Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(2):398-405. doi:10.7150/ijms.48134 This issue Cite
Review
A review of linc00673 as a novel lncRNA for tumor regulation
1. Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, China.
2. NHC Key Laboratory of Carcinogenesis, and Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, Hunan, China.
3. Department of Oral and Maxillofacial Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan, China.
Received 2020-5-13; Accepted 2020-10-28; Published 2021-1-1
Abstract
Long non-coding RNAs (LncRNAs) act as regulators and play important roles in a variety of biological processes. These regulators constitute a huge information network among genes and participate in the pathophysiological process of human diseases. Increasing evidence has demonstrated that LncRNA, as an oncogene or tumor suppressor gene, is closely related to the occurrence and development of tumors. Linc00673 is a recently discovered LncRNA molecule that is dysregulated in several solid tumors. Moreover, its genetic polymorphism is believed to affect the susceptibility of a population to the corresponding cancer species. This article summarizes the role of Linc00673 in different human cancers and its molecular mechanisms with a focus on the characteristics of Linc00673 and the existing literature on it while highlighting the future research directions for Linc00673. Linc00673 has the potential to become a feasible clinical diagnostic and prognostic marker toward providing a new molecular therapeutic target for cancer patients.
Keywords: LncRNA, Diagnosis, Prognosis, SNP, Cancer
Introduction
Cancer is a malignant disease caused by the transformation of normal cells into cancer cells. The key initiation events for the transformation include oncogene activation, telomerase deletion, and aneuploidy induction [1, 2]. The early human genome project studied genes that are abundantly expressed, evolutionarily conserved, and can encode proteins. Hence, RNAs that did not encode proteins were treated as the “dark matter” of genome [3, 4]. However, the rapid development of new sequencing technologies such as tiled arrays and high-throughput sequencing has highlighted that >98% of the RNAs transcribed from genes in the human cells are non-coding RNAs [5]. Long non-coding RNAs (LncRNAs) are longer than 200 nt and are produced by RNA polymerase ΙΙ. They can regulate the gene expression at epigenetic, transcriptional, and post-transcriptional processing levels by acting on the nucleic acid molecules and proteins by cis or trans [6], and they participate in a series of cell events such as differentiation, proliferation, invasion, apoptosis, and migration [7].
Linc00673 is located on the chromosome 17q24.3 [8], at approximately 275-kb telomeric of SOX9 [9]. This LncRNA sequence contains a conserved region with significant similarity to steroid receptor RNA activator 1 (SRA1) and is hence also known as “SRA-like non-coding RNA” (SLNCR) [10]. A 2015 article [11] was the first one to grab the attention of scientists from all across the world to Linc00673. This article reported about single nucleotide polymorphisms (SNPs) associated with pancreatic cancer susceptibility in individuals of European descent; two of these SNPs are located on Linc00673 [11]. Subsequent research on Linc00673 covered diverse cancer types. In the present paper, we have summarized the correlation between Linc00673 and clinical factors in different cancer types. Simultaneously, we have focused on the molecular mechanism of Linc00673 in regulating tumors, as well as discussed the prospects of clinical application of Linc00673. Finally, we have proposed the possible future research directions for Linc00673.
Linc00673 expression in tumor tissues as a molecular marker
Over the past few years, pan-oncogenic transcriptome analysis has been performed after thorough rearrangement and relabeling of thousands of RNA sequences retrieved from The Cancer Genome Atlas (TCGA). Reportedly, LncRNAs are often dysregulated in tumors and are associated with other cancer-related genetic modifications [12]. These LncRNAs, as oncogenes or tumor suppressors, are often involved in the regulation of malignant tumors. The current clinical research on Linc00673 covers a range of cancer types, including lung cancer [13-15], pancreatic cancer [9, 16, 17], gastric cancer [18, 19], breast cancer [20-22], liver cancer [23], thyroid cancer [24, 25], prostate cancer [26], ovarian cancer [27], tongue cancer [28], and colorectal cancer [29]. Our findings suggest that the Linc00673 expression is strongly related to the baseline clinical characteristics of patients, such as tumor size, lymph node metastasis, and the TNM Classification of Malignant Tumors stage (Table 1). Moreover, a few studies used normal tissues as control to draw the receiver operating characteristic curve. As a result, the areas under the curve of Linc00673 in lung cancer [13] and pancreatic cancer [17] were found to be 0.683 and 0.6093, respectively. In addition to the tumor tissues, the corresponding trend of Linc00673 expression was also detected in the peripheral blood [17, 30]. This study report suggests that Linc00673 may be a clinically effective biomarker for early molecular diagnosis. In addition, the overall survival of patients is an effective indicator to evaluate the clinical significance of the differential expression of LincRNA. Cancer patients can be divided into 2 groups based on the expression of Linc00673: high and low. The Kaplan-Meier survival analysis revealed that the high expression of Linc00673, which acts as a pro-oncogene, is related to adverse outcomes for patients [14, 17-19, 21-24, 27-29]. The relationship between the Linc00673 expression and clinical outcomes of cancer patients suggests that the expression level may be used as a potential prognostic indicator. The expression and related clinical characteristics of Linc00673 in different cancer tissues are presented in Table 1.
Mechanism of Linc00673 in Cancers
rs11655237 of Linc00673 increases the population's susceptibility to cancer
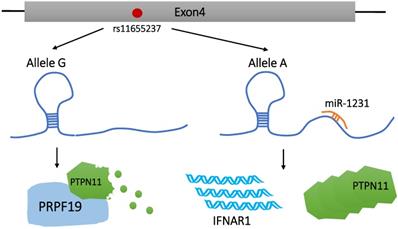
Genome-wide association study (GWAS) has identified thousands of phenotype-related DNA sequence variants toward investigating potential reasons for phenotypic differences and disease susceptibility among individuals [31, 32]. SNPs are one of the most common heritable mutations that induce DNA sequence polymorphisms at the gene level, accounting for >90% of all known polymorphisms [33]. There are 4 different variations of SNPs, namely, transitions, transversion, deletion, and insertion. Genetic mutations in protein-coding regions are often harmful or fatal, and the disease risk-related SNPs identified by GWAS are mostly located in the non-coding regions [34]. These SNPs can affect the regulation of LncRNAs via modification of the LncRNA sequence or expression level. These modifications are not immediately fatal, but can enhance susceptibility in carriers to a variety of diseases, including tumors [34, 35]. Until date, SNPs has been found to be related to susceptibility to gastric cancer [36], lung cancer [37], head and neck squamous cell carcinoma [38], breast cancer [39], colorectal cancer [40], and other malignant tumors. As illustrated in Figure 1, Linc00673, as a tumor suppressor, can increase the interaction between PTPN11 and PRPF19. However, PRPF19 belongs to the E3 ubiquitin ligase and is extremely important in promoting the degradation of PTPN11. Therefore, it inhibits the downstream carcinogenic SRC-EPK pathway and activates the STAT1-dependent tumor suppressor pathway [41].
Current studies on SNPs of Linc00673 mainly focus on rs11655237. Reportedly, rs11655237 is located on exon 4 of Linc00673 [11, 41] and 6 SNPs are involved in high-linkage disequilibrium with rs11655237 [41]. This SNP increases the susceptibility of the population to pancreatic cancer [11, 41], gastric cancer [42], hepatoblastoma [43], neuroblastoma [44, 45], and cervical cancer [46, 47] in an allele-specific manner. rs11655237 regulates the expression level of Linc00673 by forming a region that miR-1231 recognizes and binds with [41, 42, 46, 47]. Indeed, the formation of this miRNA-recognition region can be attributed to the conversion of the G allele to the A allele at rs11655237. Risk variant A blocks the normal function of Linc00673 by increasing the binding efficiency to miR-1231. As a result, Linc00673 is downregulated, PTPN11 is accumulated, and an oncogene IFNAR1 is upregulated. These changes promote the proliferation of pancreatic cancer cells and the growth of cervical tumors [41, 47]. Zhu et al. found that, among patients with cervical cancer, those carrying the A allele of rs116552337 showed worse-rate overall survival than those carrying the G allele of rs11655237 [47]. In addition, according to Haplo Reg v2, rs11655237 detected significant DNase hypersensitivity in multiple cancer cell lines and binding to transcription factors, including P300, FOXA1, FOXA2, and DNA repair protein RAD21 [11, 48]. However, this observation has not been specifically reported in the literature, which suggests that SNPs may affect the regulation of disease development by interfering with the binding of Linc00673 to transcription factors.
Interacts with RNA-binding proteins to regulate tumor development
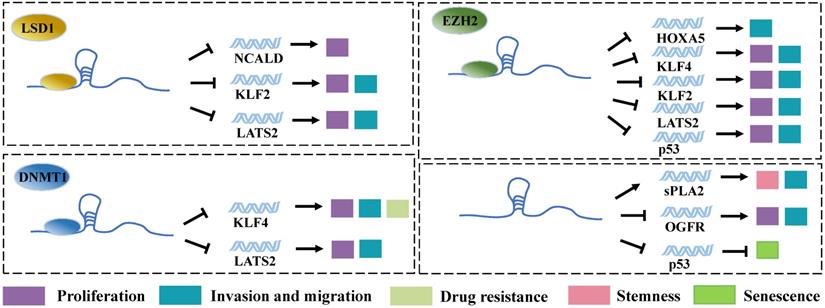
Among proteins that can be regulated by LncRNAs, transcription factors are the key ones in transcriptional regulation. In most of the typical models, gene expression control is believed to be mediated by DNA-binding proteins (DBPs). The activation of DBPs is usually regulated by signaling pathways, and their DNA-binding ability is related to sequence specificity. In contrast, RNA-binding proteins (RBPs) are often believed to be co-transcribed or post-transcribed [49]. LncRNAs exhibit different functional domains on their long sequences, which can bind to corresponding RBPs and thereby affect the stability, intracellular localization, and functioning of the respective proteins. Notably, LncRNA plays important biological functions in transcriptional regulation, epigenetics, and selective splicing through these RBPs [50]. Acting as transcription factors, SP1 [19], E2F1 [51], and YY1 [22] can bind directly to the promoter region of Linc00673 mRNA, which significantly increases the expression level of Linc00673. Moreover, the transcribed Linc00673 can bind to RBPs such as histone demethylase lysine specific demethylase 1 (LSD1) [13, 19], enhancer of Zeste Homolog 2 (EZH2) [18, 19, 24, 52], and DNA methyl-transferase 1 (DNMT1) [18, 24]. These RBPs are involved in epigenetic modifications, particularly in the methylation of genes. Thus, Linc00673 can regulate the expression of downstream genes through these 3 proteins and exert the functions of tumorigenic genes. For example, Huang et al. applied bioinformatics predictions and performed radioimmunoprecipitation assays and found that Linc00673 can bind to LSD1 and EZH2 [19]. Moreover, EZH2 and LSD1 were found to bind directly to the promoter regions of tumor suppressor genes LATS2 and KLF2, which induces the H3K27 trimethylation or H3K4 demethylation of these genes and downregulates their expression. This event ultimately promotes the proliferation, invasion, and migration of gastric cancer cells. EZH2 is a member of Polycomb Repressive Complex 2 (PRC2). This complex can inhibit the transcription of target genes by methylating the H3K27me of histone H3 [53]. DNMT1 is also a protein that is widely involved in gene methylation modification. Therefore, Linc00673 can downregulate the expression of tumor suppressor genes such as HOXA5 [52], KLF4 [18, 26], and p53 [24] through EZH2 and DNMT1. A study on lung cancer revealed that, in addition to functioning through binding proteins, Linc00673 can directly bind to the 3' untranslated region of TP53, reduce the level of p53 transcription, and cancel p53-mediated cell cycle arrest, which altogether result in the generation of cellular senescence bypass [51]. This points suggest that Linc00673 possesses certain special sequences that can directly bind to mRNA for the regulation of the expression of the target gene without needing RBPs. LSD1, another important enzyme involved in epigenetic modifications, can also assist Linc00673 in promoting tumor progression via downregulating the expression of the tumor suppressor gene NCALD [13]. In addition, the high expression level of Linc00673 is also associated with the downregulation of the expression of opioid growth factor receptor (OGFR) [27] and the upregulation of the expression of secretory phospholipase A2 (spla2) [30], although the molecular mechanisms involved in these events remain unexplored (Figure 2).
Clinical significance of Linc00673 in various cancer types
| Cancer type | Expression | Clinicopathological features | References |
|---|---|---|---|
| Lung cancer | Upregulated | Tumor size, lymphatic metastasis, TMN stage, diagnostic biomarker, ADC/SCC, poor prognosis | [13-15, 30, 52, 64] |
| Pancreatic cancer | Upregulated | Clinical stages, pathological metastasis, lower survival percent | [17] |
| Gastric cancer | Upregulated | Tumor size, advanced pathological stage, lymph node metastasis, poor prognosis, distant metastasis, TNM stage | [18, 19] |
| Mammary cancer | Upregulated | Lymph node metastasis, clinical stage, tumor size ki67 status, shorter OS | [20, 22] |
| Liver cancer | Upregulated | Advanced clinical stage, lower OS | [23] |
| Thyroid carcinoma | Upregulated | Tumor size, lymph node metastasis, worse prognosis | [24, 25] |
| Prostatic cancer | Upregulated | Tumor size, TNM stage, lymph node metastasis | [26] |
| Ovarian cancer | Upregulated | Histological subtype, FIGO stage, lymph node metastasis, shorter PFS, shorter OS, independent prognostic factors | [27] |
| Tongue cancer | Upregulated | Tumor size, invasion muscles of tongue, higher TNM stage, relapse, poor prognosis | [28] |
| Colorectal cancer | Upregulated | TNM stage, tumor size, regional lymph node metastasis, distant metastasis, lower OS | [29] |
Abbreviations: ADC/SCC, adenocarcinoma/squamous cell carcinoma; OS, overall survival; PFS, progression-free survival; FIGO, International Federation of Gynecology and Obstetrics.
Effect of rs116552337 G > A on Linc00673 function. rs116552337 G > A resulted in the formation of miR-1231-recognition binding region on Linc00673, which ultimately led to the upregulation of IFNAR1 expression and the accumulation of PTPN11.
Formation of a competitive endogenous RNA (ceRNA) network with microRNAs to regulate tumor development
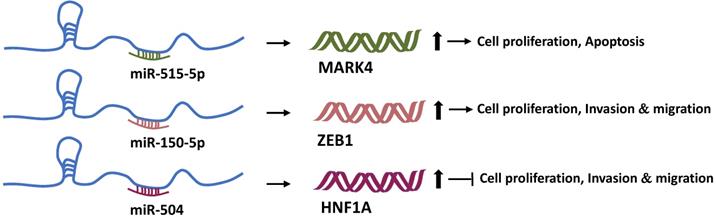
We discussed how Linc00673 regulates the transcription of downstream genes with the help of RBPs. Undoubtedly, LncRNAs can also bind to other RNA molecules based on the principle of complementary base pairing. In 2011, Pandolfi et al. of the Harvard Medical School proposed the hypothesis of ceRNA. According to this hypothesis, transcripts such as coding RNAs, pseudogenes, and non-coding RNAs can regulate each other's expression by competitively binding to “microRNA response elements” [54]. This microRNA-mediated communication method constitutes a huge regulatory network among genes. MicroRNAs (miRNAs) are known to negatively regulate the target gene expression [55]. Therefore, Linc00673 can regulate the expression of other genes through miRNAs. A past study found that Linc00673 can sponge-absorb miR-515-5p, relieve the inhibition of miR-515-5p on microtubule affinity regulating mitogen-activated protein kinase 4 (MARK4), and activate the hippo signal pathway participated by MARK4 in order to promote the proliferation of breast cancer cells [22]. In addition, Linc00673 can also competitively bind to miR-205 [23], miR-150-5p [14], and miR-504 [16]. Moreover, through miR-150-5p and miR-504, Linc00673 can regulate the expression of ZEB1 [14] and HNF1A [16], respectively, and then exert tumor-promoting or anti-tumor activity (Figure 3).
Linc00673 binds to RBPs. Linc00673 regulates the expression of downstream target gene and regulates the proliferation, migration, invasion, drug resistance, stemness, and senescence of tumor cells by binding to LSD1, E2H2, and DNMT1.
Linc00673 and mRNA competitively bind to miRNAs. Linc00673 can adsorb miR-515-5p, miR-150-5p, and miR-504, and upregulate the expression levels of MARK4, ZEB1, and HNF1A mRNA. Finally, it promotes tumor progression or inhibits tumor progression.
Binding to steroid receptors to regulate tumor progression
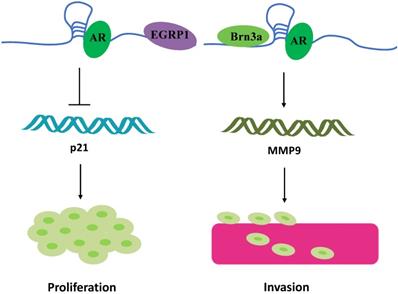
Another research direction that has attracted attention is the potential “gender preference” of Linc00673. For instance, Schmidt et al. found that Linc00673 is highly expressed in melanoma cells [10]. Meanwhile, they found a highly conserved sequence region on Linc00673 that is similar to SRA1. Brain-specific homeobox protein 3a (Brn3a) and androgen receptor (AR) bind to this conserved sequences and the adjacent sequences of Linc00673, respectively. The resultant 3-polymer model eventually promotes melanoma invasion at the transcriptional level by regulating the expression of matrix metalloproteinases 9 (MMP9) (Figure 4) [10]. Linc00673 can also recruit AR to the EGR1 genome-binding site, which is a zinc finger transcription factor, and then inhibit the expression level of p21 mRNA independent of p53, which in turn ultimately promotes the proliferation of melanoma cells (Figure 4) [56]. Therefore, blocking the combination of AR and Linc00673 can limit the invasion of melanoma mediated by Linc00673 [57]. Although Linc00673 induces AR in a ligand-independent manner, the association of AR to melanoma invasion may partially explain why male melanoma patients experience more metastasis and lower survival than the corresponding female patients. SRA1 is a bifunctional gene that produces both functional RNA SRA and corresponding proteins [58]. SRA is a non-coding RNA that specifically coactivates the transcriptional activity of steroid receptors [59, 60]. In line with its function, the expression of SRA is upregulated in steroid hormone-responsive tissue (such as in the breast, uterus, and ovary) tumors as compared with that in the normal tissues [61, 62]. Owing to the special presence of steroid hormone response tissues and the conserved sequence similar to SRA on Linc00673, Linc00673 possibly participates in the occurrence and development of cancers of the female reproductive system. Breast cancer has different subtypes, classifiable as ERα+, ERα-, and HRE2+. Abdul-rahman et al. performed a hierarchical analysis of the Affymetrix U133 Microarray data and the TCGA data set for breast cancer and found that, as compared with ER+ patients, the expression of Linc00673 in ER- patients was higher, while the high expression of LINC00673 in ER- patients was associated with adverse outcomes [21]. In addition, Linc00673 plays the role of an oncogene in ovarian cancers [27]. Based on the sequence specificity of Linc00673 and relevant past reports, Linc00673 is speculated to be involved in the regulation of tumor development through sex hormone receptors.
Linc00673 binding to steroid receptors. Linc00673 combines with AR to form trimers with EGRP1 and Brn3a, respectively, and ultimately promotes the proliferation and invasion of melanoma cells.
Linc00673 promotes the EMT process of tumor cells. The high expression of Linc00673 promotes the downregulation of E-cadherin and the upregulation of mesenchymal markers in tumor cells.
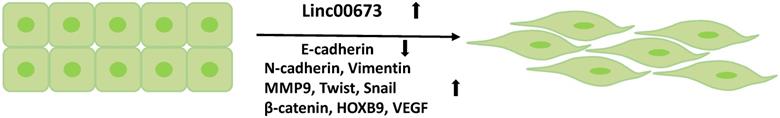
Involvement in the regulation of the epithelial-mesenchymal transition (EMT) of tumors
The EMT is associated with tumor cell invasion and metastasis. During the EMT, the expression of epithelial markers is downregulated, while that of the mesenchymal markers is upregulated. Linc00673 can promote the EMT process in tumor cells [20, 23, 25] (Figure 5) by downregulating the expression of E-cadherin and upregulating the expression of mesenchymal markers, such as N-cadherin, vimentin, snail [14], and MMP-9 [63, 64]. In addition, multiple signaling pathways are involved in the regulation of this process. Guan et al. found that Linc00673-v4-the most abundant transcription of Linc00673 in lung adenocarcinoma cells and a molecular scaffold-enhanced the interaction between DDX3 and CK1e proteins, activated the WNT/β-catenin signaling pathway, and increased the expressions of nuclear β-catenin, VEGF, Twist, HOXB9, and MMP9, which together increased the aggression of lung adenocarcinoma [64]. In addition, Linc00673 is also involved in the activation of the PI3K/AKT signaling pathway to promote glioma cell migration and invasion [65]. The PI3K/AKT signaling pathway has been implicated in the regulation of EMT in a variety of tissues, including gliomas [66-68]. Although the research reports of Linc00673 in promoting tumor progression through EMT has been well discussed, it is undeniable that Linc00673 is tissue specific. For example, in pancreatic ductal carcinoma, Linc00673 acts as a tumor suppressor gene. Arnes et al. demonstrated that silencing of Linc00673 can significantly increase the expression of MET. The upregulated MET promotes the downregulation of the epithelial markers of pancreatic cancer cells (FOXA1 and CDH1) and the upregulation of vimentin expression, which eventually promotes the EMT process and increases tumor cell migration [9].
Conclusion
Linc00673 is upregulated in several tumor tissue types; this process is accompanied with complex molecular regulatory mechanisms. Linc00673 interacts with RBPs, regulates the expression level of downstream target genes at the epigenetic level, and absorbs miRNA to participate in the construction of competitive network relationships among genes. In addition, it also participates in the regulation of SRC-EPK, WNT/β-catenin, PI3K/AKT, p53, and EMT-related signaling pathways. Finally, in such a situation, the tumor cell proliferation, migration, invasion, drug resistance, stemness maintenance, and other functions get modified. Because of its conserved sequence similar to SRA, the interaction between Linc00673 and the steroid receptor implies that an imbalanced expression of Linc00673 has a certain gender preference. This point raises a question of whether Linc00673 participates in the pathogenesis of lupus erythematosus, thyroiditis, and prostate diseases in addition to neoplastic diseases. This question deserves our consideration and further exploration. Research in prostate cancer indicates that Linc00673 is associated with resistance to paclitaxel chemotherapy drugs [26]. Moreover, Linc00673 was found to be involved in mediating the permeability of the blood-tumor barrier in the glioma as well as in promoting the killing effect of the drugs on the tumor cells [69]. Accordingly, Linc00673 may be used as a molecular target to prevent drug resistance and to provide treatment inspiration for patients resistant to chemotherapy drugs. Indeed, SNPs can also affect patients' responsiveness to chemotherapy drugs [70, 71]. Thus, whether SNPs of Linc00673 affects patients' resistance to drug treatment also needs to be considered in the future research.
Although Linc00673 is involved in the regulation of tumor progression, the biological functions of linc00673 remain to be explored. The diversification of the structure allows an RNA to perform multiple functions. For example, the maternally expressed gene 3 (MEG3) inhibits tumor growth, and all MEG3 RNA subtypes contain 3 different secondary folding motifs M1, M2, and M3 [72]. Han et al. evaluated the minimum free energy of the LncRNA AFAP1-AS1 structure and found that a stem-loop structure formed at the 91-1190 nt locus can interact with the RBP [73]. However, no researcher has focused on the structure of linc00673 until date. Therefore, it is extremely important to explore the biological functions of linc00673 in diseases by studying the structure of linc00673.
In addition to the verification of the expression level of Linc00673 in tissues, its presence in patients' blood offers a great potential to consider it as a biomarker in the real-time and dynamic monitoring of the tumor development. The proposed approach shall provide a new insight to improved clinical auxiliary diagnosis, prognosis prediction, efficacy evaluation, and individualized diagnosis and treatment. However, presently, the studies on Linc00673 are limited. The question remains whether it is present in a patient's urine, saliva, and other body fluids in addition to the peripheral blood. Moreover, whether Linc00673 participates in tumor regulation through the molecular signaling pathways related to exosomes is also worth investigating. In fact, the content of circulating tumor nucleic acid in the blood is extremely low, making it a technical challenge toward separating the free-circulating nucleic acid from the peripheral blood for enrichment proposes [74]. In addition, the level of circulating nucleic acid secreted by early tumors is low and hence cannot be monitored accurately [75]. Therefore, a convenient, economical, and sensitive detection method is necessary to promote the application of linc00673 as a “liquid biopsy” in clinical practice.
In general, as a newly discovered non-coding RNA, the precise mechanism of upstream regulation and downstream signal transduction of Linc00673 remains to be studied systematically and confirmed through in-depth research for the purpose of promoting its clinical application.
Abbreviations
LncRNAs: Long non-coding RNAs; SRA1: steroid receptor RNA activator 1; SLNCR: SRA-like non-coding RNA; SNPs: single nucleotide polymorphisms; TCGA: The Cancer Genome Atlas; GWAS: Genome-wide association study; DBPs: DNA-binding proteins; RBPs: RNA-binding proteins; LSD1: lysine specific demethylase 1; EZH2: Enhancer of Zeste Homolog 2; DNMT1: DNA methyl-transferase 1; PRC2: Polycomb Repressive Complex 2; OGFR: opioid growth factor receptor; spla2: secretory phospholipase A2; MicroRNAs: miRNAs; MARK4: mitogen-activated protein kinase 4; Brn3a: Brain-specific homeobox protein 3a; AR: androgen receptor; MMP9: matrix metalloproteinases 9; EMT: Epithelial-Mesenchymal transition; MEG3: maternally expressed gene 3.
Acknowledgements
This work was supported by grants from the Natural Science Foundation of Hunan Province (2018JJ2247) and Fundamental Research Funds for the Central South University (2019zzts319).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593-603
2. Duesberg P, Rasnick D, Li R, Winters L, Rausch C, Hehlmann R. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 1999;19:4887-906
3. Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19:R162-8
4. Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919-29
5. Diederichs S. Non-coding RNA and disease. RNA Biol. 2012;9:701-2
6. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393-407
7. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295-300
8. Tseng RC, Chang JW, Hsien FJ, Chang YH, Hsiao CF, Chen JT. et al. Genomewide loss of heterozygosity and its clinical associations in non small cell lung cancer. Int J Cancer. 2005;117:241-7
9. Arnes L, Liu Z, Wang J, Maurer C, Sagalovskiy I, Sanchez-Martin M. et al. Comprehensive characterisation of compartment-specific long non-coding RNAs associated with pancreatic ductal adenocarcinoma. Gut. 2018
10. Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ. et al. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep. 2016;15:2025-37
11. Childs EJ, Mocci E, Campa D, Bracci PM, Gallinger S, Goggins M. et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015;47:911-6
12. Acha-Sagredo A, Uko B, Pantazi P, Bediaga NG, Moschandrea C, Rainbow L. et al. Long non-coding RNA dysregulation is a frequent event in non-small cell lung carcinoma pathogenesis. Br J Cancer. 2020
13. Shi X, Ma C, Zhu Q, Yuan D, Sun M, Gu X. et al. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7:25558-75
14. Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H. et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16:118
15. Tan Q, Yu Y, Li N, Jing W, Zhou H, Qiu S. et al. Identification of long non-coding RNA 00312 and 00673 in human NSCLC tissues. Mol Med Rep. 2017;16:4721-9
16. Gong Y, Dai HS, Shu JJ, Liu W, Bie P, Zhang LD. LNC00673 suppresses proliferation and metastasis of pancreatic cancer via target miR-504/ HNF1A. J Cancer. 2020;11:940-8
17. Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res. 2018;10:2648-58
18. Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu YB. et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. 2017;8:95542-53
19. Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F. et al. Long Noncoding RNA LINC00673 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting with LSD1 and EZH2 in Gastric Cancer. Mol Ther. 2017;25:1014-26
20. Xia E, Shen Y, Bhandari A, Zhou X, Wang Y, Yang F. et al. Long non-coding RNA LINC00673 promotes breast cancer proliferation and metastasis through regulating B7-H6 and epithelial-mesenchymal transition. Am J Cancer Res. 2018;8:1273-87
21. Abdul-Rahman U, Gyorffy B, Adams BD. linc00673 (ERRLR01) is a prognostic indicator of overall survival in breast cancer. Transcription. 2018;9:17-29
22. Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang X. et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38:418
23. Zhang LG, Zhou XK, Zhou RJ, Lv HZ, Li WP. Long non-coding RNA LINC00673 promotes hepatocellular carcinoma progression and metastasis through negatively regulating miR-205. Am J Cancer Res. 2017;7:2536-44
24. Meng XF, Zhao LY, Chu XF. LncRNA LINC00673 inhibits p53 expression by interacting with EZH2 and DNMT1 in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:2075-83
25. Xia E, Bhandari A, Shen Y, Zhou X, Wang O. lncRNA LINC00673 induces proliferation, metastasis and epithelial-mesenchymal transition in thyroid carcinoma via Kruppel-like factor 2. Int J Oncol. 2018;53:1927-38
26. Jiang Z, Zhang Y, Chen X, Wu P, Chen D. Long non-coding RNA LINC00673 silencing inhibits proliferation and drug resistance of prostate cancer cells via decreasing KLF4 promoter methylation. J Cell Mol Med. 2019
27. Zheng T, Qiu J, Li C, Lin X, Tang X, Hua K. Long noncoding RNA LINC00673 promotes the proliferation and metastasis of epithelial ovarian cancer by associating with opioid growth factor receptor. Onco Targets Ther. 2019;12:6145-56
28. Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X. et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8:16621-32
29. Feng LM, Zhao DW, Li SJ, Huang J. Association of the upregulation of LncRNA00673 with poor prognosis for colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22:687-94
30. Xu W, Xu Q, Kuang D, Wang Z, Lu Q, Lin Q. et al. Long noncoding RNA SLNCR1 regulates nonsmall cell lung cancer migration, invasion and stemness through interactions with secretory phospholipase A2. Mol Med Rep. 2019;20:2591-6
31. Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA. et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet. 2017;101:5-22
32. Wang H, Lou D, Wang Z. Crosstalk of Genetic Variants, Allele-Specific DNA Methylation, and Environmental Factors for Complex Disease Risk. Front Genet. 2018;9:695
33. Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229-31
34. Gao P, Wei GH. Genomic Insight into the Role of lncRNA in Cancer Susceptibility. Int J Mol Sci. 2017 18
35. Castellanos-Rubio A, Ghosh S. Disease-Associated SNPs in Inflammation-Related lncRNAs. Front Immunol. 2019;10:420
36. Sung H, Yang HH, Hu N, Su H, Taylor PR, Hyland PL. Functional annotation of high-quality SNP biomarkers of gastric cancer susceptibility: the Yin Yang of PSCA rs2294008. Gut. 2016;65:361-4
37. O'Brien TD, Jia P, Caporaso NE, Landi MT, Zhao Z. Weak sharing of genetic association signals in three lung cancer subtypes: evidence at the SNP, gene, regulation, and pathway levels. Genome Med. 2018;10:16
38. Cardin GB, Bernard M, Bahig H, Nguyen-Tan PF, Ballivy O, Filion E. et al. Single Nucleotide Polymorphism rs6942067 Is a Risk Factor in Young and in Non-Smoking Patients with HPV Negative Head and Neck Squamous Cell Carcinoma. Cancers (Basel). 2019 12
39. Hoffman J, Fejerman L, Hu D, Huntsman S, Li M, John EM. et al. Identification of novel common breast cancer risk variants at the 6q25 locus among Latinas. Breast Cancer Res. 2019;21:3
40. Yu CY, Han JX, Zhang J, Jiang P, Shen C, Guo F, et al. A 16q22.1 variant confers susceptibility to colorectal cancer as a distal regulator of ZFP90. Oncogene. 2019
41. Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J. et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48:747-57
42. Zhao K, Zhang R, Li T, Xiong Z. Functional variants of lncRNA LINC00673 and gastric cancer susceptibility: a case-control study in a Chinese population. Cancer Manag Res. 2019;11:3861-8
43. Yang T, Li J, Wen Y, Tan T, Yang J, Pan J. et al. LINC00673 rs11655237 C>T Polymorphism Impacts Hepatoblastoma Susceptibility in Chinese Children. Front Genet. 2019;10:506
44. Zhang Z, Chang Y, Jia W, Zhang J, Zhang R, Zhu J. et al. LINC00673 rs11655237 C>T confers neuroblastoma susceptibility in Chinese population. Biosci Rep. 2018 38
45. Li Y, Zhuo ZJ, Zhou H, Liu J, Liu Z, Zhang J. et al. Additional data support the role of LINC00673 rs11655237 C>T in the development of neuroblastoma. Aging (Albany NY). 2019;11:2369-77
46. Wang Y, Luo T. LINC00673 rs11655237 Polymorphism Is Associated With Increased Risk of Cervical Cancer in a Chinese Population. Cancer Control. 2018;25:1073274818803942
47. Zhu W, Li L, Li D. Rs11655237 polymorphism of LINC00673 affects the prognosis of cervical cancer by interfering with the interaction between LINC00673 and microRNA-1231. J Cell Physiol. 2020
48. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930-4
49. Li R, Qu H, Wang S, Wei J, Zhang L, Ma R. et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34:2515-7
50. Janakiraman H, House RP, Gangaraju VK, Diehl JA, Howe PH, Palanisamy V. The Long (lncRNA) and Short (miRNA) of It: TGFbeta-Mediated Control of RNA-Binding Proteins and Noncoding RNAs. Mol Cancer Res. 2018;16:567-79
51. Roth A, Boulay K, Gross M, Polycarpou-Schwarz M, Mallette FA, Regnier M. et al. Targeting LINC00673 expression triggers cellular senescence in lung cancer. RNA Biol. 2018;15:1499-511
52. Ma C, Wu G, Zhu Q, Liu H, Yao Y, Yuan D. et al. Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget. 2017;8:32696-705
53. Healy E, Mucha M, Glancy E, Fitzpatrick DJ, Conway E, Neikes HK. et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol Cell. 2019;76:437-52 e6
54. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
55. Lai X, Eberhardt M, Schmitz U, Vera J. Systems biology-based investigation of cooperating microRNAs as monotherapy or adjuvant therapy in cancer. Nucleic Acids Res. 2019;47:7753-66
56. Schmidt K, Carroll JS, Yee E, Thomas DD, Wert-Lamas L, Neier SC. et al. The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21. Cell Rep. 2019;27:2493-507 e4
57. Schmidt K, Weidmann CA, Hilimire TA, Yee E, Hatfield BM, Schneekloth JS Jr. et al. Targeting the Oncogenic Long Non-coding RNA SLNCR1 by Blocking Its Sequence-Specific Binding to the Androgen Receptor. Cell Rep. 2020;30:541-54 e5
58. Yan Y, Cooper C, Hamedani MK, Guppy B, Xu W, Tsuyuki D. et al. The steroid receptor RNA activator protein (SRAP) controls cancer cell migration/motility. FEBS Lett. 2015;589:4010-8
59. Lanz RB, Razani B, Goldberg AD, O'Malley BW. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA). Proc Natl Acad Sci U S A. 2002;99:16081-6
60. Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A. et al. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43-7
61. Liu C, Wu HT, Zhu N, Shi YN, Liu Z, Ao BX. et al. Steroid receptor RNA activator: Biologic function and role in disease. Clin Chim Acta. 2016;459:137-46
62. Sheng L, Ye L, Zhang D, Cawthorn WP, Xu B. New Insights Into the Long Non-coding RNA SRA: Physiological Functions and Mechanisms of Action. Front Med (Lausanne). 2018;5:244
63. Wu Y, Niu Y, Leng J, Xu J, Chen H, Li H. et al. Benzo(a)pyrene regulated A549 cell migration, invasion and epithelial-mesenchymal transition by up-regulating long non-coding RNA linc00673. Toxicol Lett. 2020;320:37-45
64. Guan H, Zhu T, Wu S, Liu S, Liu B, Wu J. et al. Long noncoding RNA LINC00673-v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/beta-catenin signaling. Proc Natl Acad Sci U S A. 2019;116:14019-28
65. Zhang F, Huang Y, Wang B, Zhong C, Liu X, Ding S. LINC00673 silencing inhibits cell migration and invasion by suppressing PI3K/AKT signaling in glioma. Neuroreport. 2018;29:718-22
66. Zhong C, Li X, Tao B, Peng L, Peng T, Yang X. et al. LIM and SH3 protein 1 induces glioma growth and invasion through PI3K/AKT signaling and epithelial-mesenchymal transition. Biomed Pharmacother. 2019;116:109013
67. Luo J, Yao JF, Deng XF, Zheng XD, Jia M, Wang YQ. et al. 14, 15-EET induces breast cancer cell EMT and cisplatin resistance by up-regulating integrin alphavbeta3 and activating FAK/PI3K/AKT signaling. J Exp Clin Cancer Res. 2018;37:23
68. Meng J, Zhang XT, Liu XL, Fan L, Li C, Sun Y. et al. WSTF promotes proliferation and invasion of lung cancer cells by inducing EMT via PI3K/Akt and IL-6/STAT3 signaling pathways. Cell Signal. 2016;28:1673-82
69. Shen S, Yang C, Liu X, Zheng J, Liu Y, Liu L. et al. RBFOX1 Regulates the Permeability of the Blood-Tumor Barrier via the LINC00673/MAFF Pathway. Molecular Therapy Oncolytics. 2020;17:138-52
70. Ingle JN, Xie F, Ellis MJ, Goss PE, Shepherd LE, Chapman JW. et al. Genetic Polymorphisms in the Long Noncoding RNA MIR2052HG Offer a Pharmacogenomic Basis for the Response of Breast Cancer Patients to Aromatase Inhibitor Therapy. Cancer Res. 2016;76:7012-23
71. Chen Q, Deng X, Hu X, Guan S, He M, Wang Y. et al. Breast Cancer Risk-Associated SNPs in the mTOR Promoter Form De Novo KLF5- and ZEB1-Binding Sites that Influence the Cellular Response to Paclitaxel. Mol Cancer Res. 2019;17:2244-56
72. Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y. et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939-47
73. Han M, Gu Y, Lu P, Li J, Cao H, Li X. et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Molecular Cancer. 2020;19:26
74. Burz C, Rosca A, Pop V, Buiga R, Aldea C, Samasca G. et al. Liquid biopsy challenge and hope in colorectal cancer. Expert Rev Mol Diagn. 2019;19:341-8
75. Wu J, Hu S, Zhang L, Xin J, Sun C, Wang L. et al. Tumor circulome in the liquid biopsies for cancer diagnosis and prognosis. World J Clin Cases. 2020;10:4544-56
Author contact
![]() Corresponding authors: Wei Xiong, NHC Key Laboratory of Carcinogenesis, and Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, 410078, Hunan, China. E-mail: xiongweiedu.cn; Jianjun Yu, Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410006, Hunan, China. E-mail: dryujianjuncom.
Corresponding authors: Wei Xiong, NHC Key Laboratory of Carcinogenesis, and Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, 410078, Hunan, China. E-mail: xiongweiedu.cn; Jianjun Yu, Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410006, Hunan, China. E-mail: dryujianjuncom.

 Global reach, higher impact
Global reach, higher impact