Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(2):448-458. doi:10.7150/ijms.51297 This issue Cite
Research Paper
Heat shock factor 5 correlated with immune infiltration serves as a prognostic biomarker in lung adenocarcinoma
1. Department of Thoracic Surgery, The First Affiliated Hospital of Zhejiang University, Qingchun Road 79, Hangzhou, China, 310009.
2. Zhejiang University School of Medicine, Yuhangtang Road 866, Hangzhou, China, 310009.
3. Bone Marrow Transplantation Center, The First Affiliated Hospital of Zhejiang University, Qingchun Road 79, Hangzhou, China, 310009.
Received 2020-7-30; Accepted 2020-11-5; Published 2021-1-1
Abstract
Lung adenocarcinoma (LUAD) is the predominant subtype of lung cancer with a relatively poor prognosis. The dramatic improvements of new immunotherapy strategies have shown promising results in lung cancer patients. This study aimed to elucidate the functions of immune-associated genes in LUAD prognosis and pathogenesis by analyzing public databases. We obtained expression profiles of LUAD patients from The Cancer Genome Atlas (TCGA) database and applied the ESTIMATE algorithm to calculate immune scores and stromal scores. A series of microenvironment-related genes with prognostic value was then identified. Of note, heat shock factor 5 (HSF5) was found to be decreased in LUAD patients and positively correlated with overall survival, which was further confirmed in the Gene Expression Omnibus (GEO) database. Moreover, Gene Ontology (GO) analysis based on the correlated genes of HSF5 demonstrated that HSF5 expression was significantly associated with the immune response and inflammatory activities. Based on the Tumor IMmune Estimation Resource (TIMER) and Gene Expression Profiling Interactive Analysis (GEPIA) datasets, HSF5 expression showed strong correlations with various immune cell infiltration and diverse immune marker sets. These findings suggest that HSF5 can be used as a promising biomarker for determining prognosis and immune infiltration in LUAD patients.
Keywords: HSF5, lung adenocarcinoma, prognostic biomarker, immune infiltration, inflammatory activities
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, and lung adenocarcinoma (LUAD) represents the most prevalent subtype, which comprises approximately 40% of all lung cancer cases [1,2]. Despite the achievements in understanding the pathogenesis of this disease and the development of multidisciplinary therapies, the clinical outcomes for LUAD patients remain poor, with an overall survival rate of less than 5 years [3,4]. Therefore, there is an urgent need to discover specific prognostic factors for LUAD to predict the overall prognosis and improve the therapeutic management of patients.
Tumor cells are involved in extensive and dynamic crosstalk with the immune microenvironment, and this correlation plays crucial roles in cancer pathogenesis [5]. Evading immune destruction is one of the hallmarks of cancer [6]. The role of the immune system in lung oncogenesis is increasingly being investigated with a focus on the clinical responses to checkpoint blockade immunotherapies [7,8]. Notably, the association between the expression levels of immune markers and the response to immune-based therapy has been explored in various studies [9,10]. The immune-related markers show prognostic and predictive effects in lung cancer patients. For example, increased cytotoxic T cell lymphocytes (CTLs) appear to be associated with longer survival [11]. The use of bioinformatic technologies based on expression profiling from public databases is an effective method to better understand the immune microenvironment in LUAD patients [12,13].
Heat shock factors (HSFs) are transcription factors that mediate responses to versatile forms of physiological and environmental stimuli [14]. There are several HSF isoforms in the human genome, and studies have largely focused on HSF1 and HSF2 family members [14,15]. HSF1 has been shown to play a vital role in innate immunity and immunosenescence [16]. The deregulation of HSF activity is involved in various human diseases. For example, the compromised activation of HSF1 has been reported to be linked with the pathology of Huntington's disease and Parkinson's disease [17,18]. Research has revealed that HSF1 drives carcinogenesis [19-21]. However, HSF2 has appeared to decrease in a wide range of cancers and act as a tumor suppressor [22,23]. To date, only a few studies have focused on HSF5, and its detailed functional characterization in tumors has not been performed [24].
In the present study, we evaluated the gene expression profiles of LUAD patients from The Cancer Genome Atlas (TCGA) database and identified a series of microenvironment-related genes with prognostic value. The positive correlation between HSF5 expression and overall survival was further validated in the Gene Expression Omnibus (GEO) database. Moreover, we explored the association of HSF5 with immune response and inflammatory activities, as well as immune cell infiltration and diverse immune marker sets in LUAD. This study revealed the crucial role of HSF5 in LUAD and an underlying mechanism between HSF5 and tumor-immune interactions.
Materials and Methods
Database
The RNA-Seq dataset of LUAD patients and corresponding clinical information were obtained from the TCGA database (https://gdc.nci.nih.gov/). We adopted two datasets (GSE31210 and GSE37745) from the GEO database. The data of GSE31210 were based on GPL570 platforms (HG-U133_Plus_2 Affymetrix Human Genome U133 Plus 2.0 Array) and included 226 lung adenocarcinoma patients. The GSE37745 data were based on GPL570 platforms and included 106 lung adenocarcinoma patients. Immune scores and stromal scores were calculated by the ESTIMATE algorithm of the downloaded database.
DEG identification and functional enrichment analysis
All the LUAD patients were classified into high- and low-score groups based on their immune/stromal scores. Data analysis was performed by using the package edgeR. In this study, genes with a p value < 0.05 and | fold change | > 1.5 were defined as differentially expressed genes (DEGs). Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david-d.ncifcrf.gov/) was applied to analyze the gene functions.
Survival analysis
Kaplan-Meier plots were constructed to investigate the correlation between gene expression and the overall survival of LUAD patients. The statistical significance of the correlation was tested by a log-rank test. The online Kaplan-Meier plotter database (http://kmplot.com/analysis/) was used to verify the prognostic values of the identified genes.
Immune-associated analysis
The correlations between continuous variables were investigated by Spearman correlation analysis. Gene set variation analysis (GSVA) was conducted as previously described [25]. Gene Ontology (GO) analysis of the most related genes was constructed by Heatmap. The GO gene set was obtained from the AmiGO 2 Web portal (http://amigo.geneontology.org/amigo/landing). Inflammatory-related metagenes were selected as described previously [26,27]. The metagene expression values were determined by assessing the mean of the normalized expression values of all genes in a respective cluster [27]. The Tumor IMmune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/) was applied to estimate the abundance of immune infiltrates and the correlations between HSF5 expression and the gene markers of immune cells. The online database Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) was used to further validate the significantly correlated genes in TIMER.
Results
Identification of DEGs based on immune scores and stromal scores
The complete gene expression profiles and clinical information of 517 LUAD patients were downloaded from the TCGA database. We calculated the immune scores and stromal scores of all these patients with the ESTIMATE algorithm and plotted the distribution of the scores according to stage classifications of LUAD patients. As shown in Figure 1A, the immune scores were significantly associated with the pathologic stage, while the stromal scores displayed no statistically significant differences.
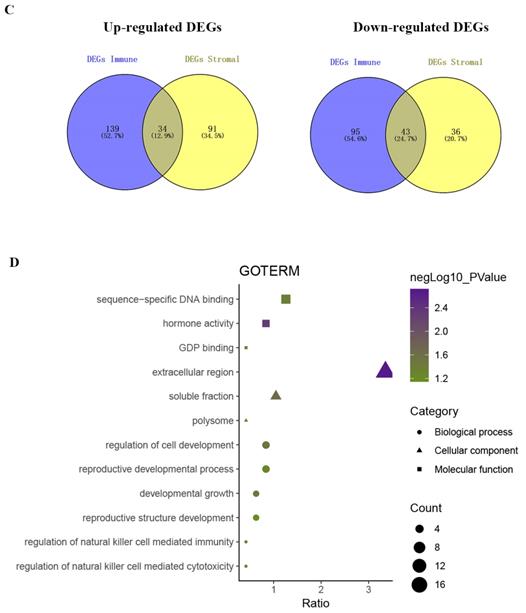
Identification of DEGs based on immune scores and stromal scores. (A) Distribution of immune scores and stromal scores for LUAD pathologic stage. (B) The Kaplan‐Meier survival curve reveals that high immune scores are associated with significantly longer overall survival. The high stromal score group showed a longer median overall survival than the low-score group, with no significant difference. (C) The commonly changed DEGs in the stromal and immune score groups (34 up- and 43 downregulated genes) were identified. (D) GO term enrichment analysis of the common DEGs.
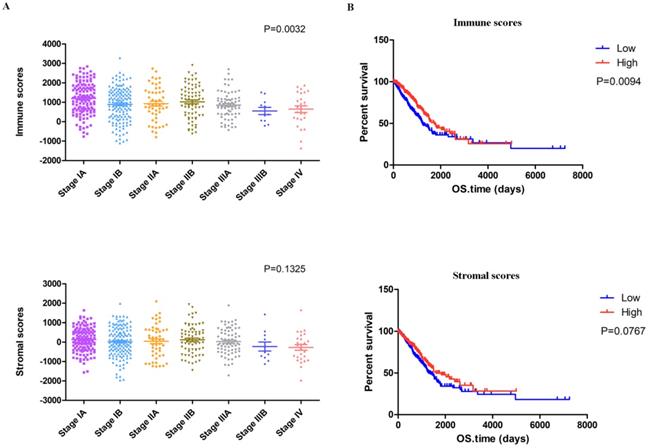
To explore the potential correlation of overall survival with immune scores and stromal scores, we classified the 517 LUAD cases into high- and low-score groups based on their scores (the top 259 scores are the high score group and the rest are the low score group). The Kaplan-Meier survival analysis (Figure 1B) demonstrated that the median overall survival of patients with high immune scores was longer than that of patients with low scores (1725 d vs. 1229 d, p = 0.0094). Moreover, patients in the high stromal score group had a longer median overall survival rate than patients in the low-score group, but with no significant difference (1600 d vs. 1293 d, p = 0.0767).
Setting p < 0.05 and | fold change | > 1.5 as the cut-off criteria, we identified 311 and 204 DEGs between the high and low immune score/stromal score groups, respectively. The integrated bioinformatic analysis revealed that 34 genes were commonly upregulated and 43 genes were commonly downregulated in the high-score group (Figure 1C). Subsequently, we conducted a functional enrichment analysis of the common DEGs with the DAVID gene annotation tool. As shown in Figure 1D, the top GO terms identified included extracellular region, soluble fraction, regulation of cell development and regulation of natural killer cell-mediated immunity.
Survival analysis of the DEGs
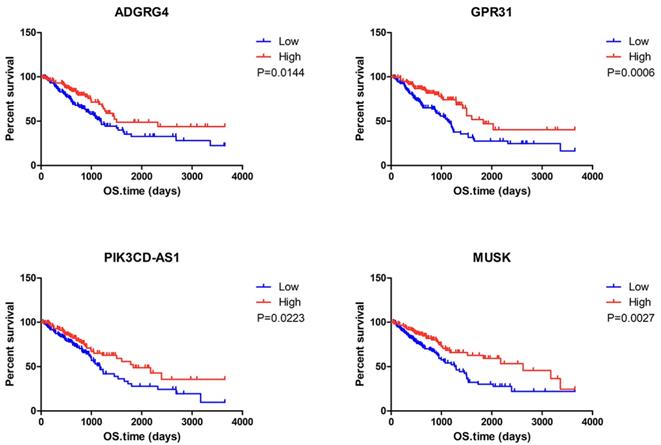
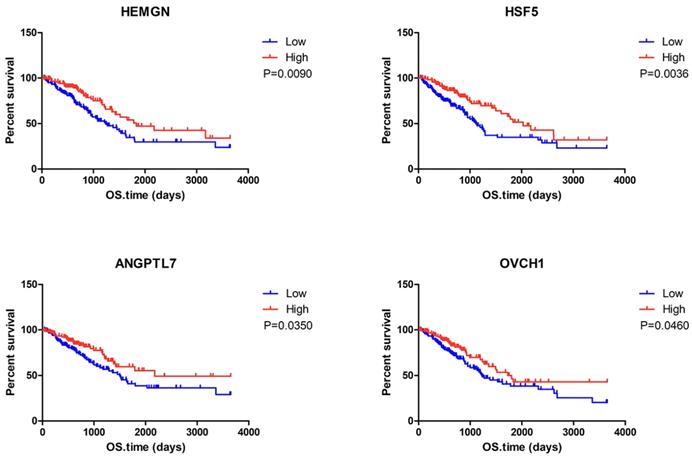
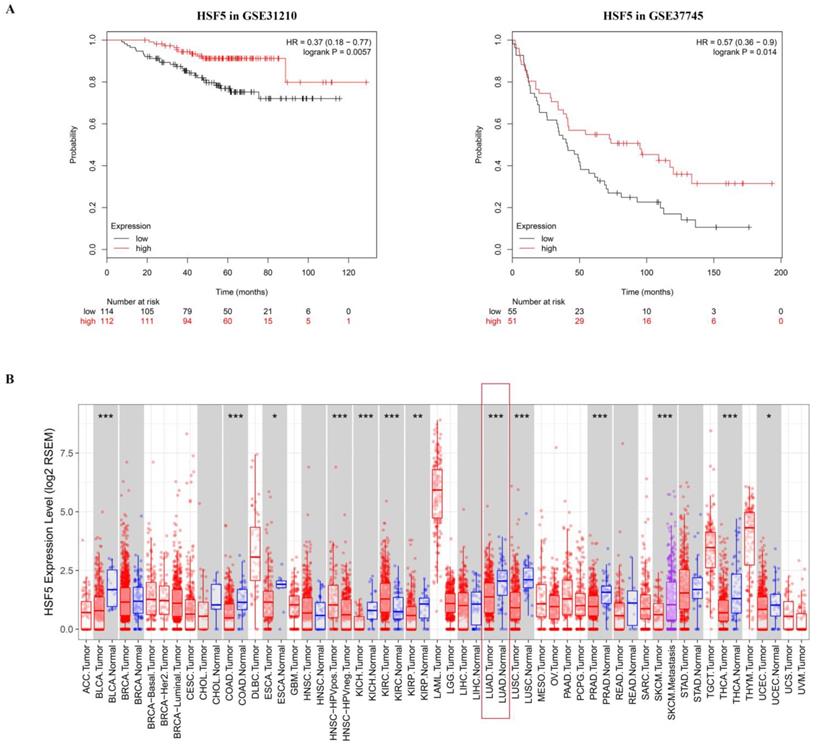
To determine the potential association of the total 77 DEGs with the overall survival of LUAD patients, we constructed Kaplan-Meier survival curves. Seventeen DEGs (8 upregulated DEGs and 9 downregulated DEGs) were significantly associated with overall survival in the log-rank test (p < 0.05). The 8 upregulated DEGs of prognostic value are shown in Figure 2. The prognostic evaluation of these genes in the Kaplan-Meier plotter database was consistent with our results (Supplementary Figure S1). Furthermore, we evaluated the prognostic potential of these genes in the GEO database (GSE31210 and GSE37745). Among the 8 upregulated DEGs, HSF5 was further confirmed to be positively associated with the overall survival of LUAD patients (Figure 3A). Additionally, we analyzed the differences in HSF5 expression in various tumor and normal tissues. As shown in Figure 3B, HSF5 expression was significantly lower in LUAD tissues than in adjacent normal tissues. Downregulated HSF5 expression was also observed in various cancers, including bladder urothelial carcinoma (BLCA), colon adenocarcinoma (COAD), kidney chromophobe (KICH), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD) and thyroid carcinoma (THCA). These results confirmed the decreased expression and prognostic value of HSF5 in LUAD.
HSF5 is associated with the immune response and inflammatory activities in LUAD
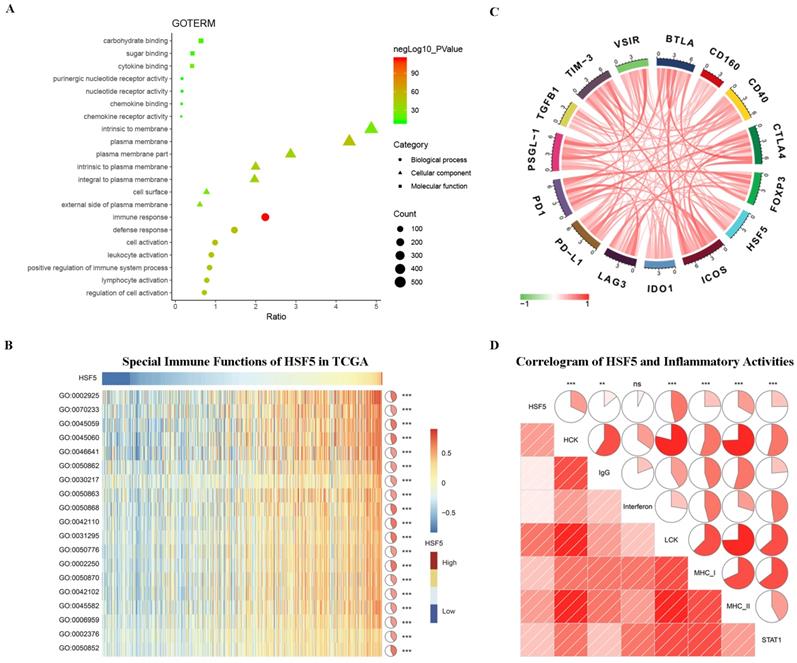
According to the above results, HSF5 may play a crucial role in the biological functions of LUAD, which has not been reported previously. To better understand the relevance and underlying mechanisms of HSF5 in LUAD, 1296 genes were screened to be significantly correlated with HSF5 based on the TCGA dataset and Spearman's correlation analysis (Spearman R > 0.3). The biological functions of these related genes were further analyzed by DAVID. GO analysis revealed that the related genes were mainly involved in the immune response, lymphocyte activation, the inflammatory response and the regulation of T cell activation (Figure 4A).
Subsequently, we performed GSVA analysis to explore the relationship between HSF5 and the immune response in LUAD (Figure 4B). The results showed that HSF5 was positively correlated with the adaptive immune response, T cell costimulation, T cell activation, the T cell receptor signaling pathway, the humoral immune response and the regulation of immune response. These results indicated that HSF5 may play an important role in the immune response, especially in T cell immunity. Additionally, we conducted a Spearman's correlation analysis on the expression of HSF5 and a variety of immune checkpoints from the TCGA dataset, such as PD-L1, PD1, CTLA-4, and IDO1. As shown in Figure 4C, HSF5 demonstrated a high correlation with ICOS and BTLA, followed by PSGL-1, CTLA4 and TIM-3.
To further understand HSF5-related inflammatory activities, we analyzed seven metagenes by a previously described method [26,27]. We found that HSF5 expression was positively associated with HCK, LCK, MHC-I, MHC-II, STAT1, and IgG but not significantly associated with interferon in the TCGA database (Figure 4D). This result revealed that HSF5 was mainly associated with the activities of macrophages, the signal transduction of T cells, B cells and antigen-presenting cells. Together, these findings indicate that HSF5 has crucial immune and inflammatory functions in LUAD.
HSF5 expression is correlated with the immune infiltration level in LUAD
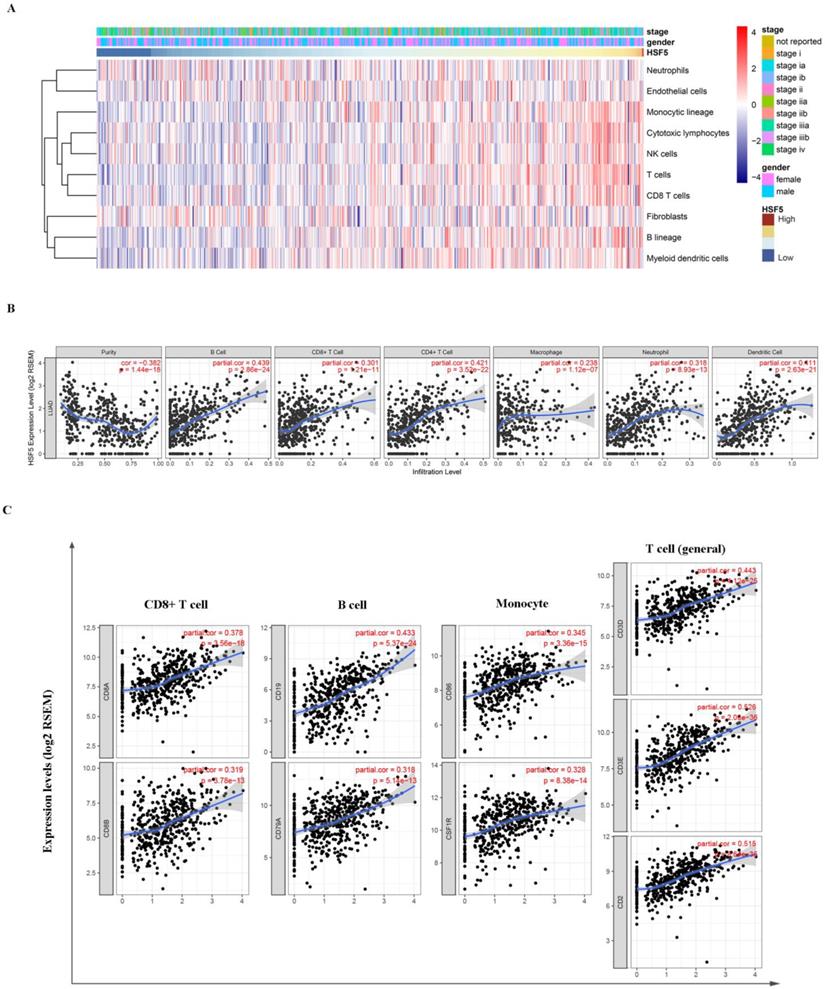
Based on the TCGA dataset, we assessed the relationship between HSF5 expression and various immune cell populations by the Microenvironment Cell Populations-counter method as previously described [28]. As shown in Figure 5A, HSF5 was significantly associated with T cells, B lineage, monocytic lineage and cytotoxic lymphocytes. In addition, we evaluated the correlations of HSF5 with immune infiltration levels in LUAD from TIMER. The result showed that the HSF5 expression level has strong positive relevance with infiltrating levels of B cells (r = 0.439, P =2.86e-24), CD8+ T cells (r = 0.301, P =1.21e-11), CD4+ T cells (r= 0.421, P = 3.52e-22), macrophages (r = 0.238, P = 1.12e-07), neutrophils (r = 0.318, P = 8.93e-13) and dendritic cells (DCs) (r =0.411, P = 2.63e-21) in LUAD (Figure 5B). These findings suggest that HSF5 plays a specific role in immune infiltration in LUAD, especially in T cells and B cells.
Correlation between individual DEG expression and the overall survival of LUAD patients in TCGA. Kaplan-Meier survival curves with the log-rank test are represented for the upregulated DEGs. ADGRG4, adhesion G protein-coupled receptor G4; GPR31,G protein-coupled receptor 31; HEMGN, hemogen; HSF5, heat shock transcription factor 5; PIK3CD-AS1, PIK3CD antisense RNA 1; MUSK, muscle associated receptor tyrosine kinase; ANGPTL7, angiopoietin like 7; OVCH1, ovochymase 1.
The HSF5 prognostic value in GEO and expression in different cancers. (A) HSF5 was further confirmed to be positively associated with the overall survival of LUAD patients in the GEO dataset (GSE31210 and GSE37745). (B) HSF5 expression levels in different types of human cancers were investigated by TIMER, red indicate the mRNA expression of HSF5 in tumor tissues; blue indicate the mRNA expression of HSF5 in normal tissues; purple indicate the mRNA expression of HSF5 in metastatic tissues (*p < 0.05, **p < 0.01, ***p < 0.001).
Correlation of HSF5 expression and immune marker sets
To validate the relationship between HSF5 and immune cells, we further estimated the correlation between HSF5 and the immune marker genes of various immune cells in LUAD based on the TIMER and GEPIA databases. We focused on the association between HSF5 and immune marker sets of diverse immune cells, including CD8+ T cells, T cells (general), B cells, monocytes, tumor-associated macrophages (TAMs), M1 and M2 macrophages, neutrophils, natural killer (NK) cells and DCs (Table 1). Specifically, we showed that CD8A and CD8B of CD8+ T cells, CD3D, CD3E, and CD2 of general T cells, CD86 and CD115 of monocytes, and CD19 and CD79A of B cells are significantly associated with HSF5 expression (Figure 5C). Subsequently, we employed the GEPIA dataset to validate the above correlations (Table 2, Supplementary Figure S2). These findings were consistent with the correlation analysis between HSF5 expression and immune cells, indicating that HSF5 plays a vital role in the immune response in the microenvironment of LUAD.
Discussion
LUAD remains one of the most aggressive and fatal tumor types despite the dramatic improvements of new therapeutic strategies [3,4]. In this study, we analyzed microenvironment-associated genes of prognostic value to LUAD based on the TCGA and GEO databases. HSF5 was found to be decreased in LUAD patients and positively correlated with overall survival. Furthermore, we demonstrated that HSF5 is significantly associated with immune response and inflammatory activities, as well as immune cell infiltration and diverse immune marker sets (Figure 6).
The ESTIMATE algorithm is designed to calculate immune and stromal scores according to gene expression data and signatures [29]. Various studies have employed this algorithm to explore the microenvironment of prostate cancer [30], colon cancer [31] and glioblastoma [32]. Here, we first assessed the infiltration of stromal and immune cells in LUAD patients based on the ESTIMATE algorithm. The immune scores were significantly correlated to the pathologic stage and overall survival of LUAD patients. Consistent with our results, a recent study revealed that a high immune score was associated with better progression-free survival (PFS) of lung cancer patients based on clinical data [33]. The immune microenvironment is widely recognized to influence lung cancer outcomes by contributing to inflammation, angiogenesis, immune modulation and the response to therapies [34,35]. Much effort has been put into exploring immune biology and developing effective immunotherapeutic strategies for lung cancer [36,37]. Thus, integrating and reanalyzing genomic profiles from public databases are important to obtain a better understanding of the immune microenvironment in LUAD.
HSF5-related immune response and inflammatory activities in LUAD. (A) The biological functions of the HSF5-related genes were investigated by GO term enrichment analysis. (B) The correlation between HSF5 and the immune response in the TCGA dataset. (C) The relationship between HSF5 and immune checkpoint members. (D) The correlation between HSF5 and inflammatory activities. ns, **, and *** indicate no significant difference, p<0.01, and p<0.001, respectively. GO: 0002925, positive regulation of humoral immune response mediated by circulating immunoglobulin. GO: 0070233, negative regulation of T cell apoptotic process. GO: 0045059, positive thymic T cell selection. GO: 0045060, negative thymic T cell selection. GO: 0046641, positive regulation of alpha-beta T cell proliferation. GO: 0050862, positive regulation of T cell receptor signaling pathway. GO: 0030217, T cell differentiation. GO: 0050863, regulation of T cell activation. GO: 0050868, negative regulation of T cell activation. GO: 0042110, T cell activation. GO: 0031295, T cell costimulation. GO: 0050776, regulation of immune response. GO: 0002250, adaptive immune response. GO: 0050870, positive regulation of T cell activation. GO: 0042102, positive regulation of T cell proliferation. GO: 0045582, positive regulation of T cell differentiation. GO: 0006959, humoral immune response. GO: 0002376, immune system process. GO: 0050852, T cell receptor signaling pathway.
HSF5 expression is correlated with the immune infiltration level in LUAD. (A) The association between HSF5 expression and immune cell populations was evaluated by the Microenvironment Cell Populations-counter method. (B) The strong positive relevance between HSF5 and infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells were confirmed in the TIMER database. (C) The correlation of HSF5 and immune marker sets of various immune cells in LUAD based on TIMER database (CD8A and CD8B of CD8+T cells, CD86 and CD115 of monocytes, CD19 and CD79A of B cells, CD3D, CD3E and CD2 of general T cells).
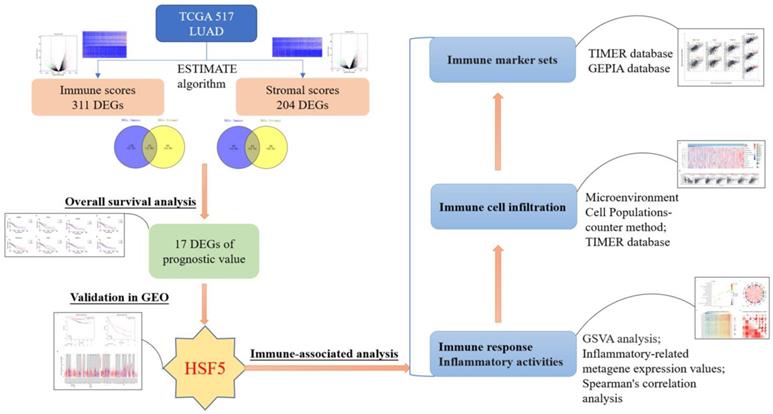
Work flow of the current study. The expression profiles of LUAD patients from TCGA database and immune scores and stromal scores, calculated from the ESTIMATE algorithm. HSF5 (microenvironment-related genes with prognostic value) was then identified, and further confirmed in the GEO database. Moreover, GO and GSVA analyses demonstrated that HSF5 expression was significantly associated with the immune response and inflammatory activities. According to TIMER and GEPIA datasets, the HSF5 expression significantly correlated with various immune cell infiltration and diverse immune marker sets.
The common DEGs between the high and low immune score/stromal score groups were identified, followed by an overall survival analysis. The results demonstrated that 17 DEGs were significantly associated with the prognosis of LUAD patients. The prognostic value of these genes was also confirmed in the Kaplan-Meier plotter database. Of note, HSF5 was further verified to be positively associated with the overall survival of LUAD patients from the GEO database. Additionally, we revealed that HSF5 expression was significantly downregulated in LUAD tissues compared with adjacent normal tissues. Moreover, lower HSF5 expression was also observed in bladder, colon, kidney, prostate and thyroid cancers based on TIMER. Consistent with this result, another HSF family member, HSF2, has been reported to be frequently decreased in several human malignancies and acts as a tumor suppressor [23]. However, the major stress-responsive factor HSF1 appears to support cancer cell growth, survival and metastasis [19,21]. HSF5 has not previously been connected to cancer, and in this study, we demonstrated the low expression and prognostic value of HSF5 in LUAD.
HSF5 belongs to the heat shock transcription factor family, which is involved in differentiation, reproduction, and stress-induced adaptation [14]. Previous studies have revealed that HSF5 plays a critical role in germ cell development and meiotic progression [24,38]. However, the functional characterization of HSF5 involved in cancer and the immune response has not been conducted. In this study, we showed that HSF5-related genes were mainly enriched in the immune response, lymphocyte activation, and the inflammatory response in LUAD. Further GSVA analysis also demonstrated that HSF5 was positively correlated with the adaptive immune response, T cell activation, the T cell receptor signaling pathway, and the regulation of the immune response. Consistently, another HSF family member, HSF1, has been reported to enable the normal function of the immune system [16,39]. These results indicated the potential role of HSF5 in the immune response, especially in T cell immunity.
Another important aspect of this study is that HSF5 expression is associated with diverse immune cell infiltration and immune marker sets in LUAD. Our analysis demonstrated that there was a strong positive correlation between HSF5 expression level and infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and DCs in LUAD. Importantly, the correlation between HSF5 expression and the marker genes of these immune cells implicates the function of HSF5 in regulating tumor immunology. In lung cancer, the prognostic and predictive significance of immune markers has been elucidated in various studies [8,40]. CD8+ T cells, CD4+ T cells and mature DCs appeared to be associated with good prognosis. Regulatory T cells (Tregs), immature DCs, and M2 macrophages were shown to be related to poor outcomes [41]. In recent years, immunotherapy has changed the therapeutic strategy and shown promising results in lung cancer patients [7,42]. Ongoing immunotherapy biomarker research is essential to develop more accurately customized immunotherapy strategies [8]. Our findings suggest that HSF5 plays an important role in the regulation of immune infiltration and may be a biomarker for immunotherapy in LUAD. The detailed function and underlying mechanism of HSF5 need to be further investigated.
Correlation analysis between HSF5 and relate genes and markers of immune cells in TIMER
| Description | Gene markers | LUAD | |||||
|---|---|---|---|---|---|---|---|
| None | Purity | Age | |||||
| Cor | P | Cor | P | Cor | P | ||
| CD8+ T cell | CD8A | 0.475 | *** | 0.378 | *** | 0.466 | *** |
| CD8B | 0.401 | *** | 0.319 | *** | 0.399 | *** | |
| T cell (general) | CD3D | 0.547 | *** | 0.443 | *** | 0.534 | *** |
| CD3E | 0.612 | *** | 0.526 | *** | 0.6 | *** | |
| CD2 | 0.608 | *** | 0.515 | *** | 0.596 | *** | |
| B cell | CD19 | 0.517 | *** | 0.433 | *** | 0.525 | *** |
| CD79A | 0.425 | *** | 0.318 | *** | 0.428 | *** | |
| Monocyte | CD86 | 0.463 | *** | 0.345 | *** | 0.46 | *** |
| CD115 (CSF1R) | 0.437 | *** | 0.328 | *** | 0.433 | *** | |
| TAM | CCL2 | 0.297 | *** | 0.197 | *** | 0.288 | *** |
| CD68 | 0.353 | *** | 0.247 | *** | 0.356 | *** | |
| IL10 | 0.403 | *** | 0.299 | *** | 0.404 | *** | |
| M1 Macrophage | INOS (NOS2) | 0.056 | 0.206 | -0.024 | 0.596 | 0.041 | 0.370 |
| IRF5 | 0.33 | *** | 0.227 | *** | 0.337 | *** | |
| COX2 (PTGS2) | -0.088 | 0.0471 | -0.083 | 0.0645 | -0.087 | 0.057 | |
| M2 Macrophage | CD163 | 0.351 | *** | 0.243 | *** | 0.354 | *** |
| VSIG4 | 0.349 | *** | 0.253 | *** | 0.348 | *** | |
| MS4A4A | 0.414 | *** | 0.306 | *** | 0.414 | *** | |
| Neutrophils | CD66b (CEACAM8) | 0.241 | *** | 0.228 | *** | 0.217 | *** |
| CD11b (ITGAM) | 0.418 | *** | 0.31 | *** | 0.418 | *** | |
| CCR7 | 0.63 | *** | 0.561 | *** | 0.623 | *** | |
| Natural killer cell | KIR2DL1 | 0.134 | * | 0.074 | 0.1 | 0.128 | * |
| KIR2DL3 | 0.139 | * | 0.043 | 0.337 | 0.165 | ** | |
| KIR2DL4 | 0.111 | 0.0121 | 0.027 | 0.555 | 0.119 | * | |
| KIR3DL1 | 0.122 | * | 0.059 | 0.189 | 0.121 | * | |
| KIR3DL2 | 0.22 | *** | 0.144 | * | 0.223 | *** | |
| KIR3DL3 | 0.024 | 0.594 | -0.003 | 0.946 | 0.019 | 0.676 | |
| KIR2DS4 | 0.144 | * | 0.058 | 0.201 | 0.154 | ** | |
| Dendritic cell | HLA-DPB1 | 0.504 | *** | 0.413 | *** | 0.487 | *** |
| HLA-DQB1 | 0.357 | *** | 0.254 | *** | 0.341 | *** | |
| HLA-DRA | 0.474 | *** | 0.376 | *** | 0.46 | *** | |
| HLA-DPA1 | 0.471 | *** | 0.379 | *** | 0.457 | *** | |
| BDCA-1 (CD1C) | 0.435 | *** | 0.369 | *** | 0.412 | *** | |
| BDCA-4 (NRP1) | 0.101 | 0.0216 | 0.05 | 0.266 | 0.09 | 0.0484 | |
| CD11c (ITGAX) | 0.451 | *** | 0.338 | *** | 0.463 | *** | |
| Th1 | T-bet (TBX21) | 0.477 | *** | 0.378 | *** | 0.465 | *** |
| STAT4 | 0.476 | *** | 0.371 | *** | 0.463 | *** | |
| STAT1 | 0.269 | *** | 0.161 | *** | 0.272 | *** | |
| IFN-γ (IFNG) | 0.326 | *** | 0.22 | *** | 0.324 | *** | |
| TNF-α (TNF) | 0.306 | *** | 0.178 | *** | 0.291 | *** | |
| Th2 | GATA3 | 0.388 | *** | 0.266 | *** | 0.38 | *** |
| STAT6 | 0.09 | 0.0416 | 0.104 | 0.0204 | 0.072 | 0.117 | |
| STAT5A | 0.489 | *** | 0.38 | *** | 0.476 | *** | |
| IL13 | 0.244 | *** | 0.178 | *** | 0.225 | *** | |
| Tfh | BCL6 | 0.019 | 0.671 | 0.007 | 0.872 | 0.003 | 0.504 |
| IL21 | 0.269 | *** | 0.206 | *** | 0.289 | *** | |
| Th17 | STAT3 | 0.01 | 0.826 | 0.03 | 0.511 | -0.001 | 0.986 |
| IL17A | 0.262 | *** | 0.199 | *** | 0.274 | *** | |
| Treg | FOXP3 | 0.505 | *** | 0.391 | *** | 0.501 | *** |
| CCR8 | 0.536 | *** | 0.429 | *** | 0.538 | *** | |
| STAT5B | 0.302 | *** | 0.295 | *** | 0.303 | *** | |
| TGFβ (TGFB1) | 0.295 | *** | 0.2 | *** | 0.28 | *** | |
| T cell exhaustion | PD-1 (PDCD1) | 0.473 | *** | 0.38 | *** | 0.466 | *** |
| CTLA4 | 0.54 | *** | 0.429 | *** | 0.542 | *** | |
| LAG3 | 0.363 | *** | 0.264 | *** | 0.369 | *** | |
| TIM-3 (HAVCR2) | 0.448 | *** | 0.329 | *** | 0.443 | *** | |
| GZMB | 0.247 | *** | 0.119 | * | 0.249 | *** | |
LUAD, lung adenocarcinoma; TAM, tumor-associated macrophage; Th, T helper cell; Tfh, Follicular helper T cell; Treg, regulatory T cell; Cor, R value of Spearman's correlation; None, correlation without adjustment. Purity, correlation adjusted by purity. Age, correlation adjusted by age. *P < 0.01; **P < 0.001; ***P < 0.0001.
Correlation analysis between HSF5 and relate genes and markers of immune cells in GEPIA
| Description | Gene markers | LUAD | |||
|---|---|---|---|---|---|
| Tumor | Normal | ||||
| R | P | R | P | ||
| CD8+ T cell | CD8A | 0.45 | *** | -0.035 | 0.79 |
| CD8B | 0.37 | *** | 0.07 | 0.6 | |
| CD8A and CD8B | 0.42 | *** | 0.014 | 0.01 | |
| T cell (general) | CD3D | 0.51 | *** | -0.071 | 0.59 |
| CD3E | 0.61 | *** | 0.12 | 0.38 | |
| CD2 | 0.6 | *** | -0.09 | 0.5 | |
| CD3D, CD3E and CD2 | 0.89 | *** | -0.04 | 0.76 | |
| B cell | CD19 | 0.49 | *** | 0.16 | 0.22 |
| CD79A | 0.4 | *** | 0.011 | 0.94 | |
| CD19 and CD79A | 0.45 | *** | 0.039 | 0.77 | |
| Monocyte | CD86 | 0.48 | *** | 0.043 | 0.75 |
| CD115 (CSF1R) | 0.46 | *** | 0.21 | 0.1 | |
| CD86 and CD115(CSF1R) | 0.48 | *** | 0.17 | 0.19 | |
LUAD, lung adenocarcinoma; tumor, correlation analysis in tumor tissue of TCGA; normal, correlation analysis in normal tissue of TCGA. ***P < 0.0001.
In conclusion, we explored the microenvironment-associated genes of prognostic value to LUAD through integrated bioinformatics analysis. We found that HSF5 was downregulated and positively correlated with the overall survival of LUAD patients.
Moreover, HSF5 is involved in the immune response and potentially contributes to the regulation of B cells, CD8+ T cells, CD4+ T cells and DCs. These findings suggest that HSF5 plays a crucial role in the immune microenvironment and as a prognostic biomarker in LUAD patients.
Supplementary Material
Supplementary figures.
Acknowledgements
The authors thank the TIMER, GEPIA, TCGA and GEO databases for the availability of the data.
Author Contributions
Rusidanmu Aizemaiti conceived and designed the study; Jie Tang obtained the datasets; Zhigang Wu performed data analysis; Haimeng Yan wrote the paper. Xiayi Lv revised the manuscript. All authors reviewed and approved the final manuscript.
Data Availability Statement
The data that support the findings of this study are available in TCGA database (https://gdc.nci.nih.gov/) and GEO database (https://www.ncbi.nlm.nih.gov/geo/, GSE31210 and GSE37745).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
2. Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB. et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2015;10:1243-60
3. Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH. et al. Cancer treatment and survivorship statistics, 2016. CA: a cancer journal for clinicians. 2016;66:271-89
4. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell death & disease. 2018;9:117
5. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883-99
6. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. cell. 2011;144:646-74 %@ 0092-8674
7. Martinez P, Peters S, Stammers T, Soria JC. Immunotherapy for the First-Line Treatment of Patients with Metastatic Non-Small Cell Lung Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2019;25:2691-8
8. Prelaj A, Tay R, Ferrara R, Chaput N, Besse B, Califano R. Predictive biomarkers of response for immune checkpoint inhibitors in non-small-cell lung cancer. European journal of cancer. 2019;106:144-59
9. Henick BS, Herbst RS, Goldberg SB. The PD-1 pathway as a therapeutic target to overcome immune escape mechanisms in cancer. Expert opinion on therapeutic targets. 2014;18:1407-20
10. Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563-7
11. Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN. et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. Journal of the National Cancer Institute. 2015 107
12. Zheng S, Luo X, Dong C, Zheng D, Xie J, Zhuge L. et al. A B7-CD28 family based signature demonstrates significantly different prognoses and tumor immune landscapes in lung adenocarcinoma. 2018; 143: 2592-601.
13. Dong Y, Liu Y, Bai H, Jiao S. Systematic assessment of the clinicopathological prognostic significance of tissue cytokine expression for lung adenocarcinoma based on integrative analysis of TCGA data. Scientific reports. 2019;9:6301
14. Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nature reviews Molecular cell biology. 2018;19:4-19
15. Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in Stress, Development, and Organismal Health. Trends in cell biology. 2017;27:895-905
16. Singh V, Aballay A. Heat shock and genetic activation of HSF-1 enhance immunity to bacteria. Cell cycle (Georgetown, Tex). 2006;5:2443-6
17. Gomez-Pastor R, Burchfiel ET, Neef DW, Jaeger AM, Cabiscol E, McKinstry SU. et al. Abnormal degradation of the neuronal stress-protective transcription factor HSF1 in Huntington's disease. Nature communications. 2017;8:14405
18. Kim E, Wang B, Sastry N, Masliah E, Nelson PT, Cai H. et al. NEDD4-mediated HSF1 degradation underlies α-synucleinopathy. Human Molecular Genetics. 2016;25:211-22
19. Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM. et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549-62
20. Dai C, Sampson SB. HSF1: Guardian of Proteostasis in Cancer. Trends in cell biology. 2016;26:17-28
21. Meng L, Gabai VL, Sherman MY. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene. 2010;29:5204-13
22. Elsing AN, Aspelin C, Bjork JK, Bergman HA, Himanen SV, Kallio MJ. et al. Expression of HSF2 decreases in mitosis to enable stress-inducible transcription and cell survival. The Journal of cell biology. 2014;206:735-49
23. Bjork JK, Akerfelt M, Joutsen J, Puustinen MC, Cheng F, Sistonen L. et al. Heat-shock factor 2 is a suppressor of prostate cancer invasion. Oncogene. 2016;35:1770-84
24. Saju JM, Hossain MS, Liew WC, Pradhan A, Thevasagayam NM, Tan LSE. et al. Heat Shock Factor 5 Is Essential for Spermatogenesis in Zebrafish. Cell reports. 2018;25:3252-61.e4
25. Li G, Wang Z, Zhang C, Liu X, Cai J, Wang Z. et al. Molecular and clinical characterization of TIM-3 in glioma through 1,024 samples. Oncoimmunology. 2017;6:e1328339
26. Wang Z, Zhang C, Liu X, Wang Z, Sun L, Li G. et al. Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology. 2016;5:e1196310
27. Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E. et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast cancer research: BCR. 2009;11:R15
28. Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome biology. 2016;17:218
29. Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature communications. 2013;4:2612
30. Shah N, Wang P, Wongvipat J, Karthaus WR, Abida W, Armenia J. et al. Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. eLife. 2017 6
31. Alonso MH, Aussó S, Lopez-Doriga A, Cordero D, Guinó E, Solé X. et al. Comprehensive analysis of copy number aberrations in microsatellite stable colon cancer in view of stromal component. British Journal of Cancer. 2017;117:421-31
32. Jia D, Li S, Li D, Xue H, Yang D, Liu Y. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging. 2018;10:592-605
33. Ojlert AK, Halvorsen AR, Nebdal D, Lund-Iversen M, Solberg S, Brustugun OT. et al. The immune microenvironment in non-small cell lung cancer is predictive of prognosis after surgery. Molecular oncology. 2019;13:1166-79
34. Wang L, Zhu B, Zhang M, Wang X. Roles of immune microenvironment heterogeneity in therapy-associated biomarkers in lung cancer. Seminars in cell & developmental biology. 2017;64:90-7
35. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B. et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nature reviews Cancer. 2019;19:9-31
36. Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C. et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169:750-65.e17
37. Qin H, Wang F, Liu H, Zeng Z, Wang S, Pan X. et al. New advances in immunotherapy for non-small cell lung cancer. American journal of translational research. 2018;10:2234-45
38. Chalmel F, Lardenois A, Evrard B, Mathieu R, Feig C, Demougin P. et al. Global human tissue profiling and protein network analysis reveals distinct levels of transcriptional germline-specificity and identifies target genes for male infertility. Human reproduction (Oxford, England). 2012;27:3233-48
39. Barna J, Csermely P, Vellai T. Roles of heat shock factor 1 beyond the heat shock response. Cellular and molecular life sciences: CMLS. 2018;75:2897-916
40. Suzuki K, Kachala SS, Kadota K, Shen R, Mo Q, Beer DG. et al. Prognostic immune markers in non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:5247-56
41. Catacchio I, Scattone A, Silvestris N, Mangia A. Immune Prophets of Lung Cancer: The Prognostic and Predictive Landscape of Cellular and Molecular Immune Markers. Translational oncology. 2018;11:825-35
42. Doroshow DB, Sanmamed MF, Hastings K, Politi K, Rimm DL, Chen L. et al. Immunotherapy in Non-Small Cell Lung Cancer: Facts and Hopes. Clinical cancer research: an official journal of the American Association for Cancer Research. 2019
Author contact
![]() Corresponding author: Xiayi Lv, Department of Thoracic Surgery, The First Affiliated Hospital of Zhejiang University, Qingchun Road 79, Hangzhou, China, 310009. E-mail: lyuxiayiedu.cn.
Corresponding author: Xiayi Lv, Department of Thoracic Surgery, The First Affiliated Hospital of Zhejiang University, Qingchun Road 79, Hangzhou, China, 310009. E-mail: lyuxiayiedu.cn.

 Global reach, higher impact
Global reach, higher impact