invest Dermatol. 2017;137(8):1757-65 Epub 2017/04/11. doi: 10.1016/j.jid.2017.03.024. PubMed PMID: 28392346
19. Najem A, Krayem M, Sales F, Hussein N, Badran B, Robert C. et al. P53 and MITF/Bcl-2 identified as key pathways in the acquired resistance of NRAS-mutant melanoma to MEK inhibition. Eur J Cancer. 2017;83:154-65 Epub 2017/07/25. doi: 10.1016/j.ejca.2017.06.033. PubMed PMID: 28738256
20. Urban P, Rabajdova M, Velika B, Spakova I, Bolerazska B, Marekova M. [The Importance of MITF Signaling Pathway in the Regulation of Proliferation and Invasiveness of Malignant Melanoma]. Klin Onkol. 29(5):347-50. Epub 2016/10/16. doi: 10.14735/amko2016347. PubMed. PMID: 27739313
21. Puri N, Ahmed S, Janamanchi V, Tretiakova M, Zumba O, Krausz T. et al. c-Met is a potentially new therapeutic target for treatment of human melanoma. Clin Cancer Res. 2007;13(7):2246-53 Epub 2007/04/04. doi: 10.1158/1078-0432.CCR-06-0776. PubMed PMID: 17404109
22. Beuret L, Flori E, Denoyelle C, Bille K, Busca R, Picardo M. et al. Up-regulation of MET expression by alpha-melanocyte-stimulating hormone and MITF allows hepatocyte growth factor to protect melanocytes and melanoma cells from apoptosis. J Biol Chem. 2007;282(19):14140-7 Epub 2007/03/21. doi: 10.1074/jbc.M611563200. PubMed PMID: 17371876
23. McGill GG, Haq R, Nishimura EK, Fisher DE. c-Met expression is regulated by Mitf in the melanocyte lineage. J Biol Chem. 2006;281(15):10365-73 Epub 2006/02/04. doi: 10.1074/jbc.M513094200. PubMed PMID: 16455654
24. Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2010;23(1):27-40 Epub 2009/12/10. doi: 10.1111/j.1755-148X.2009.00653.x. PubMed PMID: 19995375
25. Yan D, Dong XD, Chen X, Yao S, Wang L, Wang J. et al. Role of microRNA-182 in posterior uveal melanoma: regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS One. 2012;7(7):e40967. Epub 2012/08/01. doi: 10.1371/journal.pone.0040967. PubMed PMID: 22848417; PubMed Central PMCID: PMCPMC3407171
26. Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A. et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013;133(3):768-75 Epub 2012/11/16. doi: 10.1038/jid.2012.357. PubMed PMID: 23151846
27. Aida S, Sonobe Y, Tanimura H, Oikawa N, Yuhki M, Sakamoto H. et al. MITF suppression improves the sensitivity of melanoma cells to a BRAF inhibitor. Cancer Lett. 2017;409:116-24 Epub 2017/09/20. doi: 10.1016/j.canlet.2017.09.008. PubMed PMID: 28923400
28. Soura E, Eliades PJ, Shannon K, Stratigos AJ, Tsao H. Hereditary melanoma: Update on syndromes and management: Emerging melanoma cancer complexes and genetic counseling. J Am Acad Dermatol. 2016;74(3):411-20 quiz 21-2. Epub 2016/02/20. doi: 10.1016/j.jaad.2015.08.037. PubMed PMID: 26892651; PubMed Central PMCID: PMCPMC4761106
29. Riesenberg S, Groetchen A, Siddaway R, Bald T, Reinhardt J, Smorra D. et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun. 2015;6:8755. Epub 2015/11/05. doi: 10.1038/ncomms9755. PubMed PMID: 26530832; PubMed Central PMCID: PMCPMC4659938
30. Karanth SS, Gupta A, Prabhu M. Melanonychia and mucocutaneous hyperpigmentation from hydroxyurea use for the treatment of essential thrombocytosis. Singapore Med J. 2014;55(1):e7-8 Epub 2014/01/24. doi: 10.11622/smedj.2013187. PubMed PMID: 24452985; PubMed Central PMCID: PMCPMC4291920
31. Cantisani C, Kiss N, Naqeshbandi AF, Tosti G, Tofani S, Cartoni C. et al. Nonmelanoma skin cancer associated with Hydroxyurea treatment: Overview of the literature and our own experience. Dermatol Ther. 2019;32(5):e13043. Epub 2019/08/01. doi: 10.1111/dth.13043. PubMed PMID: 31364787
32. Kovacic P. Hydroxyurea (therapeutics and mechanism): metabolism, carbamoyl nitroso, nitroxyl, radicals, cell signaling and clinical applications. Med Hypotheses. 2011;76(1):24-31 Epub 2010/09/14. doi: 10.1016/j.mehy.2010.08.023. PubMed PMID: 20833482
33. Cassileth PA, Hyman GA. Treatment of malignant melanoma with hydroxyurea. Cancer Res. 1967;27(10):1843-5 Epub 1967/10/01. PubMed PMID: 6064961
34. Saito A, Fujisawa Y, Maruyama H, Nakamura Y, Ishitsuka Y, Watanabe R. et al. Subungual melanoma complicated by hydroxyurea-induced melanonychia with non-melanoma Hutchinson's sign. Eur J Dermatol. 2018;28(4):559-61 Epub 2018/06/27. doi: 10.1684/ejd.2018.3345. PubMed PMID: 29941409
35. Mnika K, Mazandu GK, Jonas M, Pule GD, Chimusa ER, Hanchard NA. et al. Hydroxyurea-Induced miRNA Expression in Sickle Cell Disease Patients in Africa. Front Genet. 2019;10:509. Epub 2019/06/25. doi: 10.3389/fgene.2019.00509. PubMed PMID: 31231425; PubMed Central PMCID: PMCPMC6568309
36. Pule GD, Mowla S, Novitzky N, Wonkam A. Hydroxyurea down-regulates BCL11A, KLF-1 and MYB through miRNA-mediated actions to induce gamma-globin expression: implications for new therapeutic approaches of sickle cell disease. Clin Transl Med. 2016;5(1):15. Epub 2016/04/09. doi: 10.1186/s40169-016-0092-7. PubMed PMID: 27056246; PubMed Central PMCID: PMCPMC4824700
37. Yakisich JS, Azad N, Venkatadri R, Kulkarni Y, Wright C, Kaushik V. et al. Digitoxin and its synthetic analog MonoD have potent antiproliferative effects on lung cancer cells and potentiate the effects of hydroxyurea and paclitaxel. Oncol Rep. 2016;35(2):878-86 Epub 2015/11/18. doi: 10.3892/or.2015.4416. PubMed PMID: 26573786; PubMed Central PMCID: PMCPMC4689486
38. Zhu S, Wurdak H, Wang Y, Galkin A, Tao H, Li J. et al. A genomic screen identifies TYRO3 as a MITF regulator in melanoma. Proc Natl Acad Sci U S A. 2009;106(40):17025-30 Epub 2009/10/07. doi: 10.1073/pnas.0909292106. PubMed PMID: 19805117; PubMed Central PMCID: PMCPMC2761329
39. Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In vivo. 2014;28(6):1005-11 Epub 2014/11/16. PubMed PMID: 25398793
40. Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S. et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117-22 Epub 2005/07/08. doi: 10.1038/nature03664. PubMed PMID: 16001072
41. Zhu Y, Wen X, Zhao P. MicroRNA-365 Inhibits Cell Growth and Promotes Apoptosis in Melanoma by Targeting BCL2 and Cyclin D1 (CCND1). Med Sci Monit. 2018;24:3679-92 Epub 2018/06/03. doi: 10.12659/MSM.909633. PubMed PMID: 29858490; PubMed Central PMCID: PMCPMC6011806
42. Calance DN, Steixner C, Gross S, Schuler-Thurner B, Knoll G, Ehrenschwender M. Hypertonicity primes malignant melanoma cells for apoptosis. Apoptosis. 2018;23(3-4):201-9 Epub 2018/02/13. doi: 10.1007/s10495-018-1446-y. PubMed PMID: 29435687
43. Wang S, Qiu L, Song H, Dang N. NPS - 2143 (hydrochloride) inhibits melanoma cancer cell proliferation and induces autophagy and apoptosis. Med Sci (Paris). 2018;34 Focus issue F1:87-93. Epub 2018/11/08. doi: 10.1051/medsci/201834f115. PubMed. PMID: 30403181
44. Vlckova K, Reda J, Ondrusova L, Krayem M, Ghanem G, Vachtenheim J. GLI inhibitor GANT61 kills melanoma cells and acts in synergy with obatoclax. Int J Oncol. 2016;49(3):953-60 Epub 2016/08/31. doi: 10.3892/ijo.2016.3596. PubMed PMID: 27572939
45. Chohan TA, Qian H, Pan Y, Chen JZ. Cyclin-dependent kinase-2 as a target for cancer therapy: progress in the development of CDK2 inhibitors as anti-cancer agents. Curr Med Chem. 2015;22(2):237-63 Epub 2014/11/12. doi: 10.2174/0929867321666141106113633. PubMed PMID: 25386824
46. Bacevic K, Lossaint G, Achour TN, Georget V, Fisher D, Dulic V. Cdk2 strengthens the intra-S checkpoint and counteracts cell cycle exit induced by DNA damage. Sci Rep. 2017;7(1):13429. Epub 2017/10/19. doi: 10.1038/s41598-017-12868-5. PubMed PMID: 29044141; PubMed Central PMCID: PMCPMC5647392
47. Yin X, Yu J, Zhou Y, Wang C, Jiao Z, Qian Z. et al. Identification of CDK2 as a novel target in treatment of prostate cancer. Future Oncol. 2018;14(8):709-18 Epub 2018/01/13. doi: 10.2217/fon-2017-0561. PubMed PMID: 29323532
48. Chauhan S, Diril MK, Lee JH, Bisteau X, Manoharan V, Adhikari D. et al. Cdk2 catalytic activity is essential for meiotic cell division in vivo. Biochem J. 2016;473(18):2783-98 Epub 2016/07/03. doi: 10.1042/BCJ20160607. PubMed PMID: 27371320
49. Czyz M. HGF/c-MET Signaling in Melanocytes and Melanoma. Int J Mol Sci. 2018 19(12). Epub 2018/12/06. doi: 10.3390/ijms19123844. PubMed PMID: 30513872; PubMed Central PMCID: PMCPMC6321285
50. Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF. et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251(4995):802-4 Epub 1991/02/15. doi: 10.1126/science.1846706. PubMed PMID: 1846706
51. Giebeler N, Schonefuss A, Landsberg J, Tuting T, Mauch C, Zigrino P. Deletion of ADAM-9 in HGF/CDK4 mice impairs melanoma development and metastasis. Oncogene. 2017;36(35):5058-67 Epub 2017/05/30. doi: 10.1038/onc.2017.162. PubMed PMID: 28553955
52. Cao HH, Cheng CY, Su T, Fu XQ, Guo H, Li T. et al. Quercetin inhibits HGF/c-Met signaling and HGF-stimulated melanoma cell migration and invasion. Mol Cancer. 2015;14:103. Epub 2015/05/15. doi: 10.1186/s12943-015-0367-4. PubMed PMID: 25971889; PubMed Central PMCID: PMCPMC4435529
53. Halaban R, Rubin JS, Funasaka Y, Cobb M, Boulton T, Faletto D. et al. Met and hepatocyte growth factor/scatter factor signal transduction in normal melanocytes and melanoma cells. Oncogene. 1992;7(11):2195-206 Epub 1992/11/01. PubMed PMID: 1331934
54. Aida S, Sonobe Y, Yuhki M, Sakata K, Fujii T, Sakamoto H. et al. MITF suppression by CH5552074 inhibits cell growth in melanoma cells. Cancer Chemother Pharmacol. 2017;79(6):1187-93 Epub 2017/04/28. doi: 10.1007/s00280-017-3317-6. PubMed PMID: 28447210
55. Simmons JL, Pierce CJ, Al-Ejeh F, Boyle GM. MITF and BRN2 contribute to metastatic growth after dissemination of melanoma. Sci Rep. 2017;7(1):10909. Epub 2017/09/09. doi: 10.1038/s41598-017-11366-y. PubMed PMID: 28883623; PubMed Central PMCID: PMCPMC5589904
56. McGann PT, Ware RE. Hydroxyurea therapy for sickle cell anemia. Expert Opin Drug Saf. 2015;14(11):1749-58 Epub 2015/09/15. doi: 10.1517/14740338.2015.1088827. PubMed PMID: 26366626; PubMed Central PMCID: PMCPMC5868345
57. Ansari SH, Lassi ZS, Khowaja SM, Adil SO, Shamsi TS. Hydroxyurea (hydroxycarbamide) for transfusion-dependent beta-thalassaemia. Cochrane Database Syst Rev. 2019;3:CD012064. Epub 2019/03/19. doi: 10.1002/14651858.CD012064.pub2. PubMed PMID: 30882896; PubMed Central PMCID: PMCPMC6421980
58. Sawant M, S C, Colah R, Ghosh K, Nadkarni A. Does HbF induction by hydroxycarbamide work through MIR210 in sickle cell anaemia patients? Br J Haematol. 2016;173(5):801-3 Epub 2015/09/08. doi: 10.1111/bjh.13642. PubMed PMID: 26344186
59. Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10(2):103-21 Epub 2015/01/15. doi: 10.1080/15592294.2014.1003746. PubMed PMID: 25587943; PubMed Central PMCID: PMCPMC4622872
Author contact
![]() Corresponding authors: Jia-Bao Zhang, E-mail: zjbedu.cn, Tel.: +86-431-8783-6551; Bao Yuan, E-mail: yuan_baoedu.cn, Tel.: +86-431-8783-6536.
Corresponding authors: Jia-Bao Zhang, E-mail: zjbedu.cn, Tel.: +86-431-8783-6551; Bao Yuan, E-mail: yuan_baoedu.cn, Tel.: +86-431-8783-6536.

 Global reach, higher impact
Global reach, higher impact