Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(2):419-431. doi:10.7150/ijms.49730 This issue Cite
Research Paper
LncRNA XIST acts as a MicroRNA-520 sponge to regulate the Cisplatin resistance in NSCLC cells by mediating BAX through CeRNA network
1. Department of Pulmonary and Critical Care Medicine, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan 250012, China.
2. Department of Pulmonary and Critical Care Medicine, Qilu Hospital of Shandong University, Jinan 250012, China.
Received 2020-6-20; Accepted 2020-11-9; Published 2021-1-1
Abstract
Background: In recent years, LncRNA acts as a member of competing endogenous RNA (ceRNA), playing an important role in drug resistance of lung cancer. The aim of this study was to identify potential biomarkers about cisplatin resistant lung cancer cells using a comprehensive ceRNA network.
Methods: GSE6410 (GPL-201) analyzed gene expression changes about cisplatin resistance in A549 NSCLC cells. GSE43249 (GPL-14613) included noncoding RNA expression profiling derived from the cisplatin resistant A549 lung cells. GEO2R, an online analysis tool, analyzed the differentially expressed mRNAs and miRNAs (DEmRNAs and DEmiRNAs). To explore the functional enrichment implication of differentially expressed mRNAs, we used the GO (Gene ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis. Through miRDB, Targetscan, Starbase and miRWalk, we found targeted miRNAs. The Kaplan-Meier curve method was used to show clinical survival analysis of targeted RNAs (P<0.05). The Starbase database predicted potential lncRNAs mediated targeted miRNAs. Eventually, the novel ceRNA network of lncRNAs, miRNAs, mRNA was constructed by cytoscape3.7.2.
Results: 118 differentially expressed mRNAs were the basis of the mediated ceRNA network. DAVID and Kaplan-Meier picked out BAX, an apoptosis regulator. Venn diagram demonstrated 8 miRNAs commonly regulating BAX. Starbase predicted lncRNA XIST mediated miRNAs. Finally, lncRNA XIST may be a useful biomarker regulating cisplatin resistance in lung cancer cells and further, we explored the BAX may effect tumor-infiltrating immune cells.
Conclusions: LncRNA XIST competitively bound to miRNA 520 in the regulation of cisplatin resistance by BAX, participating apoptosis in the p53 signaling pathway.
Keywords: ceRNA, LncRNAXIST, biomarker, BAX, apoptosis
Introduction
Lung cancer is considered to have a significant role in cancer-related deaths worldwide. It is well known that NSCLC comprises approximately 85% of all lung cancers and lung adenocarcinoma (LAC) accounts for 40% of all lung cancers all over the whole world [1].
As for NSCLC, many treatment strategies are effective and include surgical operation, chemotherapy and radiotherapy. NSCLC patients, who are treated with cisplatin, also termed cisdiamminedichloroplatinum (CDDP) or diamminedichloro platinum (DDP); may develop chemoresistance [2]. Cisplatin has been adopted for about 30 years [3], primarily acting by causing DNA damage [4]. Chemoresistance is a major problem for cancer therapy [5]. There are many resistant mechanisms examined in cancer cells, such as the P53 signaling pathway, apoptosis [6] and the cell cycle. At present, chemotherapy has been limited due to resistance [7]. However, relative biomarkers are considered absent [8]. Thus, it is high time that we should establish effective network of biomarkers to predict gene changes in cisplatin resistant NSCLC cells.
LncRNAs have been considered as oncogenes mediating tumorigenesis and chemoresistance. They may have lower expression and exist in the cytoplasm or nucleus [9]. They inhibit effects on miRNAs and mRNAs [10]. In different kinds of cancers, a large number of lncRNAs are explored to mediate cellular processes and drug-resistance, such as NSCLC, ovarian cancer [11], gastric cancer [12], pancreatic cancer [13], and breast cancer [14]. LncRNA acts as a member of competing endogenous RNAs (ceRNAs) by competitively binding targeted microRNAs (miRNAs) [15, 16]. In our study, lncRNA XIST locates on chromosome 8q24.21 and combines with the microRNA-520 to regulate the cisplatin resistance by mediating BAX through ceRNA network.
BAX [17] (ENSG00000087088), an apoptosis regulator molecule [18], is located on the chromosome 19, with 13 transcripts. BAX is a member of Bcl2 family [19]. DAVID database and Kaplan-Meier curve were used to analyze the GSE6410 DEmRNAs. Next, through four databases, miR-525-5P, miR-4640-3p, miR-214-3P, miR-520a-5p all regulated the BAX. Comparing with GSE43249 (GPL-14613), we decided to explore miR-520 (survival curve P<0.01). In this present novel, the ceRNA network of lncRNA-miRNA-mRNA about cisplatin resistance in NSCLC was created through RNA sequencing data from the GEO database.
Materials and Methods
Data collection and microarray analysis
We used GSE6410 (6 samples mRNAs) and GSE43249 (6 samples miRNAs) [20] about cisplatin resistant non-small cell lung cancer (NSCLC) cells from the GEO database [20] (https://www.ncbi.nlm.nih.gov/gds/?term=). The inclusion criteria were as follows. Firstly, our goal was to find mRNA and noncoding RNA expression changes about cisplatin resistance in A549 NSCLC cells from GEO database. Through some key words, such as mRNA, noncoding RNA, cisplatin, lung cancer, we screened targeted datasets. Next, for better comparison, we used data including A549 cisplatin-sensitive and cisplatin-resistant non-small cell lung cancer cell lines. The exclusion criteria were incomplete data, which didn't have control group. First, DEmRNAs were analyzed by online software GEO2R. There were 8793 DEmRNAs in GSE6410, and |Log 2 FC|>1 and the P <0.05 [21] were two screening criteria. These 118 DEmRNAs were the basis of ceRNA network. GraphPad-Prism 8.4.0 (https://www. graphpad.com) [22] was used to show volcano maps and heat maps. To normalize the samples, we showed Box diagram vividly. The Independent two-samples T-test proved that samples were representative by SPSS Statistics 17.0.
Functional enrichment analysis of DEmRNAs
The DAVID database 6.8 (https://david.ncifcrf.gov/) [23] and Metascape (http://metascape.org) [24] were used to find the function and pathways of 118 DEmRNAs. As to GO-BP (biological process), sorted by P value <0.05, 5 counts (CDKN1A, BTG2, BAX, MDM2, and GADD45A) were most significant in participating DNA damage response, p53 signaling pathway. To GO-CC (Cellular component), calcium channel complex was numerous. Steroid binding accounted most in the GO-MF (molecular function). All genes in the genome have been used as the enrichment background. Through the Kaplan Meier-plotter software (https://kmplot.com) [24], we finally decided to study the targeted mRNA BAX, an apoptosis regulator molecule, which participated in the P53 signaling pathway [25].
Explore potential miRNAs
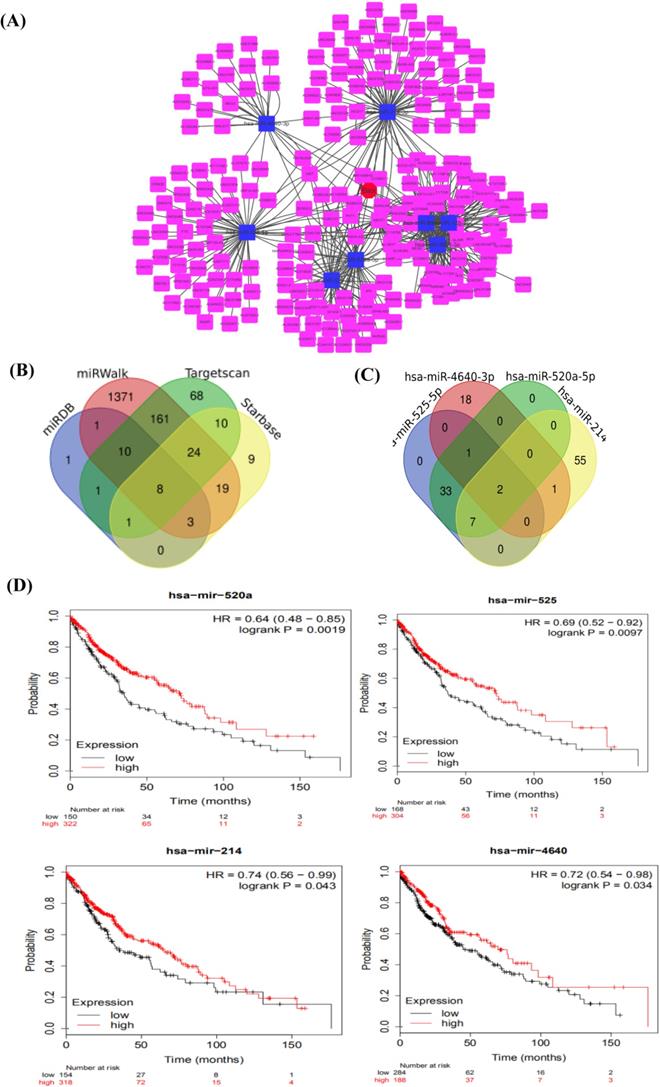
According to the targeted molecule BAX, we found miRNAs, which commonly regulated BAX from miRDB [26] (http://www.mirdb.org), Targetscan [26] (http://www.targetscan.org), Starbase [27] (https://starbase.sysu.edu.cn/), miRWalk (http://mirwalk.umm.uniheidelberg.de/) [28]. Then, we showed 8 miRNAs regulating BAX in only four databases by Venn software, such as hsa-miR-3681-3p, hsa-miR-766-5p, hsa-miR-525-5p, hsa-miR-4640-3p, hsa-miR-128-3p, hsa-miR-216a-3p, hsa-miR-520a-5p, hsa-miR-214-3p. But these miRNAs were not all related to the survival of patients. Later, 4miRNAs (hsa-miR-4640, hsa-miR-520a-5p, hsa-miR-214-3p, hsa-miR-525-5p) were obviously related to overall survival [29] (P<0.05) by the Kaplan Meier plotter software (https://kmplot.com) [24].
The discovery of LncRNA XIST in cisplatin resistant NSCLC cells
Identification of potential lncRNA about 4 miRNAs (hsa-miR-4640, hsa-miR-520a-5p, hsa-miR-214-3p, and hsa-miR-525-5p) was the critical step. The Starbase database (https://starbase.sysu.edu.cn/) [27] was used to separately predict potential lncRNAs for 4 miRNAs. Two lncRNAs XIST, MIR29B2CHG were found to regulate common 4 miRNAs by the Dram-Venn-Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) [30] database. According to the present articles, lncRNA XIST has some researches in the cisplatin resistant NSCLC cells. The expression of lncRNA-XIST was significantly higher in NSCLC tumor tissues and cisplatin resistant A549 LAC cells [31]. At the moment, research has found that lncRNA XIST was upregulated and induced chemoresistance by combing with miR-29c participating in the DNA repair pathway [32]. However, the ceRNA network mediated by lncRNA XIST remained poorly understood. Thus, our study explored lncRNAXIST to predict the prognosis of NSCLC patients. This molecule may be a potential target for the cisplatin resistant lung cancer cells.
Construction of lncRNAs-miRNAs-mRNAs network in cisplatin resistant NSCLC cells
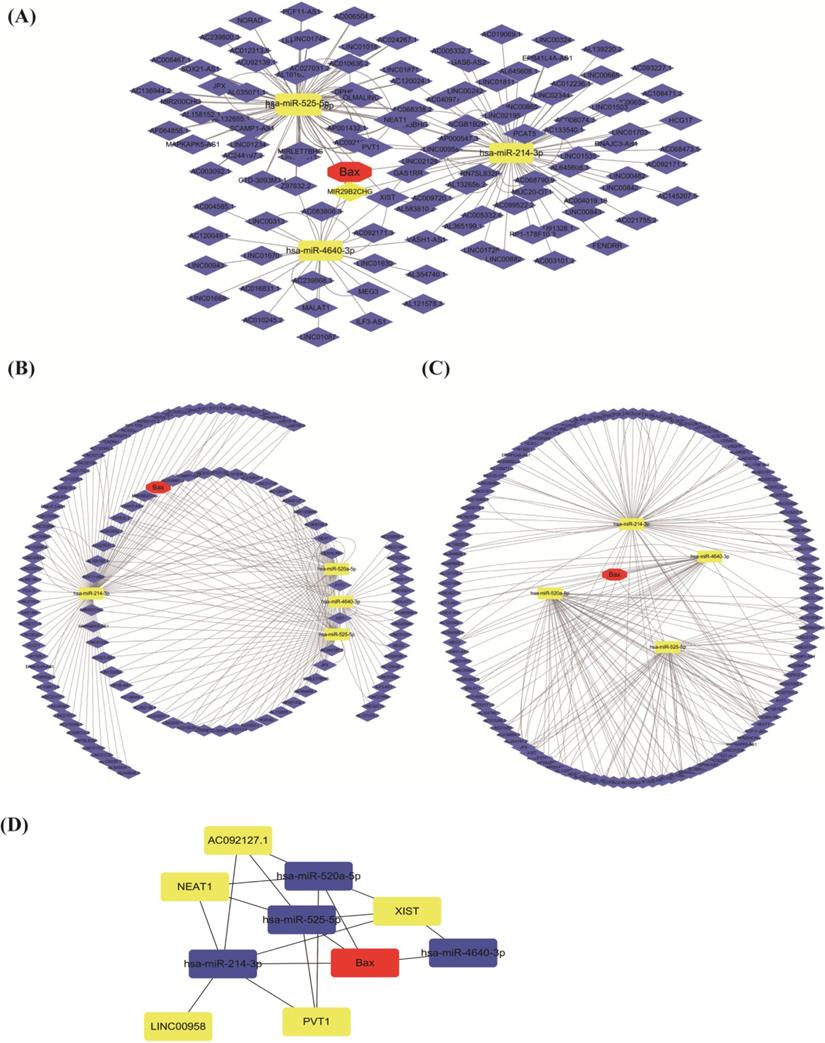
PPI networks can provide information on the molecular mechanism. The same to us, lncRNA-miRNAs-BAX network were established by cytoscape3.7.2 software obviously (http://www.cytoscape.org/) [24]. Every type of RNAs all represented different nodes and the relationships between these genes were considered to be edges. In this circumstance, BAX, 4miRNAs, lncRNA XIST were all visualized in this ceRNA family. LncRNAXIST regulated the miRNAs by BAX, participating in P53 signaling pathway in cisplatin resistant NSCLC (Figure 1).
Database validation of the most likely miRNA
Contrarily, through the database miRcode [33] (http://www.mircode.org), we predicted potential miRNA by lncRNA XIST. An miRNA family including miRNA 520 and miRNA 214 were also expressed in the ceRNA network. To recognize the differentially expressed miRNAs, GEO2R, an online software was used to conclude the primary data in GSE43249 (P value <0.05 and |Log2FC| >1). Besides, according to the differentially expressed miRNAs in the GSE43249 (GPL14613), we found out miRNA520 in NSCLC. This further proved the importance of miRNA520 in the cisplatin resistant NSCLC cells. Thus lncRNA XIST, miRNA520, BAX were considered to be the key molecules, mediating the apoptosis in the P53 signaling pathway.
Results
Differentially expressed mRNAs in NSCLC
Through comparison and screening, we chose GSE6410 (GPL201) including mRNAs about cisplatin resistant cancer cells. GEO2R, an online software analyzed 8793 DEmRNAs from the samples (absolute Log Fold change>1 and p-value <0.05). There were 118 DEmRNAs, including 80 up-regulated genes, 38 down-regulated genes. In order to show DEmRNAs, we used the volcano maps and heat maps (Figure 2A, 2B). We chose top 25 down regulated and up regulated mRNAs (Table 1).
Enrichment Analysis and pathway of DEmRNAs
We used the Metascape (http://metascape.org), an online software, to figure out the functions and pathways of 118 DEmRNAs. The most significant term was shown to represent the cluster (Figure 2C). The p53 signaling pathway had numerous counts, including the targeted biomarker BAX. To show pathways of DEmRNAs, the enriched terms were constructed as a network. The different colors were classified by cluster ID, and the common cluster ID was closely related to each other (Figure 2D).
The detailed process of constructing a ceRNA network about lncRNAs- miRNAs - mRNA.
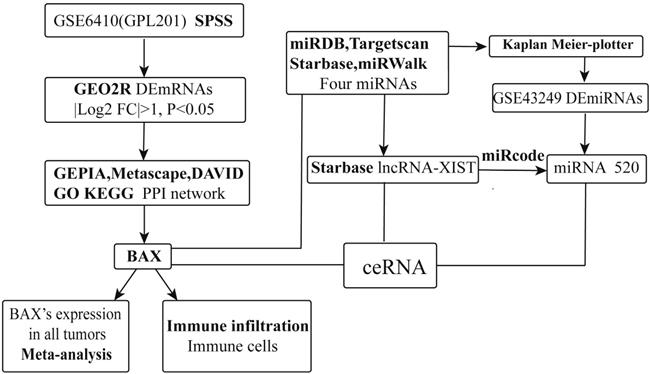
The map and enrichment analysis of DERNAs. (A) Volcano map of differentially expressed mRNAs in cisplatin sensitive-resistant NSCLC, red represents up-regulated mRNAs,  is the most obvious in mRNAs,
is the most obvious in mRNAs,  is considered to be down-regulated significantly; (B) Heat map of DEmRNAs in NSCLC. The red represents more expression in samples. (C) GO enrichment results of 114 (118) DEmRNAs, P53 pathway is remarkable. The size of the rectangle represents the number of enrichment analysis participants. (D) Network of enriched terms: colored by cluster ID, the red is the molecule BAX. All nodes are typically close to each other.
is considered to be down-regulated significantly; (B) Heat map of DEmRNAs in NSCLC. The red represents more expression in samples. (C) GO enrichment results of 114 (118) DEmRNAs, P53 pathway is remarkable. The size of the rectangle represents the number of enrichment analysis participants. (D) Network of enriched terms: colored by cluster ID, the red is the molecule BAX. All nodes are typically close to each other.
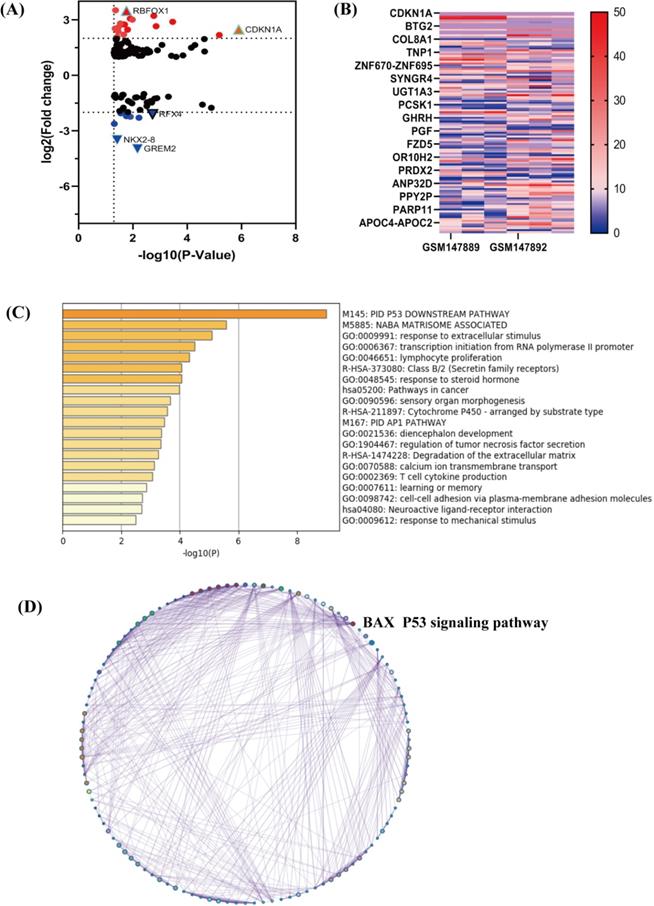
At the same time, the top 5 terms of enrichments were listed as follows (Table 2). Through the GO and KEGG analysis, the terms of P53 signaling pathway were typical. DEmRNAs may take part in this pathway to enhance cisplatin resistance in NSCLC cells. We selected the terms having the best p-values from each of the 20 clusters to show the relationships between the terms. Through the Cytoscape, we constructed the network where each node was an enriched term. Firstly, the network was colored by cluster ID (Figure 3A) and then by p-value (Figure 3B). We can identify that DEmRNAs were interrelate with each other. PPI networks can provide information on the molecular mechanism underlying cellular activity. In our study, a PPI network of differentially expressed mRNAs in NSCLC provided 4 mRNAs (CDKN1A, BAX, MDM2, GADD45A), which were associated with each other tightly (Figure 3C). They were all key biomarkers in the P53 signaling pathway.
The differentially expressed mRNAs in the NSCLC, top 25 down and up regulated mRNAs
| Gene.symbol | logFC | P-value |
|---|---|---|
| IGLL1 | -1.5812651 | 0.00002736 |
| KCNMA1 | -1.1513622 | 0.00048796 |
| COL15A1 | -1.2280541 | 0.00129318 |
| SEPT5-GP1BB | -1.2577165 | 0.00158965 |
| PPBP | -1.4687682 | 0.00202229 |
| RFX4 | -2.0975544 | 0.00189187 |
| SLIT3 | -1.387397 | 0.0022444 |
| NR1H4 | -1.4536816 | 0.00299161 |
| F2RL3 | -1.6589632 | 0.00308639 |
| GMDS | -1.0053824 | 0.00390183 |
| ATXN1 | -1.1879763 | 0.00431833 |
| HTR7 | -1.8627858 | 0.00541648 |
| IL12A | -2.289003 | 0.00596437 |
| REG1CP | -6.3655607 | 0.00114347 |
| ZBTB48 | -1.1855046 | 0.00714396 |
| GREM2 | -3.9641684 | 0.00679631 |
| UGT1A3 | -1.1995722 | 0.00804717 |
| SLC25A21 | -1.5112204 | 0.01079803 |
| HTN1 | -2.2367784 | 0.01239841 |
| TNS1 | -1.431632 | 0.01284794 |
| IFIH1 | -1.380793 | 0.01468609 |
| CELA2B | -2.0812906 | 0.01586124 |
| IL5 | -2.1994291 | 0.0172905 |
| MYOT | -1.9866327 | 0.01536891 |
| CDKN1A | 2.4718188 | 0.00000126 |
| FAS | 2.1834746 | 0.00000654 |
| SESN1 | 1.289292 | 0.00002265 |
| FDXR | 1.965755 | 0.00002315 |
| PPM1D | 1.6388142 | 0.00005177 |
| BTG2 | 1.4970197 | 0.00006986 |
| DDB2 | 1.0777424 | 0.00014334 |
| CYP39A1 | 1.0008984 | 0.00025609 |
| XPC | 1.056704 | 0.00037856 |
| MDM2 | 1.6441189 | 0.00084947 |
| GADD45A | 1.3567254 | 0.00123572 |
| COL8A1 | 2.6565612 | 0.00139461 |
| CYP4F11 | 2.8957083 | 0.00033948 |
| TNP1 | 1.2582715 | 0.00236534 |
| ATF3 | 1.3156861 | 0.0024596 |
| RGS9 | 1.2250779 | 0.00281468 |
| BAX | 1.4006089 | 0.00303574 |
| ZNF670-ZNF695 | 1.1936179 | 0.00394263 |
| ADGRE2 | 1.3658147 | 0.00435895 |
| NRXN3 | 1.0653136 | 0.00465841 |
| RYR1 | 1.4004822 | 0.00621905 |
| SYNGR4 | 1.0586675 | 0.00599735 |
| MLNR | 3.2174996 | 0.00169798 |
| FOXE3 | 1.4357355 | 0.00719558 |
| SPIN2A | 1.4427327 | 0.00800466 |
Identification of targeted molecule BAX
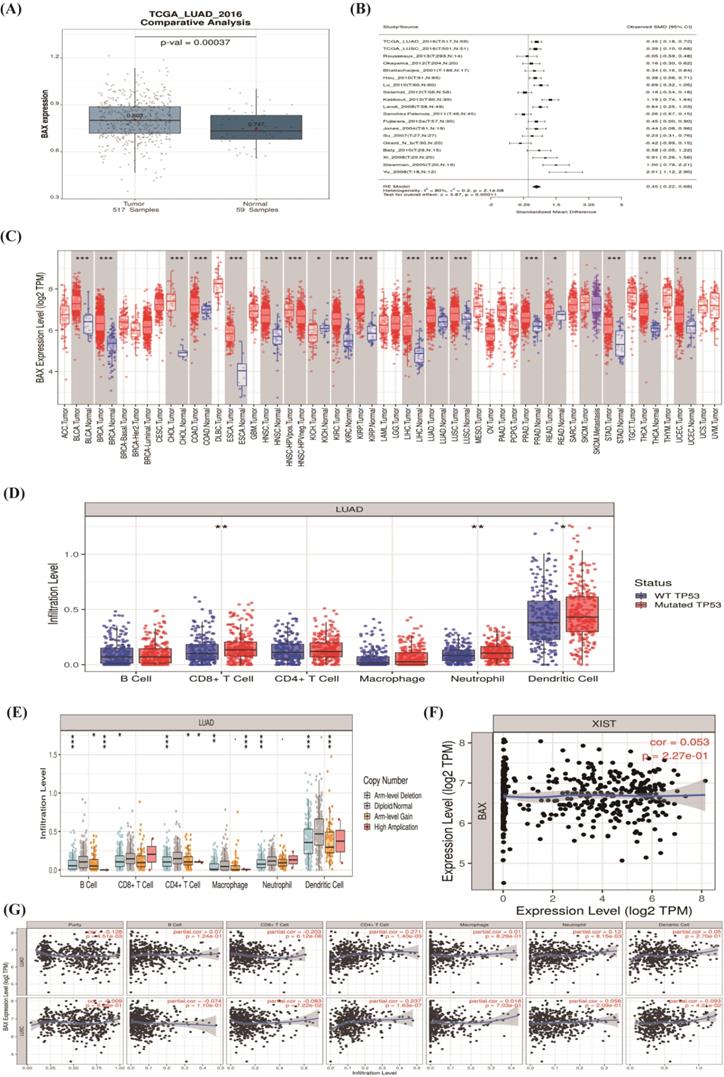
Through the function enrichment analysis and pathway, we found that the counts in the P53 signaling pathway were most typical. Four mRNAs, CDKN1A, BAX, MDM2, and GADD45A; attracted our interest deeply. BAX may regulate the cisplatin resistance in NSCLC. We utilized a t-test to illustrate that the samples were representative (P-value=0.013) by SPSS17.0 [34]. We used the box diagram to prove that GSE6410 was normalized (Figure 4A). In the NSCLC, the expression of BAX was highly showed in resistant lung cancer cells (Figure 4B). The Kaplan Meier-plotter software (https://kmplot.com) was used to find the survival analysis of BAX (P<0.01) (Figure 4C). By the GEPIA database (http://gepia.cancer-pku.cn/index.html), BAX's expression Profiling was higher in the LAC and LUSC cancer cells (Figure 4D, 4E). As for cancers, different stages represent people's prognosis. Through our study, we proved that BAX had little impact on patients' staging (Figure 4F). Some genes which were similarly with BAX, maybe up-regulated or down-regulated on chromosome (Figure 4G). From the graph, some genes were considered to be over-expressed typically on the chromosome 14.
Representative enriched terms."Log10(P)" represents the value of different terms
| GO | Category | Description | Log10(P) |
|---|---|---|---|
| M145 | Canonical Pathways | PID P53 DOWNSTREAM PATHWAY | -8.99 |
| M5885 | Canonical Pathways | NABAMATRISOME ASSOCIATED | -5.58 |
| GO:0009991 | Biological Processes | Response to extracellular stimulus | -5.09 |
| GO:0006367 | Biological Processes | RNA polymerase II promoter | -4.5 |
| GO:0046651 | Biological Processes | Lymphocyte proliferation | -4.32 |
Searching of potential lncRNAXIST-miRNAs-BAX
As is shown above, the BAX may be a useful molecule to cisplatin resistance. We chose the BAX as the important molecule to find miRNAs through different databases, including miRDB (http://www.mirdb.org); Targetscan (http://www.targ-etscan.org); Starbase (https://starbase.sysu.edu.cn/); and, miRWalk (http://mirwalk.umm.uni-heidelberg.de/). 8 miRNAs, miR-3681-3p, miR-766-5p, miR-525-5p, miR-4640-3p, miR-128-3p, miR-216a-3p, miR-520a-5p, and miR-214-3p were expressed commonly by the Dram-Venn-Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) (Figure 5B). The ceRNA network of lncRNA XIST-8 miRNAs-BAX were showed in the map (Figure 5A) and we used the cytoHubba to choose the Hub gene. In the NSCLC, we predicted the survival of 8 miRNAs by Kaplan-Meier-Plotter, but only 4 miRNAs significantly affected quality of people's life and longevity (Figure 5D). Using the starbase, we found lncRNAs of these 4 miRNAs and selected out together expressed lncRNA (Figure 5C). LncRNA XIST and MIR29B2CHG may be the targeted ceRNAs. Combining with some articles reported in recent years, lncRNA XIST has some researches in cisplatin sensitive-resistant NSCLC cells. We considered that lncRNA XIST may act as a miRNA sponge to lead cisplatin resistance. At the same time, we combined the GSE43249's differentially expressed miRNAs, 2 miRNAs were included, such as miR-520, miR-525. miRcode database was used to verify miRNAs from lncRNA XIST. MiR-520, miR-525 were targeted miRNAs of lncRNA XIST. In the present study, lncRNA XIST acted as a member of ceRNA, competitively bound miRNA520 to regulate the BAX, apoptosis associated X, in the P53 signaling pathway.
The correlation between hub genes and immune cell infiltration
In order to verify the accuracy of the data, we used the The Cancer Genome Atlas (TCGA) database through Lung cancer explore (LCE) database (http://lce.biohpc.swmed.edu/lungcancer/index.php#page-top). The expression of BAX was higher in 517 tumor samples than 59 normal samples (P=0.0037) (Figure 6 A). In different types of lung cancer, we found BAX's expression was significant through meta-analysis (Figure 6B). At the moment, BAX expression level was commonly higher in all tumors through the TIMER database (https://cistrome.shinyapps.io/timer/) (Figure 6C). Details were given in Table 3. In recent years, the tumor immune infiltration has taken important roles in cancer. The mutation of TP53 and BAX all influence immune cells; such as the B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell (P<0.05) (Figure 6D, 6E). Our results revealed positive correlations between BAX and immune cells in LAC and LUSC (Figure 6G). LncRNA XIST and targeted molecule BAX's expressions were positive correlation (cor=0.053, P=2.27e-01) (Figure 6F).
Network of enriched terms by metascape software. (A) Colored by cluster ID, nodes are different terms. (B) Colored by p-value. The significant p-value represents that terms contain more genes. (C) PPI network and MCODE components identified in the gene lists. The red represents mRNAs have good correlation and mediate in the P53 signaling pathway, including the targeted molecule BAX.
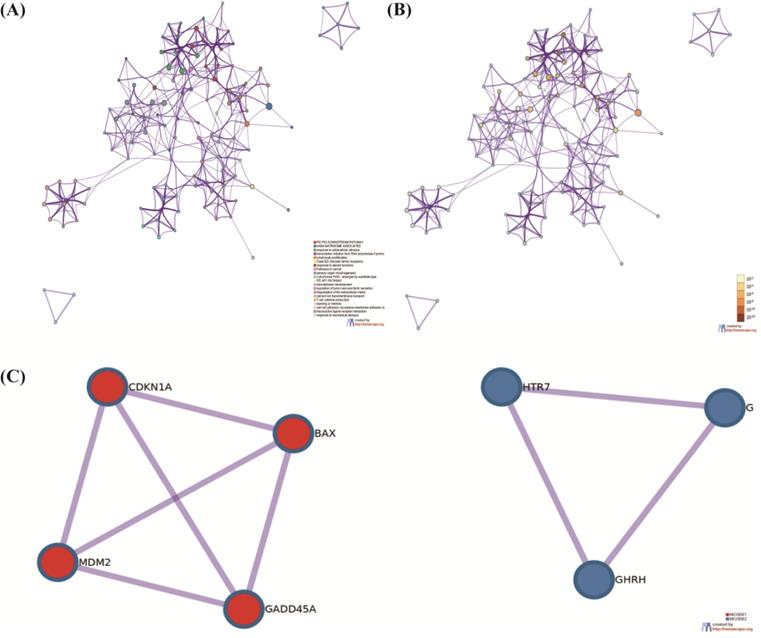
The expression of targeted molecule BAX. (A) The box diagram demonstrates the samples of cisplatin sensitive-resistant samples are standardized. (B) BAX is up-regulated obviously in cisplatin resistant NSCLC cells. Detailed research of BAX in some online databases. (C) BAX is associated with overall survival (P<0.05) in NSCLC. (D,E) The expression of BAX in LAC and LUSC is higher than normal cells. (F) BAX has little impact on patient's staging. (G) On the chromosome 14, some genes, associated with BAX are considered to be over-expressed typically.
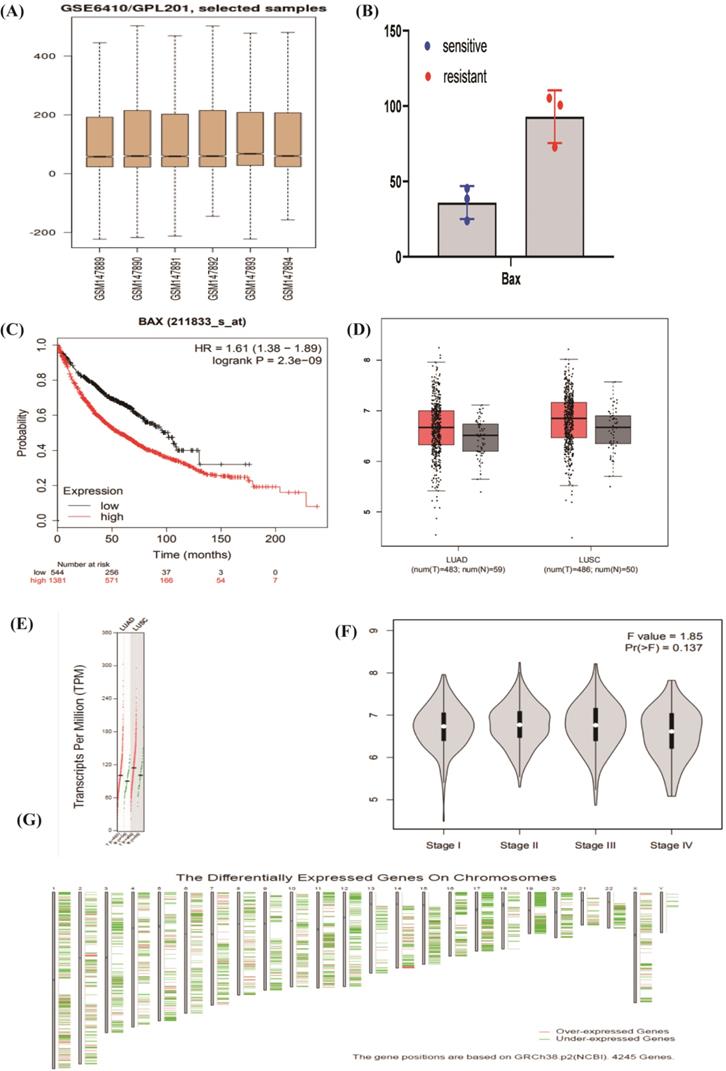
The construction of ceRNA in NSCLC. (A)The ceRNA network of lncRNA-8miRNAs-BAX, red is the BAX, blue is miRNAs, pink is lncRNAs. (B) The combination of miRNAs and lncRNAs. Four kinds of databases are mixed to explore miRNAs. (C) Looking for the common lncRNA of 4miRNAs. Two common lncRNAs are XIST and MIR29B2CHG. (D) The OS (overall survival) analysis of 4 miRNAs in NSCLC patients using Kaplan Meier curve (P<0.05).
The correlation between BAX and immune cell infiltration. (A)The expression of BAX in 517 tumor samples than 59 normal samples (P=0.0037). (B) BAX's expression was significant through meta-analysis. (C) BAX expression level was commonly higher in all tumors. (D,E) The mutation of TP53 and BAX all influence immune cells (P<0.05). (F) LncRNA XIST and targeted molecule BAX's expressions were positive correlation (cor=0.053,P=2.27e-01). (G) The positive correlations between BAX and immune cells in LAC and LUSC.
The differential expression of BAX between tumor and adjacent normal tissues across all TCGA tumors
| Tumor | Nor | p |
|---|---|---|
| BLCA.Tumor | BLCA.Normal | 2.95E-08 |
| BRCA.Tumor | BRCA.Normal | 2.70E-47 |
| CHOL.Tumor | CHOL.Normal | 4.29E-08 |
| COAD.Tumor | COAD.Normal | 0.000245234 |
| ESCA.Tumor | ESCA.Normal | 1.48E-07 |
| HNSC-HPVpos.Tumor | HNSC-HPVneg.Tumor | 5.08E-08 |
| HNSC.Tumor | HNSC.Normal | 1.35E-19 |
| KICH.Tumor | KICH.Normal | 0.01868825 |
| KIRC.Tumor | KIRC.Normal | 1.07E-30 |
| KIRP.Tumor | KIRP.Normal | 2.24E-17 |
| LIHC.Tumor | LIHC.Normal | 1.28E-22 |
| LUAD.Tumor | LUAD.Normal | 1.03E-05 |
| LUSC.Tumor | LUSC.Normal | 0.000144932 |
| PRAD.Tumor | PRAD.Normal | 6.39E-14 |
| READ.Tumor | READ.Normal | 0.010291591 |
| SKCM.Tumor | SKCM.Metastasis | 0.341891447 |
| STAD.Tumor | STAD.Normal | 4.40E-09 |
| THCA.Tumor | THCA.Normal | 1.00E-28 |
| UCEC.Tumor | UCEC.Normal | 7.93E-09 |
Discussions
Lung cancer represents a common cancer all over the world and lung adenocarcinoma (LAC) is the most numerous type, with high diversity [35]. At present, there are many treatments for different types of cancer, such as surgical operation, chemotherapy, and so on. With the development of molecule-targeted drugs, the molecule-targeted treatment of tumors has been widely agreed. However, due to various reasons, a large number of patients are not sensitive to drugs. Because of complex molecular mechanisms, multiple drug resistance may lead chemotherapeutic failure for lung cancer patients [36].
DDP is a significant chemotherapeutic drug in lung cancer. Thus, the potential mechanism of chemical resistance remains necessary to determine [37]. Therefore the further understanding of resistant theory is helpful for choosing chemotherapy drugs [38].
In recent years, lncRNAs have been explored through functional analysis [39]. More importantly, lncRNA acts as a member of ceRNA, participating in many functions in the field of cancer, such as protein modification, cell proliferation. Cell apoptosis, a classic signaling pathway, participates in cancer Proliferation and drug resistance.
Our study explored the role of potential lncRNA XIST, acting as a ceRNA of miR520 by regulating BAX involving in the P53 signaling pathway. Through the enrichment analysis of 118 DEmRNAs, we found that the targeted molecule BAX had higher expression in DDP. Four online databases were used to identify potential miRNAs mediated BAX. LncRNA XIST regulated four valuable miRNAs by starbase database.
The present findings clearly demonstrate that the expression of long non-coding RNA (lncRNA XIST) is up-regulated in lung cancer cells, and it maybe involve in the cell proliferation and TGF-β1-mediated apoptosis [38]. And lncRNAXIST may have some functions by downregulating miRNA-144. Compared with previous studies, the present study eventually constructed ceRNA [40] network of lncRNA XIST-miRNAs-BAX. We selected top 10 hub RNAs with higher degree through the cytoHubba (Figure 7A-D).
The implications of the research were providing some new treatment ideas. We found that molecules BAX, miRNA520, lncRNA XIST have been regulated closely each other. In addition, cisplatin can induce cell death by engaging endogenous apoptotic signaling that activates mitochondrial apoptosis [8]. We found that induction of p53 promoted apoptosis, autophagy, and cell sensitivity to cisplatin treatment [41]. Our study provided new insight into the mechanisms underlying chemoresistance of lung cancer [42]. Our study found an important role for LncRNA XIST in cisplatin resistance, acting as a ceRNA regulatory pathway in NSCLC that may be effective for preventing chemoresistance. In the future, researchers can use these promising targeted biomarkers to avoid cisplatin resistance.
For different tumors, the relations of long noncoding RNA (lncRNA) and microRNA influence cell process, cell growth, and drug resistance [43, 44]. We all know that miRNAs regulate their genes to perform different functions [45]. For lung cancer, miRNAs have numerous functions, such as tumor cell proliferation and progression, inflammation [46]. A previous study has showed that miR‑1284 influences apoptosis of lung tumorigenesis [47]. BAX is associated with apoptosis in cancer cells through some reports, and it participates in the classic P53 signal pathway. We predicted different molecules through individual databases. This study firstly explored 8 potential miRNAs. Later after analysis, only 4 miRNAs were associated with patients' survival (P<0.05). Combining with the GSE43249 and miRcode database, miR520 was a potential molecule by regulating BAX in cisplatin sensitive-resistant NSCLC cells. Through the TIMER database [48, 49], we explored the BAX may effect tumor-infiltrating immune cells (B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell).
Immunotherapy has achieved unprecedented success in the treatment of cancer [50]. In our research, targeted biomarkers BAX and XIST all influenced immune cells, such as the B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell (P<0.05). The interaction between these RNAs may have an important regulatory role in the immune infiltration in NSCLC, thereby affecting the patient's prognosis and immunotherapy efficacy.
The ceRNA network of lncRNA XIST-miRNAs-BAX by cytoscape. (A, B, C) The red represents the BAX, the yellow is miRNAs and the blue is lncRNAs. (D) Through the cytoHubba, we select top 10 RNAs with higher degree. Different colors show different RNAs.
Through our study, potential molecules maybe provided for DDP. In some way, this can reduce cisplatin resistance to chemotherapy and be benefit of clinical NSCLC patients. Of course, there are certain limitations in present study. These associated molecules were predicted only in theoretical aspects. At present, the corresponding experimental verification is lacking. At the same time, the research of molecular mechanism was not deep enough. More importantly, the present novel indicated that lncRNAXIST-miRNA 520-BAX influence cisplatin resistance in NSCLC cells.
Conclusion
In conclusion, BAX and LncRNA XIST were up-regulated in NSCLC patients. LncRNA XIST may act as a miRNA520 sponge by regulating BAX, associated apoptosis X, through activating in the P53 signaling pathway to affect cisplatin resistance. To summary, based on our data, this may enrich the effective therapeutic methods for cisplatin resistant NSCLC patients.
Abbreviations
LncRNA: Long non-coding RNA; GO: Gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; DAVID: The annotation, visualization and integrated discovery database; NSCLC: Non-small cell lung cancer; LAC: Lung adenocarcinoma; |Log2FC|: Absolute Log Fold change; TCGA: The Cancer Genome Atlas; LUSC: Lung squamous cell carcinoma.
Acknowledgements
Ethics Committee Approval and Patient Consent
RNA sequencing data originated from GEO database, thus, no ethics committee certification was required.
Availability of data and materials
All data are available from the sources listed in the manuscript - GEO database (https://www.ncbi.nlm.nih.gov/gds/?term=).
Author Contributions
TTL and RL, XL conceived and designed the study. XJZ, CH and JPL collected the literature. TTL draft the manuscript, YQQ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Major Scientific and Technological Innovation Project of Shandong Province (2018CXGC1212), the Science and Technology Foundation of Shandong Province (2014GSF118084), the CSCO-Qilu Cancer Research Fund (Y-Q201802-014), the Medical and Health Technology Innovation Plan of Jinan City (201805002) and the National Natural Science Foundation of China (81372333).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A. et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44(8):900-903
2. Tian LJ, Wu YP, Wang D, Zhou ZH, Xue SB, Zhang DY, Wei YG, Liu W. Upregulation of Long Noncoding RNA (lncRNA) X-Inactive Specific Transcript (XIST) is Associated with Cisplatin Resistance in Non-Small Cell Lung Cancer (NSCLC) by Downregulating MicroRNA-144-3p. Med Sci Monit. 2019;25:8095-8104
3. Rose MC, Kostyanovskaya E, Huang RS. Pharmacogenomics of cisplatin sensitivity in non-small cell lung cancer. Genomics Proteomics Bioinformatics. 2014;12(5):198-209
4. Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265-7279
5. Wang L, Shang X, Feng Q. LncRNA TATDN1 contributes to the cisplatin resistance of non-small cell lung cancer through TATDN1/miR-451/TRIM66 axis. Cancer Biol Ther. 2019;20(3):261-271
6. Espina M, Corte-Rodriguez M, Aguado L, Montes-Bayon M, Sierra MI, Martinez-Camblor P, Blanco-Gonzalez E, Sierra LM. Cisplatin resistance in cell models: evaluation of metallomic and biological predictive biomarkers to address early therapy failure. Metallomics. 2017;9(5):564-574
7. He L, Luo L, Zhu H, Yang H, Zhang Y, Wu H, Sun H, Jiang F, Kathera CS, Liu L. et al. FEN1 promotes tumor progression and confers cisplatin resistance in non-small-cell lung cancer. Mol Oncol. 2017;11(6):640-654
8. Wang A, Ning Z, Lu C, Gao W, Liang J, Yan Q, Tan G, Liu J. USP22 Induces Cisplatin Resistance in Lung Adenocarcinoma by Regulating gammaH2AX-Mediated DNA Damage Repair and Ku70/Bax-Mediated Apoptosis. Front Pharmacol. 2017;8:274
9. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775-1789
10. Yuan Y, Haiying G, Zhuo L, Ying L, Xin H. Long non-coding RNA LINC00339 facilitates the tumorigenesis of non-small cell lung cancer by sponging miR-145 through targeting FOXM1. Biomed Pharmacother. 2018;105:707-713
11. Li Z, Niu H, Qin Q, Yang S, Wang Q, Yu C, Wei Z, Jin Z, Wang X, Yang A. et al. lncRNA UCA1 Mediates Resistance to Cisplatin by Regulating the miR-143/FOSL2-Signaling Pathway in Ovarian Cancer. Mol Ther Nucleic Acids. 2019;17:92-101
12. Li M, Zhang YY, Shang J, Xu YD. LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis. Eur Rev Med Pharmacol Sci. 2019;23(10):4185-4191
13. Yin F, Zhang Q, Dong Z, Hu J, Ma Z. LncRNA HOTTIP Participates in Cisplatin Resistance of Tumor Cells by Regulating miR-137 Expression in Pancreatic Cancer. Onco Targets Ther. 2020;13:2689-2699
14. Campos-Parra AD, Lopez-Urrutia E, Orozco Moreno LT, Lopez-Camarillo C, Meza-Menchaca T, Figueroa Gonzalez G, Bustamante Montes LP, Perez-Plasencia C. Long Non-Coding RNAs as New Master Regulators of Resistance to Systemic Treatments in Breast Cancer. Int J Mol Sci. 2018 19(9)
15. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666-681
16. Yoon JH, You BH, Park CH, Kim YJ, Nam JW, Lee SK. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 2018;417:47-57
17. Fathy M, Fawzy MA, Hintzsche H, Nikaido T, Dandekar T, Othman EM. Eugenol Exerts Apoptotic Effect and Modulates the Sensitivity of HeLa Cells to Cisplatin and Radiation. Molecules. 2019 24(21)
18. Wei X, Zhou Y, Qiu J, Wang X, Xia Y, Sui L. Low expression of TUG1 promotes cisplatin sensitivity in cervical cancer by activating the MAPK pathway. J BUON. 2019;24(3):1020-1026
19. Li R, Ding C, Zhang J, Xie M, Park D, Ding Y, Chen G, Zhang G, Gilbert-Ross M, Zhou W. et al. Modulation of Bax and mTOR for Cancer Therapeutics. Cancer Res. 2017;77(11):3001-3012
20. Ruiz EJ, Diefenbacher ME, Nelson JK, Sancho R, Pucci F, Chakraborty A, Moreno P, Annibaldi A, Liccardi G, Encheva V. et al. LUBAC determines chemotherapy resistance in squamous cell lung cancer. J Exp Med. 2019;216(2):450-465
21. Li B, Wang L, Li Z, Wang W, Zhi X, Huang X, Zhang Q, Chen Z, Zhang X, He Z. et al. miR-3174 Contributes to Apoptosis and Autophagic Cell Death Defects in Gastric Cancer Cells by Targeting ARHGAP10. Mol Ther Nucleic Acids. 2017;9:294-311
22. Huang WC, Kuo KT, Wang CH, Yeh CT, Wang Y. Cisplatin resistant lung cancer cells promoted M2 polarization of tumor-associated macrophages via the Src/CD155/MIF functional pathway. J Exp Clin Cancer Res. 2019;38(1):180
23. Li Q, Zhang LY, Wu S, Huang C, Liu J, Wang P, Cao Y. Bioinformatics Analysis Identifies MicroRNAs and Target Genes Associated with Prognosis in Patients with Melanoma. Med Sci Monit. 2019;25:7784-7794
24. Zhou J, Hui X, Mao Y, Fan L. Identification of novel genes associated with a poor prognosis in pancreatic ductal adenocarcinoma via a bioinformatics analysis. Biosci Rep. 2019 39(8)
25. Kong L, Hao Q, Wang Y, Zhou P, Zou B, Zhang YX. Regulation of p53 expression and apoptosis by vault RNA2-1-5p in cervical cancer cells. Oncotarget. 2015;6(29):28371-28388
26. Zhang B, Liu Y, Zhang J. Silencing of miR-19a-3p enhances osteosarcoma cells chemosensitivity by elevating the expression of tumor suppressor PTEN. Oncol Lett. 2019;17(1):414-421
27. Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39(Database issue):D202-209
28. Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One. 2018;13(10):e0206239
29. Chiang YC, Chang MC, Chen PJ, Wu MM, Hsieh CY, Cheng WF, Chen CA. Epigenetic silencing of BLU through interfering apoptosis results in chemoresistance and poor prognosis of ovarian serous carcinoma patients. Endocr Relat Cancer. 2013;20(2):213-227
30. O'Brien RM. A consistent and general modified Venn diagram approach that provides insights into regression analysis. PLoS One. 2018;13(5):e0196740
31. Sun W, Zu Y, Fu X, Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep. 2017;38(6):3347-3354
32. Du P, Zhao H, Peng R, Liu Q, Yuan J, Peng G, Liao Y. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci Rep. 2017 37(5)
33. Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062-2063
34. Xie S, Tu Z, Xiong J, Kang G, Zhao L, Hu W, Tan H, Tembo KM, Ding Q, Deng X. et al. CXCR4 promotes cisplatin-resistance of non-small cell lung cancer in a CYP1B1-dependent manner. Oncol Rep. 2017;37(2):921-928
35. Arima C, Kajino T, Tamada Y, Imoto S, Shimada Y, Nakatochi M, Suzuki M, Isomura H, Yatabe Y, Yamaguchi T. et al. Lung adenocarcinoma subtypes definable by lung development-related miRNA expression profiles in association with clinicopathologic features. Carcinogenesis. 2014;35(10):2224-2231
36. Fang Z, Chen W, Yuan Z, Liu X, Jiang H. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed Pharmacother. 2018;101:536-542
37. Xavier CP, Pesic M, Vasconcelos MH.Understanding Cancer Drug Resistance by Developing, Studying Resistant Cell Line Models. Curr Cancer Drug Targets. 2016;16(3):226-237.
38. Wang X, Zhang G, Cheng Z, Dai L, Jia L, Jing X, Wang H, Zhang R, Liu M, Jiang T. et al. Knockdown of LncRNA-XIST Suppresses Proliferation and TGF-beta1-Induced EMT in NSCLC Through the Notch-1 Pathway by Regulation of miR-137. Genet Test Mol Biomarkers. 2018;22(6):333-342
39. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661-5667
40. Xu YH, Tu JR, Zhao TT, Xie SG, Tang SB. Overexpression of lncRNA EGFRAS1 is associated with a poor prognosis and promotes chemotherapy resistance in nonsmall cell lung cancer. Int J Oncol. 2019;54(1):295-305
41. Zhang X, Qi Z, Yin H, Yang G. Interaction between p53 and Ras signaling controls cisplatin resistance via HDAC4- and HIF-1alpha-mediated regulation of apoptosis and autophagy. Theranostics. 2019;9(4):1096-1114
42. Sun J, Pan LM, Chen LB, Wang Y. LncRNA XIST promotes human lung adenocarcinoma cells to cisplatin resistance via let-7i/BAG-1 axis. Cell Cycle. 2017;16(21):2100-2107
43. Li H, Shi H, Gao M, Ma N, Sun R. Long non-coding RNA CASC2 improved acute lung injury by regulating miR-144-3p/AQP1 axis to reduce lung epithelial cell apoptosis. Cell Biosci. 2018;8:15
44. Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H. et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74(24):7573-7582
45. Liu HN, Qie P, Yang G, Song YB. miR-181b inhibits chemoresistance in cisplatin-resistant H446 small cell lung cancer cells by targeting Bcl-2. Arch Med Sci. 2018;14(4):745-751
46. Lei L, Huang Y, Gong W. miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep. 2013;30(6):2897-2902
47. Li J, Jin H, Yu H, Wang B, Tang J. miRNA1284 inhibits cell growth and induces apoptosis of lung cancer cells. Mol Med Rep. 2017;16(3):3049-3054
48. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77(21):e108-e110
49. Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S. et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174
50. Finn OJ. A Believer's Overview of Cancer Immunosurveillance and Immunotherapy. J Immunol. 2018;200(2):385-391
Author contact
![]() Corresponding author: Yi-Qing Qu, Department of Pulmonary and Critical Care Medicine, Qilu hospital of Shandong, University, Jinan 250012, China. Tel.: +86 531 8216 9335; Fax: +86 531 8296 7544; E-mail: quyiqingedu.cn.
Corresponding author: Yi-Qing Qu, Department of Pulmonary and Critical Care Medicine, Qilu hospital of Shandong, University, Jinan 250012, China. Tel.: +86 531 8216 9335; Fax: +86 531 8296 7544; E-mail: quyiqingedu.cn.

 Global reach, higher impact
Global reach, higher impact