Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(13):1897-1908. doi:10.7150/ijms.46091 This issue Cite
Research Paper
Retinal dehydrogenase 5 (RHD5) attenuates metastasis via regulating HIPPO/YAP signaling pathway in Hepatocellular Carcinoma
1. Department of Oncology, First Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou (510407), China.
2. The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou 510080, Guangdong, P. R. China.
3. The Sixth Affiliated Hospital, Sun Yat-Sen University, Guangzhou 510080, Guangdong, P. R. China.
4. Department of Chinese Medicine, the Third Affiliated Hospital, Sun Yat-Sen University, Guangzhou 510630, Guangdong, P. R. China.
5. Department of Radiation Oncology, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, 250117, P. R. China.
*These authors contributed equally to this study.
Received 2020-3-17; Accepted 2020-6-18; Published 2020-7-19
Abstract
Retinal dehydrogenase 5 (RDH5) is an important enzyme in the visual cycle. Several studies have reported that the RDH family may play crucial roles in tumor prognosis. However, the role of RDH5 in tumor prognosis is still unclear. We examined the mRNA level of RDH5 by using q-PCR in hepatocellular carcinoma (HCC) and adjacent non-cancerous tissues. The proliferation rate of HCC cells was detected by MTS assay, and the invasive ability was examined by transwell and scratch wound assays. The YAP protein localization and expression were visualized by immunofluorescence in two different cell lines. CpG islands in the promoter region were predicted by using the methprimer database. Clinical characteristics of a patient cohort data came from The Cancer Genome Atlas database. RDH5 was significantly downregulated in hepatocellular carcinoma tissues, and low RDH5 expression was associated with metastasis and poor patient prognosis. Functional assays revealed that the RDH5 promoter is methylated in HCC cell lines. Moreover, overexpressing RDH5 can suppress metastasis by reversing the epithelial-mesenchymal transition (EMT) process, and RDH5 also inhibits cell proliferation in HCC cell lines. Furthermore, suppressing RDH5 can activate the Hippo/YAP signaling pathway and promote the nuclear translocation of YAP. Clinical data demonstrated that RDH5 is an independent prognostic factor in HCC. In our study, we provided the first evidence that RDH5 plays a crucial role in suppressing proliferation and metastasis, and the RDH5 promoter is methylated in hepatocellular carcinoma. And as an important regulator, RDH5 can suppress the Hippo/YAP signaling pathway. Taken together, it revealed that RDH5 might be a potential therapeutic target in HCC patients.
Keywords: Retinal dehydrogenase 5, Hepatocellular carcinoma, Metastasis, Yes-associated protein
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common primary malignancy and the third leading cause of cancer-related mortality in the world [1, 2]. In East Asia, especially China, the hepatitis B virus (HBV) infection is the main etiological factor for HCC [3]. Nowadays, metastasis is the most common reason for poor prognosis in HCC patients [4, 5]. Although surgical resection [6], liver transplantation [7, 8], and other clinical methods are used to treat HCC, a large number of patients with long distant metastasis have not opportunity for radical resection. Therefore, there is an urgent need to find valuable biomarkers and potential therapeutic targets in HCC patients.
Currently, serum alpha-fetoprotein (AFP) [9, 10], imaging technologies [11], and histology [12] are three predominant methods for HCC screening. However, clinically, the limitation of AFP testing is lacking high sensitivity and specificity, especially in patients without typical AFP changes. Imaging technologies can only effectively diagnose the disease when the tumor is huge, and it failed to detect tumors that smaller than 1 cm in diameter. Histological diagnosis depends on the availability of tissue biopsy samples, and this method carries the risk of implantation metastasis.
Retinal dehydrogenase 5 (RDH5) belongs to the RDH enzyme family, and it is an important enzyme in the visual cycle that catalyzes the oxidation of 11-cis-retinol to 11-cis-retinal [13, 14]. Human RDH5 is encoded by the RDH5 gene, which maps to chromosome 12q13-q14 and encodes a 32 kDa protein with 318 amino acids [15, 16]. According to the literature, the expression dysregulation, nonsense mutations, and DNA methylation of RDH5 could easily result in night blindness. Some reports have demonstrated RDH5 was dysregulated in many different cancers. In colorectal carcinoma, RDH5 was significantly decreased in the mRNA level, leading to diminished all-trans-retinoic acid biosynthesis and thus contributing to the progression of colorectal cancer by affecting cell growth and differentiation [17]. Exome sequencing of early gastric carcinoma (EGC) patient samples revealed that RDH5 mutations might be involved in early carcinogenesis [18]. Another study reported RDH5 were hypomethylated and overexpressed in papillary thyroid carcinoma compared to matched non-neoplastic adjacent tissues [19]. However, whether RDH5 expression is involved in HCC recurrence and metastasis remains unclear.
The Hippo pathway is an evolutionarily conserved signaling module that plays a critical role in liver size control and tumorigenesis [20, 21]. In humans, the Hippo pathway is a kinase cascade wherein macrophage-stimulating protein 1/2 (MST1/2) and large tumor suppressor kinase 1/2 (LATS1/2), it could be phosphorylated and activated, and result in the phosphorylation and inactivation of Yes-associated protein (YAP) [22]. When YAP is retained in the cytoplasm, it is degraded rapidly; when it enters the nucleus, it transcriptionally activates anti-apoptosis genes [23]. Nevertheless, the link between RDH5 and the Hippo pathway has not been reported.
In this study, we aimed to document the clinical significance of RDH5 and identify potential biomarkers for metastatic HCC patients and cell lines. Every effort was made to identify a novel therapeutic target for HCC patients.
Materials and methods
Cell lines and cell culture
The human hepatocellular carcinoma cell lines Hep 3B and SK-Hep-1 were obtained from the Institute of Biochemistry and Cell Biology (Shanghai, China) and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS at 37°C and 5% CO2. The cancer cell lines were authenticated using short tandem repeat profiling, and the cells were not cultured for more than two months.
Patients and specimens
Fifteen matched human HCC tissues and adjacent non-cancerous tissues were collected and analyzed by quantitative real-time PCR. These samples were obtained by surgical resection from patients with HCC in May and July of 2016 and stored immediately in liquid nitrogen for subsequent analysis [24]. All of our studies were approved by the committee of the Ethical Review of Research at Sun Yat-Sen University Cancer Center.
Experimental Animals
Totally 10 specific pathogen-free (SPF) C57BL/6 mice (male; weight, 23±2 g) were purchased from the Animal Experimental Center of the Guangdong province. The Diethylnitrosamine (N-nitrosodiethylamine, DEN, CAS: 55-18-5) was purchased from Sigma, and the reserpine injection was purchased from the Aladdin (R101672). All of the C57BL/6 mice accepted 25 mg/kg DEN with intraperitoneal injection in the 14 days, and another DEN injection performed in the 30 days for inducing HCC have been developed, it will take 10 months to develop HCC. The “3R” principles with humanism care to the experimental animals was performed strictly and all of our studies obtained approval from the research ethics committee at Sun Yat-Sen University.
Plasmid and small interfering RNA (siRNA) transfections
RDH5 was cloned into the pcDNA3.1 vector. All transfections were performed using Lipofectamine 2000 according to the manufacturer's instructions. The empty pcDNA3.1 vector was used as the control, pcDNA3.1-RDH5 and vector control were transfected into the SK-Hep-1 cell. The targets of the RDH5 siRNAs were 5'-AAAUGAGCUACAUCCCGCCUCAG-3' (si#1) and 5'-ACCCUGUUGGAUAUCACUGAUTT-3' (si#2). The negative control (NC) siRNA was purchased from KEYGENTEC. All transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. The gene-silencing or plasmid transfection efficacy was evaluated 48 h after transfection by qPCR and immunoblotting.
Immunoblotting
Cells were lysed in RIPA buffer containing a protease (TargetMol, CC0001) and phosphatase (TargetMol, CC0004). Protein concentrations were assessed using the BCA Protein Assay Kit (Pierce Biotechnology). Equal amounts of protein mixed with sample loading buffer (5X; Beyotime) were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). After blocking with 5% skim milk or BSA in Tris-buffered saline Tween-20 (TBST), the membranes were incubated with primary antibodies overnight at 4°C and then with horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 h. The protein bands were visualized using a chemiluminescence kit (Pierce). GAPDH was used as a control for equal protein loading. The primary antibodies included anti-RDH5 (Abcam, ab200197), anti-GAPDH (Proteintech, 60004-1-1g), anti-YAP1 (Proteintech, 13584-1-AP), and anti P-YAP antibodies (CST, 13008), anti-mouse and anti-rabbit peroxidase-conjugated secondary antibodies were used [25, 26]. All experiments were independently repeated at least three times.
Real-time quantitative PCR analysis
The expression level of each target gene was determined by RT-PCR. Total mRNA was isolated from hepatocellular carcinoma tissues using Trizol reagent (Invitrogen, CA, USA). The internal control for measuring the RDH5 expression level was GAPDH. The relative expression levels of the target genes were estimated by calculating two to the power of ΔCt (Ct of GAPDH minus Ct of the target gene), and the experiments were repeated three times [27, 28]. The following PCR primer sequences were used in our research:
- RDH5, forward, 5'-CTGTGACCAACCTGGAGAGTCT-3',
- RDH5, reverse, 5'-GATGCGCTGTTGCATTTTCAGGT-3',
- GAPDH, forward, 5'-GTCTCCTCTGACTTCAACAGCG-3',
- GAPDH, reverse, 5'-ACCACCCTGTTGCTGTAGCCAA-3'.
Immunofluorescence
For immunofluorescence analysis of cultured cells, 3×104 HCC cells were seeded on coverslips, fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature, blocked with 5% BSA in PBS + 0.1% Triton X-100 (PBST), incubated with the primary antibody at 4°C overnight, washed with PBST, and incubated with secondary antibody at room temperature. Mounting medium containing 4',6-diamidino-2-phenylindole (DAPI) was placed on the slides, and the staining was imaged using an OLYMPUS FV1000 microscope [29].
DNA methylation
Hep 3B and SK-Hep-1 cells were treated for 48 h with 0, 2, 5, and 10 mM 5-Aza. After 2 or 3 days, cells were harvested, and total RNA was extracted from the cultured cell lines. GAPDH was used as the loading control.
Cell growth curve
MTS assay was performed to determine cancer cell growth and viability. Briefly, 1000 cells/200 ml of medium were seeded into a 96-well plate (Corning) and cultured without FBS to maintain synchronization and avoid too rapid growth in one week. At various time points after seeding, the cells in each well were stained with 20 ml of MTS (Promega, G3580) for 3 h, and the OD490 was determined with a microplate reader. All experiments were independently repeated at least three times.
Scratch wound assay
Cells were digested after transfection 48 hrs and seeded in a 6-well plate. A scratch wound assay was performed by generating a wound in the center of each well in a 6-well plate with a sterile 200 µl pipette tip. The unattached cells were removed by washing with PBS, and serum-free medium or medium with 3% FBS was added. Subsequently, cells were observed with an inverted microscope at 0, 20, or 24 h.
The Cancer Genome Atlas (TCGA) data analysis
Publicly available human HCC datasets in the TCGA were analyzed. The mRNA expression profiles downloaded from the TCGA portal (https://cancergenome.nih.gov/) included 49 non-neoplastic adjacent tissue samples and 374 tissue samples. Excluding those with unavailable clinicopathological features, 307 cases were included in the cohort to analyze the clinical characteristics [24].
Statistical analysis
All data in this study were evaluated with SPSS 21.0 software (SPSS Inc., Chicago, USA) and GraphPad Prism (GraphPad software). All data are shown as the mean ± standard deviation. The results were evaluated using the Student's t-test. Survival analysis was performed using the Kaplan-Meier method and log-rank test. The relevance of the relationship between RDH5 expression and clinicopathological parameters was determined using the chi-square test. Cox proportional hazards regression models were constructed to evaluate the data, and p<0.05 indicated a statistically significant difference.
Results
RDH5 is downregulated in HCC tissues
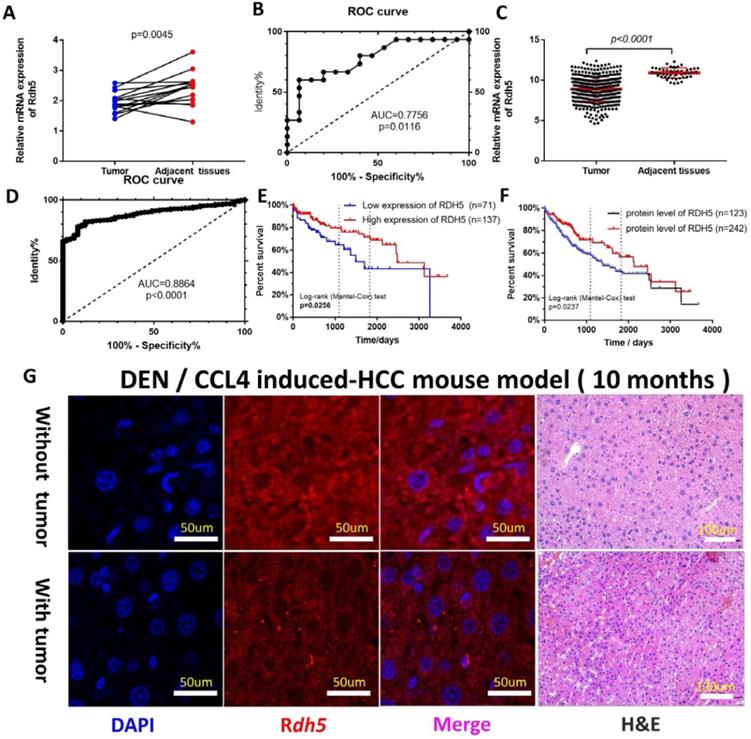
To investigate the possible role of RDH5 in HCC progression, a real-time quantitative PCR assay was used to detect gene expression in 15 paired tumor tissues and adjacent non-cancerous tissues. The results showed RDH5 was highly expressed in adjacent non-cancerous tissues than in HCC tissues (p=0.0045) (Fig. 1A). The Receiver operating characteristic (ROC) curves were used to estimate the value of RDH5 in mRNA expression level, which yielded an area under the curve (AUC) of 0.7756 (Fig. 1C). Similar results from the TCGA database was shown that the expression of RDH5 was significantly downregulated in tumor tissues (p<0.001) (Fig. 1B), with an area under the curve of 0.8864 (n=422) (Fig. 1D). Protein expression was visualized by immunofluorescence, which shown that downregulating RDH5 in DEN and CCL4 induced-mouse hepatocellular carcinoma (Fig. 1G). Therefore, we hypothesized that RDH5 plays a critical role in HCC tumorigenesis and progression.
RDH5 is down-regulation in HCC tissues and its expression is associated with overall survival. In our cohort (A) and TCGA data (B), mRNA expression of RDH5 is significantly higher in adjacent tissues than HCC tissues. The area under the curve of the ROC curve is 0.7756 in our tissue samples (C) and 0.8864 in the TCGA database (D) respectively, p<0.05. Overall survival in mRNA expression (E). or the protein level (F), by Kaplan-Meier analysis. (G) RDH5 is down-regulation in mouse HCC tissues by Immunofluorescence technique.
Correlation of RDH5 expression with clinicopathological features in HCC
| Characteristics | Cases | RDH5 expression | p value | |
|---|---|---|---|---|
| High expression | Low expression | |||
| Gender | 0.481 | |||
| Female | 93 | 54 | 39 | |
| Male | 204 | 139 | 65 | |
| Age, years | 0.230 | |||
| ≤60 | 138 | 90 | 48 | |
| >60 | 168 | 103 | 65 | |
| Child Pugh Classification | 0.324 | |||
| A | 182 | 115 | 67 | |
| B | 16 | 9 | 7 | |
| C | 10 | 9 | 1 | |
| AFP | 0.812 | |||
| Negative | 176 | 112 | 64 | |
| Positive | 130 | 81 | 49 | |
| Fibrosis | 0.794 | |||
| Non-Fibrosis | 51 | 29 | 22 | |
| portal-Fibrosis | 27 | 16 | 11 | |
| Fibrosis-speta | 21 | 16 | 5 | |
| Nodular-Fibrosis | 6 | 4 | 2 | |
| Cirrhosis | 57 | 38 | 19 | |
| Grade | 0.998 | |||
| G1 | 39 | 24 | 15 | |
| G2 | 151 | 95 | 56 | |
| G3 | 100 | 63 | 37 | |
| G4 | 11 | 7 | 4 | |
| M Stage | 0.023* | |||
| M0 | 226 | 151 | 75 | |
| M1 | 70 | 42 | 38 | |
| N Stage | 0.139 | |||
| N0 | 214 | 140 | 74 | |
| N1-2 | 87 | 49 | 38 | |
| T Stage | 0.764 | |||
| I | 160 | 97 | 63 | |
| II | 71 | 45 | 26 | |
| III | 63 | 43 | 20 | |
| IV | 11 | 7 | 4 | |
| Clinical Stage | 0.555 | |||
| I-II | 215 | 136 | 79 | |
| III-IV | 71 | 47 | 24 | |
| Vascular history | 0.639 | |||
| None | 176 | 107 | 69 | |
| Micro | 73 | 49 | 24 | |
| Macro | 11 | 7 | 4 | |
| Year of diagnosis | 0.694 | |||
| Before 2010 | 135 | 83 | 52 | |
| After 2010 | 161 | 110 | 61 | |
The data are reported as number. P-value were obtained from the chi-square test.
RDH5 is associated with the metastasis and prognosis in HCC patients
As the increased RDH5 mRNA level in adjacent non-cancerous tissues were clearly shown, it was necessary to elucidate the correlation between RDH5 expression and clinical survival. To further determine whether the downregulation of RDH5 is associated with poor prognosis in HCC patients, 307 HCC specimens were analyzed for clinicopathological features: 63% (193 of 306) of HCC patients had low RDH5 expression, and the remaining 37% (113 of 306) had high RDH5 expression. The clinical characteristics of these patients are shown in Table 1. Clinicopathological parameters included gender, age, Child-Pugh classification, AFP, fibrosis, grade, TNM stage, clinical-stage, vascular stage, and year of diagnosis. Only the M stage was relevant to RDH5 expression, and lower expression of RDH5 was associated with much more metastasis in HCC patients.
To further explore the relationship between RDH5 expression and survival, we conducted an overall survival analysis by the Kaplan-Meier method. We found out in the low RDH5 level group, the estimated 3- and 5-year OS rates were 64.62% and 43.54%, respectively, for the high RDH5 level group was 79.70% and 71.46%, respectively (Fig. 1E). Disease-free survival was not affected by RDH5 expression (data not shown). In the cohort collected from the human protein alter (THPA), the results showed that patients with high RDH5 expression had a longer mean survival time than patients with low RDH5 expression, and lower level of the protein RDH5 may be contributed to the poor prognosis in overall survival (Fig. 1F). Taken together, these analyses revealed that a high RDH5 level in HCC significantly correlated with poor patient outcomes.
RDH5 suppresses cell migration by reversing epithelial-mesenchymal transition in hepatocellular carcinoma cell lines
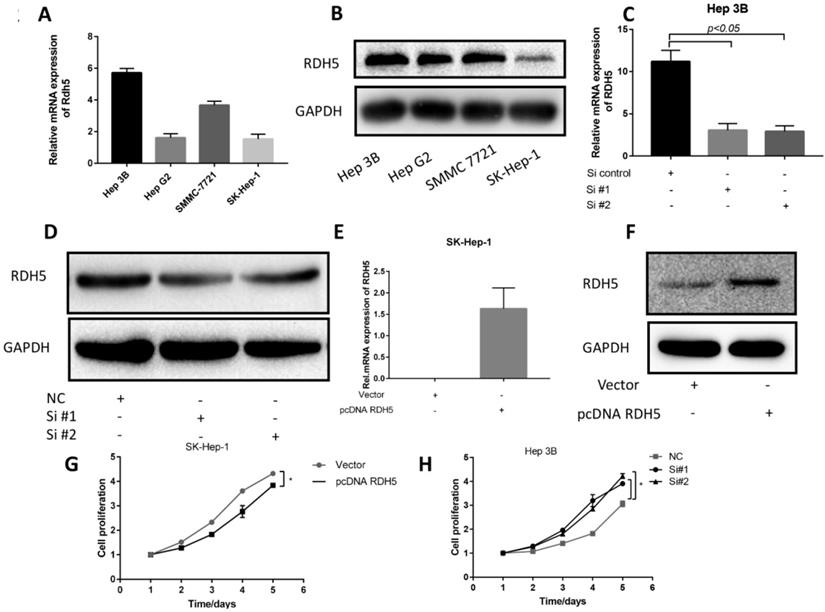
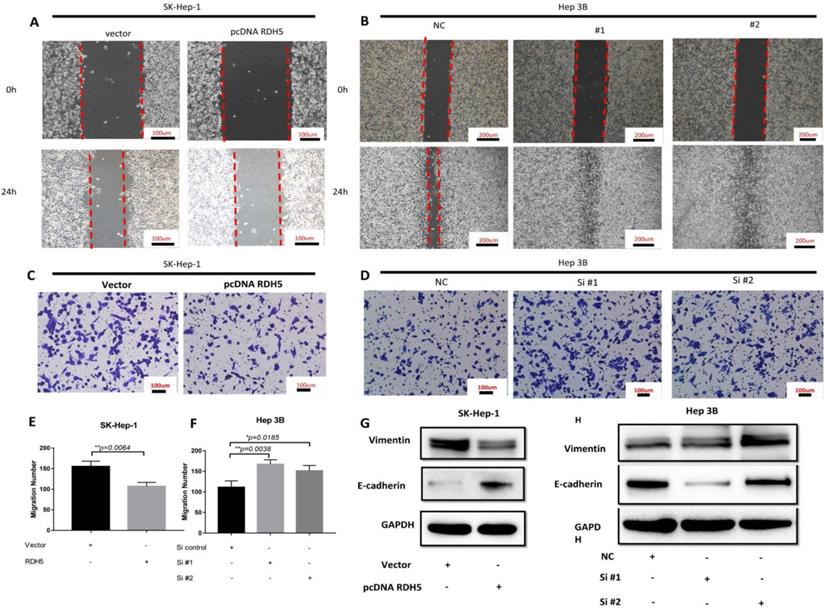
Upon investigating the mRNA and protein levels of RDH5 in HCC cell lines, decreased RDH5 expression was observed in Hep 3B and SMMC-7721 cells compared with SK-Hep-1 and HepG2 cells (Fig. 2A, B). To explore the relationship between RDH5 and metastasis in vitro, we transiently overexpress RDH5 in SK-Hep-1 cells and silence RDH5 in Hep 3B cells (Fig. 2C-F). We generated cell growth curves, which revealed that HCC cell proliferation was suppressed by overexpressing RDH5 and increased by silencing RDH5 (Fig. 2G, H). We applied to wound healing and transwell assays to investigate the migratory ability of Hep 3B and SK-Hep-1 cells and found that overexpressing RDH5 dramatically inhibited wound healing ability (Fig. 3A, B) and reduced the cell migration ability (Fig. 3C-H). Furthermore, we performed the western blotting analysis to investigate epithelial-mesenchymal transition (EMT) relevant markers. Overexpressing RDH5 resulted in decreased levels of mesenchymal markers such as vimentin and elevated levels of epithelial proteins such as E-cadherin (Fig. 3G). Silencing RDH5 resulted in increased levels of mesenchymal marker vimentin and decreased levels of the epithelial protein E-cadherin in Hep 3B cells (Fig. 3H). These results proved that RDH5 can inhibit cell migration by reversing epithelial-mesenchymal transition in vitro.
RDH5 expression in the HCC cell lines and promoted proliferation in HCC cell lines. (A) The mRNA level of RDH5 (normalized to GAPDH) in 4 HCC cell lines was confirmed by qPCR, showing that RDH5 was significantly lower in Hep G2 and SK-Hep-1 than Hep 3B and SMMC-7721. (B) The protein level of RDH5 in 4 HCC cell lines was confirmed by immunoblotting. Overexpression of RDH5 in SK-Hep-1 was determined by qPCR(C) and immunoblotting (D), and GAPDH was used as the loading control. The transient suppression of RDH5 in Hep 3B was determined by qPCR (E) and by immunoblotting (F).GAPDH was used as the loading control. Up-regulation of RDH5 expression in SK-Hep-1 suppressed cell proliferation ability by MTS assay (G), cell proliferation ability was promoted after RHD5 was silenced in Hep 3B (H).
RDH5 expression is regulated by DNA methylation in Hepatocellular carcinoma cells
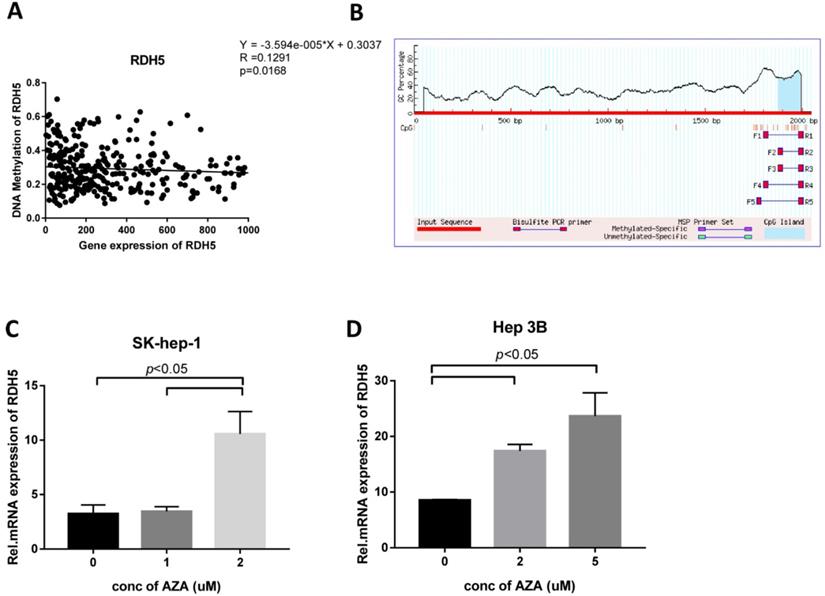
To explore the role of DNA methylation in RDH5 expression, CpG islands in the promoter region were predicted by using http://www.urogene.org/methprimer/, and RDH5 was shown to be silenced by DNA methylation in the TCGA analysis (Fig. 4A, B). Interestingly, the inhibition of DNA methyltransferase by 5-aza-2-deoxycytidine (DAC) at concentrations of 0, 1, 2 and 5 µM suppressed DNA methylation levels and increased the expression of RDH5 in Hep 3B and SK-Hep-1 cells (Fig. 4C, D). The increase of RDH5 expression after treatment with DAC was consistent with the baseline DNA methylation levels in the Hepatocellular carcinoma cell line.
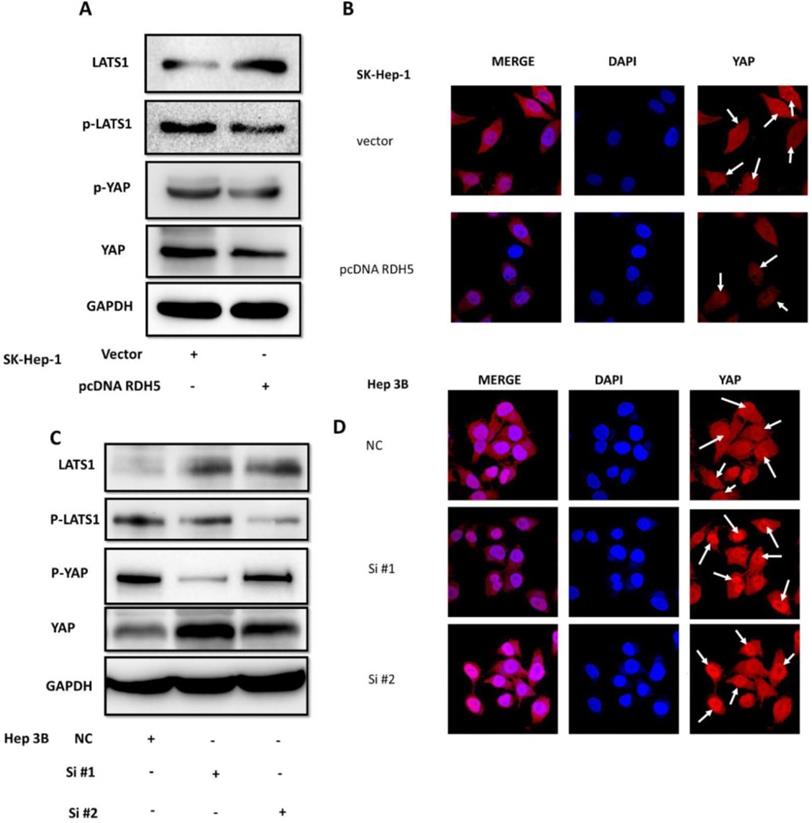
RDH5 suppresses the EMT process via inhibiting Hippo/YAP signaling pathway in hepatocellular carcinoma
Yes-associated protein (YAP) is an important mediator of cell differentiation, proliferation, and angiogenesis. Upon examination of the role of YAP in the RDH5-mediated inhibition of HCC motility, we found that overexpression of RDH5 could increase phospho-LATS1 levels and result in degradation of downstream YAP compared with the vector control (Fig. 5A). After silencing RDH5 with siRNA in Hep 3B cells, phospho-LATS1 levels were suppressed, and the total YAP level was increased (Fig. 5C). Protein localization and expression were visualized by immunofluorescence, which showed that downregulating RDH5 in Hepatocellular carcinoma could activate the Hippo/YAP signaling pathway and promote the nuclear translocation of the crucial protein YAP in Hep 3B cells (Fig. 5D); the upregulation of RDH5 by plasmid transfection into SK-Hep-1 cells suppressed the protein expression of YAP, which remained in the cytoplasm rather than translocating into the nucleus (Fig. 5B).
RDH5 promoted cell migration by reversing the epithelial-mesenchymal transition. A Wound-scratch assay was performed to determine the effect of RDH5 on cell migration. (A) Overexpression of RDH5 in SK-Hep-1 inhibited the migration ability, and (B) transient suppression of RDH5 in Hep 3B dramatically promoted the wound healing ability. Up-regulation of RDH5 expression in SK-Hep-1 suppressed the migration ability by transwell assay (C, E), suppression of RDH5 in Hep 3B promoted migration ability by transwell assay (D, F) The levels of mesenchymal marker such as vimentin was decreased and level of epithelial protein E-cadherin was elevated in the RDH5-overexpressing SK-Hep-1 cell (G), the levels of epithelial marker E-cadherin was decreased and level of mesenchymal protein vimentin was elevated in the RDH5-silencing Hep 3B (H).
The low-level RDH5 expression is an independent, unfavorable prognostic role in hepatocellular carcinoma
To investigate whether RDH5 is an independent factor in hepatocellular carcinoma in the entire study population, univariate and multivariate Cox analyses were conducted using SPSS version 21; Table 2 presents the results. The univariate analysis showed that seven factors, including new neoplasms, M stage, N stage, T stage (T3 and T4), tumor invasion, clinical stage (III-IV), and lower RDH5 expression were poor prognostic factors for OS in hepatocellular carcinoma patients. According to the multivariate analysis, M stage, clinical stage (III-IV) and downregulation of RDH5 level were independent risk markers and associated with shorter overall survival.
Discussion
The RDH5 gene encodes 11-cis retinol dehydrogenase, which is the key enzyme in the oxidation of 11-cis retinol to 11-cis retina. Low expression or mutation of the gene might contribute to retinitis pigmentosa-caused eye diseases. Although few histological studies have reported the relationship between RDH5 expression and cancer, some articles have reported that the dysregulation of RDH5 was associated with colorectal cancer [17], gastric cancer [18], and thyroid carcinoma [30]. In our analysis, the expression of RDH5 was also associated with the clinicopathological feature of metastasis of hepatocellular carcinoma. On the other hand, higher RDH5 gene expression indicated longer survival by Kaplan-Meier survival analysis. Univariate Cox analysis demonstrated that new neoplasm, M stage, N stage, T stage (T3 and T4), tumor invasion, clinical stage (III-IV), and lower RDH5 expression can play prognostic roles in overall survival in hepatocellular carcinoma. The multivariate Cox analysis confirmed that RDH5 expression, M stage, and clinical-stage play independent prognostic roles in overall survival in hepatocellular carcinoma. Another report demonstrated microarray, RNA-Sequence, and EST database analyses identified 32 genes involved in all-trans-retinoic acid generation in normal colon and tumor tissues, including genes in the RDH, ADH, AKR, and CYP families.
Interestingly, Kang et al. [18] reported that RDH5 is mutated in early gastric cancer and may be involved in early carcinogenesis; however, we found that the mutation rate of this gene was low in HCC (data not shown). Another large-scale experimental approach was used to assess CpG islands and promoter regions, and the findings showed DNA methylation of RDH5 and gene expression in thyroid carcinoma [30]. Pyrosequencing and RT-PCR showed that the RDH5 gene is hypomethylated and overexpressed in papillary thyroid carcinoma tissues compared with matched non-neoplastic adjacent tissues, indicating that DNA methylation may regulate the expression of the RDH5 gene. Based on these results, we explored the DNA methylation of RDH5. After predicting the DNA methylation profile using an online biomedical tool, we found that the RDH5 gene has a CpG island in the promoter domain within the untranslated region (UTR). Besides, we analyzed data in the TCGA database and found that DNA methylation may cause the dysregulation of RDH5 gene expression, and data showed that cells were treated with 5-Aza confirmed the previous results. This study is the first time to indicate that DNA methylation affects RDH5 expression in hepatocellular carcinoma.
RDH5 expression is regulated by DNA methylation (A) RDH5 mRNA expression was positively correlated with RDH5 DNA methylation in the TCGA database, and (B) CpG islands in the promoter region was predicted in the biomedical website. SK-Hep-1(C) and Hep 3B (D) cells were subjected to a 72-h treatment with 0,1,2,5 doses of 5-Aza. After RNA extraction, RDH5 mRNA was quantified by qPCR.
RDH5 inhibits HCC cellular migration through suppressing Hippo/YAP pathway and changes the location of YAP. Western blotting analysis of LATS1, p-LATS1, YAP, p-YAP expression levels after overexpression of RDH5 in SK-Hep-1 (A) and silencing of RDH5 in Hep 3B (C). Up-regulation of RDH5 suppress YAP protein into the nucleus in SK-Hep-1(the White arrow) (B), Down-regulation of RHD5 in hepatocellular carcinoma could promoting critical protein YAP step into the nucleus in Hep 3B (the White arrow) (D).
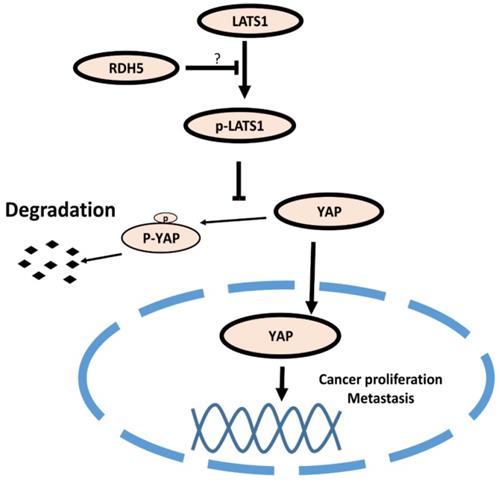
Hepatocellular carcinoma is one of the most malignant tumors in the world, about 50% of affected individuals live in China, and HBV infection is the primary risk factor [29]. Owing to poor prognosis and recurrence, HCC is the 3rd-leading cause of cancer-related mortality [3]. Numerous biomarkers have been reported in recent years, but there are no effective therapeutic targets for clinical treatment. Therefore, it is an urgent need to identify reliable biomarkers and therapeutic targets to monitor HCC progression. When the RDH5 gene was overexpressed, the HCC cell line SK-Hep-1 partially lost metastasis ability and showed decreased cell proliferation. Nevertheless, gene-silencing in the HCC cell line Hep 3B contributed significantly to cell metastasis and proliferation. Immunoblotting demonstrated that RDH5 can decrease EMT-associated protein markers, such as E-cadherin and vimentin. We confirmed that low RDH5 expression promotes metastasis by affecting EMT and cell proliferation.
Model of the RDH5 mechanism in HCC. RDH5 promoting the (LATS1/2) can be phosphorylated and activated, phosphorylating and inactivating Yes-associated Protein, suppressing YAP into the nucleus.
Univariate and multivariate COX analysis for RDH5 and survival of HCCs
| Parameters | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Gender | ||||
| Male | Reference | |||
| Female | 0.78 (0.50-1.20) | 0.260 | ||
| Age, years | ||||
| ≤60 | Reference | |||
| >60 | 1.74 (1.10-2.74) | 0.260 | ||
| Child Pugh Classification | ||||
| A | Reference | |||
| B | 1.18 (0.42-3.29) | 0.751 | ||
| C | 2.25 (0.31-16.47) | 0.425 | ||
| Grade | ||||
| G1 | Reference | |||
| G2 | 0.86 (0.45-1.65) | 0.648 | ||
| G3 | 1.09 (0.56-2,13) | 0.806 | ||
| G4 | 1.49 (0.48-4.68) | 0.490 | ||
| AFP | ||||
| Negative | Reference | |||
| Positive | 1.35 (0.88-2.07) | 0.167 | ||
| New Neoplasm | ||||
| No | Reference | |||
| Yes | 0.58 (0.36-0.93) | 0.025* | ||
| M Stage | ||||
| M0 | Reference | |||
| M1 | 2.17 (1.39-3,37) | 0.001* | 2.09 (1.13-3.89) | 0.019* |
| Embedded | ||||
| No | Reference | |||
| Yes | 0.49 (0.31-74) | 0.001* | ||
| N Stage | ||||
| N0 | Reference | |||
| N1 | 1.88 (1.21-2.94) | 0.005* | ||
| T Stage | ||||
| T1 | Reference | |||
| T2 | 1.45 (0.84-2.51) | 0.186 | ||
| T3 | 2.32 (1.38-3.90) | 0.002* | ||
| T4 | 6.32 (2.77-14.45) | 0.000* | ||
| Clinical Stage | ||||
| I | Reference | |||
| II | 1.50 (0.84-2.69) | 0.169 | ||
| III | 2.30 (1.34-3.94) | 0.002* | 3.03 (1.62-5.67) | 0.001* |
| IV | 4.57 (1.39-15.09) | 0.013* | 4.91 (1.08-22.31) | 0.040* |
| Vascular invasion | ||||
| No | Reference | |||
| Yes | 1.33 (0.90-1.96) | 0.159 | ||
| RDH5 | ||||
| High expression | Reference | |||
| Low expression | 1.78 (1.15-2.72) | 0.007* | 1.94 (1.15-3.28) | 0.013* |
NOTE: The entire clinicopathological variables lists in the table were included in the univariate and multivariate analysis.
Abbreviation: 95%CI, 95%confidence interval.
Many studies have demonstrated that the Hippo pathway plays a critical signaling role in liver size control and tumorigenesis. Nuclear YAP may contribute to hepatocellular carcinoma metastasis [31]. When RDH5 was overexpressed in SK-Hep-1 cells, total YAP protein levels decreased. Conversely, silencing RDH5 in Hep 3B cells increased the total YAP protein levels. To confirm the YAP protein localization in the cell, an immunofluorescence assay was performed, and the results indicated that silencing RDH5 promoted YAP activation in the nucleus and that overexpressing RDH5 promoted YAP inactivation by phosphorylating. In our research, we first showed that low expression of RDH5 in hepatocellular carcinoma could activate the Hippo/YAP signaling pathway and promote YAP translocation into the nucleus.
Collectively, our research showed that RDH5 was dysregulated in hepatocellular carcinoma patients, and this dysregulation can indicate a worse prognosis. First, low RDH5 expression was associated with metastasis in hepatocellular carcinoma tissues. Univariate and multivariate Cox analyses identified three factors, M stage, clinical stage (III-IV), and downregulation of RDH5 expression, as independent risk factors associated with shorter overall survival. In our research, we found that higher gene expression of RDH5 inhibited metastasis and cell proliferation. In addition, we found that RDH5 affected metastasis by regulating the Hippo/YAP signaling pathway.
In summary, our study is the first to elucidate the critical roles of RDH5 in regulating the HIPPO/YAP signaling pathway in hepatocellular carcinoma. Moreover, RDH5 severs as an independent prognostic factor for HCC. We believe that RDH5 is a potential molecular target for predicting, preventing, and treating HCC metastasis. Despite many innovations demonstrated in our research, there are some limitations as well. First, we did not obtain enough tissue samples to perform immunohistochemistry and did not acquire sufficient evidence from animal experiments. Second, we found out that RDH5 works in HIPPO/YAP signaling pathway, but we didn't discover the direct evidence to prove how to regulate in this pathway and the detailed molecular mechanism is still unclear, and further studies are needed.
Supplementary Material
Supplementary tables.
Acknowledgements
The research was supported by the National Natural Science Foundation of China (Grant No. 81673903, 81373500, 81773162).
Ethics approval and consent to participate
The Ethics Committees of the First Affiliated Hospital, Sun Yat-sen University approved the study.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Moon AM, Weiss NS, Beste LA, Su F, Ho SB, Jin GY. et al. No Association Between Screening for Hepatocellular Carcinoma and Reduced Cancer-Related Mortality in Patients With Cirrhosis. Gastroenterology. 2018;155:1128-39.e6
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115-32
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
4. Ting CT, Li WC, Chen CY, Tsai TH. Preventive and therapeutic role of traditional Chinese herbal medicine in hepatocellular carcinoma. Journal of the Chinese Medical Association: JCMA. 2015;78:139-44
5. Hu H, Luo S-J, Cao Z-r, Wu Y, Mo Z, Wang Y. et al. Depressive Disorder promotes Hepatocellular Carcinoma metastasis via upregulation of ABCG2 gene expression and maintenance of self-renewal. Journal of Cancer. 2020;11:5309-17
6. Vigano L, Conci S, Cescon M, Fava C, Capelli P, D'Errico A. et al. Liver resection for hepatocellular carcinoma in patients with metabolic syndrome: A multicenter matched analysis with HCV-related HCC. Journal of hepatology. 2015;63:93-101
7. Foerster F, Mittler J, Darstein F, Heise M, Marquardt JU, Worns MA. et al. Recipient liver function before liver transplantation influences post-transplantation survival in patients with HCC. European journal of internal medicine. 2018;55:57-65
8. Hsu CC, Chen CL, Wang CC, Lin CC, Yong CC, Wang SH. et al. Combination of FDG-PET and UCSF Criteria for Predicting HCC Recurrence After Living Donor Liver Transplantation. Transplantation. 2016;100:1925-32
9. Jin J, Zhang XY, Shi JL, Xue XF, Lu LL, Lu JH. et al. Application of AFP whole blood one-step rapid detection kit in screening for HCC in Qidong. American journal of cancer research. 2017;7:1384-8
10. Luo P, Wu S, Yu Y, Ming X, Li S, Zuo X. et al. Current Status and Perspective Biomarkers in AFP Negative HCC: Towards Screening for and Diagnosing Hepatocellular Carcinoma at an Earlier Stage. Pathology oncology research. POR. 2019
11. Choi BI, Lee JM. Advancement in HCC imaging: diagnosis, staging and treatment efficacy assessments: imaging diagnosis and staging of hepatocellular carcinoma. Journal of hepato-biliary-pancreatic sciences. 2010;17:369-73
12. Faletti R, Cassinis MC, Fonio P, Bergamasco L, Pavan LJ, Rapellino A. et al. Multiparametric Gd-EOB-DTPA magnetic resonance in diagnosis of HCC: dynamic study, hepatobiliary phase, and diffusion-weighted imaging compared to histology after orthotopic liver transplantation. Abdominal imaging. 2015;40:46-55
13. Liu X, Liu L, Li H, Xu F, Jiang R, Sui R. RDH5 retinopathy (fundus albipunctatus) with preserved rod function. Retina (Philadelphia, Pa). 2015;35:582-9
14. Skorczyk-Werner A, Pawlowski P, Michalczuk M, Warowicka A, Wawrocka A, Wicher K. et al. Fundus albipunctatus: review of the literature and report of a novel RDH5 gene mutation affecting the invariant tyrosine (p.Tyr175Phe). Journal of applied genetics. 2015;56:317-27
15. Makiyama Y, Ooto S, Hangai M, Ogino K, Gotoh N, Oishi A. et al. Cone abnormalities in fundus albipunctatus associated with RDH5 mutations assessed using adaptive optics scanning laser ophthalmoscopy. American journal of ophthalmology. 2014;157:558-70.e1 -4
16. Wang NK, Chuang LH, Lai CC, Chou CL, Chu HY, Yeung L. et al. Multimodal fundus imaging in fundus albipunctatus with RDH5 mutation: a newly identified compound heterozygous mutation and review of the literature. Documenta ophthalmologica Advances in ophthalmology. 2012;125:51-62
17. Kropotova ES, Zinovieva OL, Zyryanova AF, Dybovaya VI, Prasolov VS, Beresten SF. et al. Altered expression of multiple genes involved in retinoic acid biosynthesis in human colorectal cancer. Pathology oncology research: POR. 2014;20:707-17
18. Kang G, Hwang WC, Do IG, Wang K, Kang SY, Lee J. et al. Exome sequencing identifies early gastric carcinoma as an early stage of advanced gastric cancer. PloS one. 2013;8:e82770
19. Beltrami CM, Dos Reis MB, Barros-Filho MC, Marchi FA, Kuasne H, Pinto CAL. et al. Integrated data analysis reveals potential drivers and pathways disrupted by DNA methylation in papillary thyroid carcinomas. Clinical epigenetics. 2017;9:45
20. Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. Journal of hepatology. 2015;63:1491-501
21. Meng DF, Sun R, Peng LX, Huang YS, Yang Q, Luo DH. et al. A comparison of weekly versus 3-weekly cisplatin during concurrent chemoradiotherapy for locoregionally advanced nasopharyngeal carcinoma using intensity modulated radiation therapy: a matched study. J Cancer. 2018;9:92-9
22. Wu H, Wei L, Fan F, Ji S, Zhang S, Geng J. et al. Integration of Hippo signalling and the unfolded protein response to restrain liver overgrowth and tumorigenesis. Nature communications. 2015;6:6239
23. Cheng Y, Hou T, Ping J, Chen T, Yin B. LMO3 promotes hepatocellular carcinoma invasion, metastasis and anoikis inhibition by directly interacting with LATS1 and suppressing Hippo signaling. Journal of experimental & clinical cancer research: CR. 2018;37:228
24. Hu H, Xu L, Chen Y, Luo S-J, Wu Y-z, Xu S-H. et al. The Upregulation of Trophinin-Associated Protein (TROAP) Predicts a Poor Prognosis in Hepatocellular Carcinoma. Journal of Cancer. 2019;10:957-67
25. Xu L, Huang TJ, Hu H, Wang MY, Shi SM, Yang Q. et al. The developmental transcription factor IRF6 attenuates ABCG2 gene expression and distinctively reverses stemness phenotype in nasopharyngeal carcinoma. Cancer letters. 2018;431:230-43
26. Qiang YY, Li CZ, Sun R, Zheng LS, Peng LX, Yang JP. et al. Along with its favorable prognostic role, CLCA2 inhibits growth and metastasis of nasopharyngeal carcinoma cells via inhibition of FAK/ERK signaling. Journal of experimental & clinical cancer research: CR. 2018;37:34
27. Meng DF, Xie P, Peng LX, Sun R, Luo DH, Chen QY. et al. CDC42-interacting protein 4 promotes metastasis of nasopharyngeal carcinoma by mediating invadopodia formation and activating EGFR signaling. Journal of experimental & clinical cancer research: CR. 2017;36:21
28. Xu L, Hu H, Zheng LS, Wang MY, Mei Y, Peng LX. et al. ETV4 is a theranostic target in clear cell renal cell carcinoma that promotes metastasis by activating the pro-metastatic gene FOSL1 in a PI3K-AKT dependent manner. Cancer letters. 2020;482:74-89
29. Rouleau C, Pores Fernando AT, Hwang JH, Faure N, Jiang T, White EA. et al. Transformation by Polyomavirus Middle T Antigen Involves a Unique Bimodal Interaction with the Hippo Effector YAP. Journal of virology. 2016;90:7032-45
30. Kim TS, Maeda A, Maeda T, Heinlein C, Kedishvili N, Palczewski K. et al. Delayed dark adaptation in 11-cis-retinol dehydrogenase-deficient mice: a role of RDH11 in visual processes in vivo. The Journal of biological chemistry. 2005;280:8694-704
31. Tao Y, Shan L, Xu X, Jiang H, Chen R, Qian Z. et al. Huaier Augmented the Chemotherapeutic Sensitivity of Oxaliplatin via Downregulation of YAP in Hepatocellular Carcinoma. J Cancer. 2018;9:3962-70
Author contact
![]() Corresponding authors: Prof. Li-zhu Lin, Department of Oncology, First Affiliated Hospital of Guangzhou University of Chinese Medicine, E-mail: linlizhuedu.cn; Prof. Shi-Jun Zhang, Department of Chinese Medicine, the First Affiliated Hospital, Sun Yat-Sen University, Guangzhou 510080, Guangdong, P. R. China, E-mails: 2806973376com or zhsjun1967com.
Corresponding authors: Prof. Li-zhu Lin, Department of Oncology, First Affiliated Hospital of Guangzhou University of Chinese Medicine, E-mail: linlizhuedu.cn; Prof. Shi-Jun Zhang, Department of Chinese Medicine, the First Affiliated Hospital, Sun Yat-Sen University, Guangzhou 510080, Guangdong, P. R. China, E-mails: 2806973376com or zhsjun1967com.

 Global reach, higher impact
Global reach, higher impact