Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(7):949-959. doi:10.7150/ijms.33820 This issue Cite
Research Paper
Ensemble Feature Learning to Identify Risk Factors for Predicting Secondary Cancer
1. Department of Computer Science, University of Tsukuba, Tsukuba, Japan
2. Center for Artificial Intelligence Research, University of Tsukuba, Tsukuba, Japan
3. Division of Radiation Oncology, Far Eastern Memorial Hospital, New Taipei City, Taiwan
4. Faculty of Medicine, School of Medicine, National Yang-Ming University, Taipei, Taiwan
Received 2019-2-3; Accepted 2019-4-24; Published 2019-6-7
Abstract
Background: In recent years, the development and diagnosis of secondary cancer have become the primary concern of cancer survivors. A number of studies have been developing strategies to extract knowledge from the clinical data, aiming to identify important risk factors that can be used to prevent the recurrence of diseases. However, these studies do not focus on secondary cancer. Secondary cancer is lack of the strategies for clinical treatment as well as risk factor identification to prevent the occurrence.
Methods: We propose an effective ensemble feature learning method to identify the risk factors for predicting secondary cancer by considering class imbalance and patient heterogeneity. We first divide the patients into some heterogeneous groups based on spectral clustering. In each group, we apply the oversampling method to balance the number of samples in each class and use them as training data for ensemble feature learning. The purpose of ensemble feature learning is to identify the risk factors and construct a diagnosis model for each group. The importance of risk factors is measured based on the properties of patients in each group separately. We predict secondary cancer by assigning the patient to a corresponding group and based on the diagnosis model in this corresponding group.
Results: Analysis of the results shows that the decision tree obtains the best results for predicting secondary cancer in the three classifiers. The best results of the decision tree are 0.72 in terms of AUC when dividing the patients into 15 groups, 0.38 in terms of F1 score when dividing the patients into 20 groups. In terms of AUC, decision tree achieves 67.4% improvement compared to using all 20 predictor variables and 28.6% improvement compared to no group division. In terms of F1 score, decision tree achieves 216.7% improvement compared to using all 20 predictor variables and 80.9% improvement compared to no group division. Different groups provide different ranking results for the predictor variables.
Conclusion: The accuracies of predicting secondary cancer using k-nearest neighbor, decision tree, support vector machine indeed increased after using the selected important risk factors as predictors. Group division on patients to predict secondary cancer on the separated models can further improve the prediction accuracies. The information discovered in the experiments can provide important references to the personality and clinical symptom representations on all phases of guide interventions, with the complexities of multiple symptoms associated with secondary cancer in all phases of the recurrent trajectory.
Keywords: secondary cancer, risk factors, class imbalance, patient heterogeneity, spectral clustering, ensemble learning
Introduction
Cancer has become the second leading cause of death globally, which is characterized as a heterogeneous disease consisting of many different subtypes [1-3]. From the report of the World Health Organization (WHO), there are an estimated 9.6 million deaths due to cancer in 2018 [4]. Recently, the development and diagnosis of secondary cancer have become the main concern of cancer survivors [5-7]. In contrast to primary cancer which refers to initial cancer a person experiences, secondary cancer refers to either metastasis from primary cancer, or different cancer unrelated to primary cancer [8]. Compared to people with the same age and gender who have never had cancer, cancer survivors have an increased chance of developing secondary cancer. It is important for cancer survivors to be aware of the risk factors for secondary cancers and maintain good follow-up health care [9-11]. Furthermore, the literature shows that secondary cancer should be predicted with regard to their personal risk factors and clinical symptoms [12-15].
Over the years, many statistical methods have been developed to extract knowledge from the clinical data, to identify important risk factors that can be used to prevent the recurrence of diseases [16,17]. Tseng et al. [18] utilize five classification techniques to rank the importance of risk factors for diagnosing ovarian cancer. Liang et al. [19] combine five feature selection methods with support vector machine to develop predictive models for recurrence of hepatocellular carcinoma. However, the studies in [18] and [19] do not consider the class imbalance problem and the heterogeneity between patients. Similarly, for most existing studies, some do not deal with the class imbalance problem [18], some do not consider the heterogeneity between patients [20], and as far as we know, none focuses on secondary cancer. The presence of class imbalance is a problem in medical diagnosis, in which the abnormal instances are only a small percentage compared to a large number of normal ones. Especially for secondary cancer, class imbalance is an inevitable problem. For a dataset with class imbalance, machine learning methods are biased towards the majority class and the learned information are mostly from the normal instances, which lead to poor accuracy for identifying the rare abnormal instances. On the other hand, patient heterogeneity is also an important issue to consider. The diagnosis on the basis of data analysis results may not always suitable to a specific patient, given the biological variability among individuals [20,21].
In this study, we propose an effective ensemble feature learning method to identify the risk factors for predicting secondary cancer by considering class imbalance and patient heterogeneity. An oversampling method is utilized to deal with the class imbalance problem in secondary cancer. We divide the patients into some heterogeneous groups, and then identify the risk factors and construct a diagnosis model for each patient group for a more accurate prediction. To the best of our knowledge, this kind of methodology has never been proposed and applied for secondary cancer data analysis.
Material and Methods
Samples
The dataset of samples we studied in this paper are provided by the Chung Shan Medical University Hospital, Jen-Ai Hospital, and Far Eastern Memorial Hospital. It mainly contains four types of cancers: breast cancer, maternal cancer, colorectal cancer, head, and neck cancer, where the percentage of secondary cancer patients are 1.7%, 1.8%, 3.6% and 7.9%, respectively. Totally, 11380 patients have ever suffered from primary cancer, among which 458 (4%) patients suffered from secondary cancer. The two classes (no suffering from secondary cancer and suffering from secondary cancer) are highly unbalanced. We analyze the predictor variables to find what variables are associated with the risk factors for secondary cancer. The 20 predictor variables analyzed in this paper are based on the decision of the cancer expert committee, which is considered to be potentially relevant to secondary cancer. They include Age; Body Mass Index (BMI); 8 variables related to the status of cancer which are Primary Site (referred to the type of primary cancer), Histology, Behavior Code, Differentiation, Tumor Size, Pathologic Stage, Surgical Margin, Surgical; 7 variables related to radiological and chemical treatments which are Radiotherapy (RT), Radiotherapy (RT) surgery, Sequence of Local regional Therapy and Systemic Therapy, Dose to clinical target volumes (CTV)_High, Number to clinical target volumes (CTV)_High, Dose to clinical target volumes (CTV)_Low, Number to clinical target volumes (CTV)_Low; 3 variables related to lifestyle which are: Smoking, Betel Nut, Drinking. The analysis allows for a better understanding of which variables are more fundamental to secondary cancer.
Method design
Firstly, we divide the training data into some heterogeneous groups by using spectral clustering [22,23,24] and learn the training data in each group separately. In each group, we apply the Synthetic minority oversampling technique (SMOTE) [25] as the oversampling method to generate synthetic data in the minority class for class balance. Then, ensemble feature learning is performed to identify the risk factors and construct a diagnosis model for each group. In the testing process, each test data is first assigned to a group in the training dataset and then tested the result on the corresponding model.
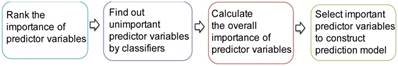
The procedure of ensemble feature learning mainly consists of four stages, as shown in Figure 1.
(1) Rank the importance of predictor variables. We use 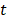 -test to rank the importance of predictor variables according to their
-test to rank the importance of predictor variables according to their 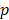 values. Lower
values. Lower 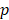 -value denotes more importance. We set the weight of predictor variables based on the ranking results. For a predictor variable
-value denotes more importance. We set the weight of predictor variables based on the ranking results. For a predictor variable 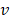 with rank order
with rank order 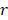 , its weight is set as
, its weight is set as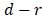 , where
, where 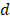 is the number of predictor variables.
is the number of predictor variables.
(2) Find out the unimportant predictor variables. We utilize three classifiers, i.e., k-nearest neighbor (kNN) [26], Decision Tree (DT) [27] and Support Vector Machine (SVM) [28], to classify the samples by increasing the predictor variables based on the ranking result. The predictor variables that do not increase the prediction accuracy are considered to be unimportant. The weights of unimportant predictor variables are set to 0.
(3) Calculate the overall importance of predictor variables. For different classifiers, the unimportant predictor variables may be different. We calculate the overall importance of predictor variables as the average weight of using the three classifiers.
(4) Select important predictor variables to construct a prediction model. We increase the number of predictor variables from 1 to 20 based on the overall importance in descending order. The combination of predictor variables obtaining the best prediction accuracy is selected for model construction. For example, if the three most important predictor variables obtain the best prediction accuracy, they will be selected for model construction. Beyond the prediction accuracy, we also consider the comments of clinical physicians.
Procedure of ensemble feature learning
Statistical analysis
All statistical analyses are performed using Matlab 9.4.0 (R2018a) on Mac OS X 10.14.2 (18C54) with core i5 CPU and 8GB ram. We apply the AUC (Area Under Curve) [29] and 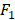 score [30] to evaluate the performance of the proposed method. AUC and
score [30] to evaluate the performance of the proposed method. AUC and 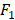 score are two useful metrics for imbalanced datasets. AUC is the area under the curve of a ROC graph, which compares the Sensitivity vs (1-Specificity). Each point on the ROC curve represents a different choice for that true/false threshold.
score are two useful metrics for imbalanced datasets. AUC is the area under the curve of a ROC graph, which compares the Sensitivity vs (1-Specificity). Each point on the ROC curve represents a different choice for that true/false threshold. 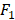 score is a harmonic mean of precision and recall for a specific threshold. AUC evaluates a model independently of the choice of threshold, whereas
score is a harmonic mean of precision and recall for a specific threshold. AUC evaluates a model independently of the choice of threshold, whereas 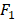 score is a measure for a particular model at a particular threshold. In general, AUC evaluates the test power (for best tests nearly 1).
score is a measure for a particular model at a particular threshold. In general, AUC evaluates the test power (for best tests nearly 1). 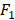 score evaluates how reliable a sensitive test is in the positive decision (nearly 1 for best tests).
score evaluates how reliable a sensitive test is in the positive decision (nearly 1 for best tests).
We use the toolbox of Matlab to run the three classifiers, i.e., kNN, DT and SVM. The spectral clustering algorithm is performed as the algorithm in [24]. The training data and test data are 80% and 20%, respectively. We create cross-validation partition for the dataset using Matlab function “cvpartition”. For SMOTE, the number of increased samples is ranged from 1 to 15 times of the samples in the minority class, the number of nearest neighbors is ranged from 3 to 13, and the best result is recorded for the following steps. All experiments were repeated 10 times and the average results are reported.
Results
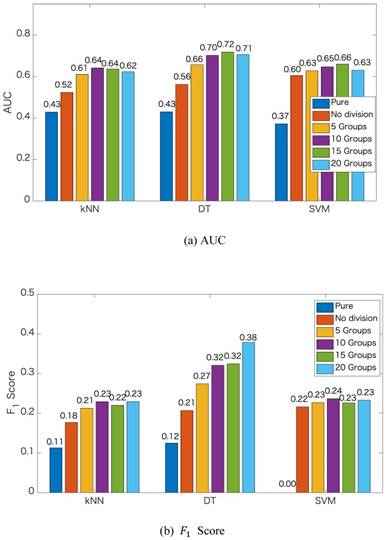
We apply the proposed method to learn the risk factors and predict secondary cancer. The number of divided groups is ranged from 1 to 20. Note that the number of divided groups being 1 is just the case that we apply ensemble feature learning without group division. The results of the prediction accuracies using the three classifiers, i.e., kNN, DT and SVM, are shown in Figure 2. Figure 2 shows the results in terms of AUC and 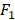 score, respectively. From the results, we can see that ensemble feature learning with group division performs better than ensemble feature learning without group division. DT obtains the best results in the three classifiers. The best results of DT are 0.72 in terms of AUC when dividing into 15 groups, and 0.38 in terms of
score, respectively. From the results, we can see that ensemble feature learning with group division performs better than ensemble feature learning without group division. DT obtains the best results in the three classifiers. The best results of DT are 0.72 in terms of AUC when dividing into 15 groups, and 0.38 in terms of 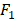 score when dividing into 20 groups. The performance of DT shows an upward trend as the number of divided groups increases, while the performance improvements of kNN and SVM are not significant when dividing into more than 3 groups.
score when dividing into 20 groups. The performance of DT shows an upward trend as the number of divided groups increases, while the performance improvements of kNN and SVM are not significant when dividing into more than 3 groups.
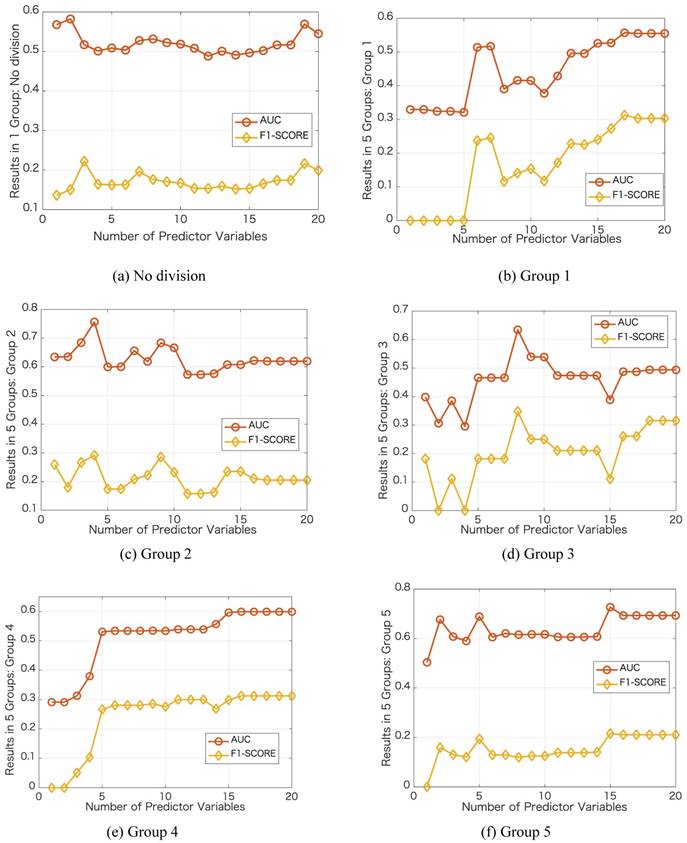
Next, we show the ranking results based on the importance of the 20 predictor variables in the cases of with and without group division using the DT classifier. For the case of group division, we show the ranking results in each group when dividing into 5 groups. The divided 5 groups are denoted as group 1, group 2, group 3, group 4, and group 5, respectively. As shown in Table 1, different groups provide different ranking results for the predictor variables. In the case of no group division, the top 5 important predictor variables are Primary Site, Pathologic Stage, Age, Surgical Margin, and Histology. In the case of group division, Primary Site, Pathologic Stage, and Surgical Margin are among the top 5 important predictor variables in each group. Age is among the top 3 important predictor variables in four groups. From the ranking results in Table 1, Primary Site, Pathologic Stage, Age, Surgical Margin are the four most critical risk factors in groups 2, 3, 5 and the case of no group division.
Results of the prediction accuracies using three classifiers
Ranking results of the importance in the 20 predictor variables for 4 types of cancers
| Rank | No division | 5 Groups | ||||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group5 | ||
| 1 | Primary Site | Pathologic Stage | Surgical Margin | Primary Site | Primary Site | Primary Site |
| 2 | Pathologic Stage | Primary Site | Pathologic Stage' | Pathologic Stage | Pathologic Stage | Pathologic Stage |
| 3 | Age | Surgical Margin | Age | Age | Age | Age |
| 4 | Surgical Margin | Surgical | Primary Site | Surgical Margin | Smoking | Surgical Margin |
| 5 | Histology | Histology | Histology | Smoking | Surgical Margin | Smoking |
| 6 | Drinking | Dose to clinical target volumes (CTV)_Low | Surgical | Number to clinical target volumes (CTV) _Low | Drinking | Drinking |
| 7 | Betel Nut | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _Low | Histology | Betel Nut | Betel Nut |
| 8 | Radiotherapy (RT) | Age | Betel Nut | Drinking | Number to clinical target volumes (CTV) _Low | Histology |
| 9 | Smoking | Tumor Size | Tumor Size | Betel Nut | Dose to clinical target volumes (CTV)_High | Number to clinical target volumes (CTV) _Low |
| 10 | Behavior Code | Dose to clinical target volumes (CTV)_High | Drinking | Dose to clinical target volumes (CTV)_Low | Histology | Dose to clinical target volumes (CTV)_High |
| 11 | Sequence of Local regional Therapy and Systemic Therapy | Betel Nut | Smoking | Dose to clinical target volumes (CTV)_High | Differentiation | Differentiation |
| 12 | Body Mass Index (BMI) | Drinking | Dose to clinical target volumes (CTV)_Low | Surgical | Number to clinical target volumes (CTV) _High | Number to clinical target volumes (CTV) _High |
| 13 | Number to clinical target volumes (CTV) _High | Differentiation | Dose to clinical target volumes (CTV)_High | Tumor Size | Surgical | Surgical |
| 14 | Differentiation | Radiotherapy (RT) surgery | Body Mass Index (BMI) | Body Mass Index (BMI) | Tumor Size | Tumor Size |
| 15 | Dose to clinical target volumes (CTV)_High | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Number to clinical target volumes (CTV) _High | Body Mass Index (BMI) | Body Mass Index (BMI) |
| 16 | Dose to clinical target volumes (CTV)_Low | Body Mass Index (BMI) | Differentiation | Differentiation | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy |
| 17 | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _High | Radiotherapy (RT) surgery | Sequence of Local regional Therapy and Systemic Therapy | Dose to clinical target volumes (CTV)_Low | Dose to clinical target volumes (CTV)_Low |
| 18 | Radiotherapy (RT) surgery | Smoking | Radiotherapy (RT) | Behavior Code | Radiotherapy (RT) | Radiotherapy (RT) |
| 19 | Tumor Size | Behavior Code | Number to clinical target volumes (CTV) _High | Radiotherapy (RT) surgery | Behavior Code | Behavior Code |
| 20 | Surgical | Radiotherapy (RT) | Behavior Code | Radiotherapy (RT) | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery |
We further investigate the performance in each group by varying the number of predictor variables. We show the results in Figure 3 with the same case in Table 1, i.e., dividing into 5 groups and no group division using DT classifier. In each group, we increase the number of predictor variables from 1 to 20 based on their importance ranking results. Taking the no division case as an example, we first use Primary Site as the predictor variable and then use Primary Site and Pathologic Stage as the two predictor variables. For the no division case, the results do not change obviously as the number of predictor variables varies. For the case of dividing into 5 groups, in each group, the results change obviously as the number of predictor variables varies. Using a certain number of the important predictor variables, the results can be improved significantly. For the best results in terms of AUC, the number of predictor variables used in the no division case is 2, and the numbers of predictor variables used in the group division case are 17, 4, 8,16, 15, respectively.
Results of the prediction accuracies by varying the number of predictor variables
Finally, to show the effectiveness of the proposed method, we also show the prediction results of the pure kNN, pure DT and pure SVM that are without ensemble feature learning. We compare the prediction results of the pure methods to that of the proposed method dividing into different numbers of groups, i.e., 1 group (no division), 5 groups, 10 groups, 15 groups, and 20 groups. The comparison results in terms of AUC and 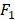 score are shown in Figures 4. From Figure 4, we can see that the accuracies of predicting secondary cancer using kNN, DT and SVM indeed increase after ensemble feature learning to select the important risk factors as the predictors. Group division to predict secondary cancer on the separated models can further improve the prediction accuracies. Note that the
score are shown in Figures 4. From Figure 4, we can see that the accuracies of predicting secondary cancer using kNN, DT and SVM indeed increase after ensemble feature learning to select the important risk factors as the predictors. Group division to predict secondary cancer on the separated models can further improve the prediction accuracies. Note that the 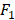 score of the pure SVM is 0. After ensemble feature learning selecting the important risk factors as the predictors, the
score of the pure SVM is 0. After ensemble feature learning selecting the important risk factors as the predictors, the 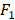 score is improved to be larger than 0.22. DT obtains better results than kNN and SVM. The improvements by group division are more significant with the DT method.
score is improved to be larger than 0.22. DT obtains better results than kNN and SVM. The improvements by group division are more significant with the DT method.
Discussion
Whether or not a patient will have secondary cancer depends on many different things [18]. In this study, we learn the importance of 20 predictor variables related to secondary cancer for four types of cancer. To the best of our knowledge, this is the first study that utilizes machine learning methods to learn the risk factors and construct the prediction model for secondary cancer.
Based on the data characteristics, i.e., class imbalance and patient heterogeneity, we use an oversampling method to increase the samples in the minority class and use spectral clustering to divide the samples into some groups. Spectral clustering is an efficient clustering algorithm, with the performance being superior to that of traditional clustering methods, such as K-means. Compared to no group division in which all patients using only one diagnosis model, group division constructs separated diagnosis models for the patients in different groups. The patients in a group are more similar than the patients in other groups, and they use a diagnosis model. Thus, using the models constructed from the groups has higher precision accuracy than using the model constructed from all samples. That is the reason why group division can improve the accuracy of predicting secondary cancer.
Since for different types of cancers, the ranking results for the predictor variables are different. We also show the ranking results of the importance in the 19 predictor variables (excluding the predictor variable of Primary Site) for each type of cancer. Similar to Table 1, Tables 2, 3, 4 and 5 show the ranking results for the four types of cancers, respectively. In no group division case, Age, Pathologic Stage, and Surgical Margin are the three most critical risk factors for maternal cancer, colorectal cancer, head, and neck cancer. For breast cancer, Pathologic Stage, Histology and Surgical Margin are the three most critical risk factors in no group division case. In the group division case, different groups provide different ranking results for the predictor variables. For colorectal cancer, head and neck cancer, Age, Pathologic Stage, and Surgical Margin are the three most critical risk factors in no group division case and remain in the five most critical risk factors in group division case. For breast cancer and maternal cancer, some important predictor variables in no group division case do not remain the same level of importance in group division case, e.g., in Table 3, age is the most critical risk factor in no group division case, however age is ranked 12 in Group 1 in group division case. One of the reasons is that the patients have similar ages. Another reason is that the number of patients suffering from secondary cancer is only 3. To obtain more samples suffering from secondary cancer to train the diagnosis models, we analyze the four types of cancers together in the experiments.
Comparison of the prediction accuracies
Ranking results of the importance in the 19 predictor variables for breast cancer
| Rank | No division | 5 Groups | ||||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group5 | ||
| 1 | Pathologic Stage | Number to clinical target volumes (CTV) _High | Surgical Margin | Surgical Margin | Surgical Margin | Surgical Margin |
| 2 | Histology | Dose to clinical target volumes (CTV)_Low | Pathologic Stage | Histology | Smoking | Smoking |
| 3 | Surgical Margin | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _High | Pathologic Stage | Histology | Pathologic Stage |
| 4 | Body Mass Index (BMI) | Pathologic Stage | Dose to clinical target volumes (CTV)_Low | Number to clinical target volumes (CTV) _High | Number to clinical target volumes (CTV) _High | Histology |
| 5 | Age | Dose to clinical target volumes (CTV)_High | Body Mass Index (BMI) | Dose to clinical target volumes (CTV)_Low | Dose to clinical target volumes (CTV)_Low | Number to clinical target volumes (CTV) _High |
| 6 | Number to clinical target volumes (CTV) _High | Age | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _Low | Pathologic Stage | Body Mass Index (BMI) |
| 7 | Betel Nut | Body Mass Index (BMI) | Age | Smoking | Number to clinical target volumes (CTV) _Low | Betel Nut |
| 8 | Dose to clinical target volumes (CTV)_Low | Surgical Margin | Dose to clinical target volumes (CTV)_High | Body Mass Index (BMI) | Body Mass Index (BMI) | Drinking |
| 9 | Behavior Code | Surgical | Tumor Size | Age | Betel Nut | Dose to clinical target volumes (CTV)_Low |
| 10 | Number to clinical target volumes (CTV) _Low | Tumor Size | Betel Nut | Betel Nut | Surgical | Number to clinical target volumes (CTV) _Low |
| 11 | Dose to clinical target volumes (CTV)_High | Histology | Differentiation | Dose to clinical target volumes (CTV)_High | Dose to clinical target volumes (CTV)_High | Surgical |
| 12 | Tumor Size | Betel Nut | Surgical | Surgical | Drinking | Dose to clinical target volumes (CTV)_High |
| 13 | Differentiation | Differentiation | Histology | Drinking | Age | Age |
| 14 | Drinking | Radiotherapy (RT) | Radiotherapy (RT) | Differentiation | Radiotherapy (RT) | Differentiation |
| 15 | Smoking | Smoking | Drinking | Tumor Size | Tumor Size | Tumor Size |
| 16 | Radiotherapy (RT) | Drinking | Smoking | Radiotherapy (RT) | Differentiation | Radiotherapy (RT) |
| 17 | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Sequence of Local regional Therapy and Systemic Therapy |
| 18 | Radiotherapy (RT) surgery | Behavior Code | Behavior Code | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Behavior Code |
| 19 | Surgical | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Behavior Code | Behavior Code | Radiotherapy (RT) surgery |
Limitations and futures studies
Since there is no existing study using machine learning methods to predict secondary cancer, we have no idea about which kind of machine learning methods are the most suitable. In this study, we try some widely used classification methods for secondary cancer prediction, i.e., k-nearest neighbor (kNN), Linear Discriminant Analysis (LDA), Quadratic Discriminant Analysis (QDA), Decision Tree (DT) and Support Vector Machine (SVM), and Naïve Bayes. kNN, DT and SVM obtain better results than other methods. Thus, we apply kNN, DT and SVM in our method for ensemble learning. From the results, we find that DT has better performance than the other two classifiers. That may be because DT uses a tree-like model of decisions, which has similar consideration of group division. Therefore, group division can future improves the performance of DT, especially when the number of divided groups increases. We just try the division of 20 groups, we do not know if increasing the number of divided groups can further improve the performance. In the future, we will try more methods to predict secondary cancer and investigate the optimal number of division groups.
Ranking results of the importance in the 19 predictor variables for maternal cancer
| Rank | No division | 5 Groups | ||||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group5 | ||
| 1 | Age | Surgical Margin | Surgical Margin | Surgical Margin | Surgical Margin | Pathologic Stage |
| 2 | Pathologic Stage | Smoking | Smoking | Smoking | Pathologic Stage | Surgical Margin |
| 3 | Surgical Margin | Pathologic Stage | Pathologic Stage | Pathologic Stage | Drinking | Drinking |
| 4 | Body Mass Index (BMI) | Histology | Age | Drinking | Sequence of Local regional Therapy and Systemic Therapy | Age |
| 5 | Histology | Body Mass Index (BMI) | Histology | Age | Age | Sequence of Local regional Therapy and Systemic Therapy |
| 6 | Betel Nut | Drinking | Body Mass Index (BMI) | Histology | Smoking | Smoking |
| 7 | Number to clinical target volumes (CTV) _High | Betel Nut | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Histology | Histology |
| 8 | Dose to clinical target volumes (CTV)_Low | Number to clinical target volumes (CTV) _High | Betel Nut | Body Mass Index (BMI) | Number to clinical target volumes (CTV) _High | Body Mass Index (BMI) |
| 9 | Number to clinical target volumes (CTV) _Low | Sequence of Local regional Therapy and Systemic Therapy | Dose to clinical target volumes (CTV)_Low | Number to clinical target volumes (CTV) _High | Dose to clinical target volumes (CTV)_Low | Betel Nut |
| 10 | Smoking | Differentiation | Drinking | Dose to clinical target volumes (CTV)_Low | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _High |
| 11 | Differentiation | Dose to clinical target volumes (CTV)_Low | Number to clinical target volumes (CTV) _High | Betel Nut | Body Mass Index (BMI) | Dose to clinical target volumes (CTV)_Low |
| 12 | Radiotherapy (RT) surgery | Age | Differentiation | Surgical | Differentiation | Differentiation |
| 13 | Behavior Code | Surgical | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _Low | Surgical | Number to clinical target volumes (CTV) _Low |
| 14 | Radiotherapy (RT) | Number to clinical target volumes (CTV) _Low | Surgical | Radiotherapy (RT) | Betel Nut | Surgical |
| 15 | Drinking | Dose to clinical target volumes (CTV)_High | Radiotherapy (RT) | Differentiation | Radiotherapy (RT) | Radiotherapy (RT) |
| 16 | Tumor Size | Tumor Size | Dose to clinical target volumes (CTV)_High | Dose to clinical target volumes (CTV)_High | Dose to clinical target volumes (CTV)_High | Dose to clinical target volumes (CTV)_High |
| 17 | Dose to clinical target volumes (CTV)_High | Radiotherapy (RT) | Tumor Size | Tumor Size | Tumor Size | Tumor Size |
| 18 | Sequence of Local regional Therapy and Systemic Therapy | Behavior Code | Behavior Code | Behavior Code | Behavior Code | Behavior Code |
| 19 | Surgical | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery |
On the other hand, from the dataset, we learn the types of original cancer and which patient has secondary cancer. However, we do not learn about the types of secondary cancer. Learning the types of secondary cancer is useful for therapeutics and preventive [31]. This is also one of the future research directions of this study.
Conclusion
The present study shows a proposed method using ensemble feature learning to identify the risk factors for predicting secondary cancer by considering class imbalance and patient heterogeneity. In the proposed method, we divide the training data into some heterogeneous groups and construct a diagnosis model for each group for a more accurate prediction. Analysis of the results shows that the accuracies of predicting secondary cancer indeed increased after using the selected important risk factors as predictors. Group division to predict secondary cancer on the separated models can further improve the prediction accuracies. Our results can provide important references to the personality and clinical symptom representations on all phases of guide interventions, with the complexities of multiple symptoms associated with secondary cancer in all phases of the recurrent trajectory.
Ranking results of the importance in the 19 predictor variables for colorectal cancer
| Rank | No division | 5 Groups | ||||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group5 | ||
| 1 | Age | Pathologic Stage | Pathologic Stage | Pathologic Stage | Pathologic Stage | Pathologic Stage |
| 2 | Pathologic Stage | Surgical Margin | Surgical Margin | Surgical Margin | Surgical Margin | Age |
| 3 | Surgical Margin | Smoking | Smoking | Age | Age | Surgical Margin |
| 4 | Betel Nut | Drinking | Drinking | Smoking | Smoking | Smoking |
| 5 | Histology | Age | Age | Drinking | Drinking | Drinking |
| 6 | Dose to clinical target volumes (CTV)_Low | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy |
| 7 | Number to clinical target volumes (CTV) _High | Body Mass Index (BMI) | Body Mass Index (BMI) | Betel Nut | Betel Nut | Body Mass Index (BMI) |
| 8 | Body Mass Index (BMI) | Betel Nut | Betel Nut | Number to clinical target volumes (CTV) _Low | Body Mass Index (BMI) | Betel Nut |
| 9 | Radiotherapy (RT) | Histology | Histology | Body Mass Index (BMI) | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _Low |
| 10 | Smoking | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _Low | Histology | Histology | Dose to clinical target volumes (CTV)_Low |
| 11 | Drinking | Dose to clinical target volumes (CTV)_Low | Differentiation | Dose to clinical target volumes (CTV)_Low | Differentiation | Histology |
| 12 | Number to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _High | Number to clinical target volumes (CTV) _High | Differentiation | Tumor Size | Number to clinical target volumes (CTV) _High |
| 13 | Dose to clinical target volumes (CTV)_High | Surgical | Dose to clinical target volumes (CTV)_Low | Number to clinical target volumes (CTV) _High | Dose to clinical target volumes (CTV)_Low | Differentiation |
| 14 | Radiotherapy (RT) surgery | Differentiation | Surgical | Radiotherapy (RT) | Radiotherapy (RT) | Tumor Size |
| 15 | Differentiation | Radiotherapy (RT) | Radiotherapy (RT) | Tumor Size | Number to clinical target volumes (CTV) _High | Surgical |
| 16 | Behavior Code | Dose to clinical target volumes (CTV)_High | Tumor Size | Surgical | Surgical | Radiotherapy (RT) |
| 17 | Tumor Size | Tumor Size | Dose to clinical target volumes (CTV)_High | Dose to clinical target volumes (CTV)_High | Dose to clinical target volumes (CTV)_High | Dose to clinical target volumes (CTV)_High |
| 18 | Sequence of Local regional Therapy and Systemic Therapy | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Behavior Code | Behavior Code | Behavior Code |
| 19 | Surgical | Behavior Code | Behavior Code | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery |
Ranking results of the importance in the 19 predictor variables for head and neck cancer
| Rank | No division | 5 Groups | ||||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group5 | ||
| 1 | Age | Pathologic Stage | Age | Pathologic Stage | Age | Pathologic Stage |
| 2 | Pathologic Stage | Age | Pathologic Stage | Age | Pathologic Stage | Age |
| 3 | Surgical Margin | Surgical Margin | Surgical Margin | Surgical Margin | Surgical Margin | Surgical Margin |
| 4 | Dose to clinical target volumes (CTV)_Low | Smoking | Smoking | Smoking | Smoking | Drinking |
| 5 | Histology | Drinking | Drinking | Drinking | Body Mass Index (BMI) | Smoking |
| 6 | Betel Nut | Body Mass Index (BMI) | Body Mass Index (BMI) | Body Mass Index (BMI) | Drinking | Body Mass Index (BMI) |
| 7 | Body Mass Index (BMI) | Betel Nut | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy | Sequence of Local regional Therapy and Systemic Therapy |
| 8 | Number to clinical target volumes (CTV) _Low | Sequence of Local regional Therapy and Systemic Therapy | Betel Nut | Betel Nut | Number to clinical target volumes (CTV) _Low | Betel Nut |
| 9 | Number to clinical target volumes (CTV) _High | Number to clinical target volumes (CTV) _Low | Histology | Number to clinical target volumes (CTV) _Low | Dose to clinical target volumes (CTV) _Low | Number to clinical target volumes (CTV) _Low |
| 10 | Drinking | Histology | Number to clinical target volumes (CTV) _Low | Tumor Size | Betel Nut | Dose to clinical target volumes (CTV) _Low |
| 11 | Differentiation | Dose to clinical target volumes (CTV) _Low | Dose to clinical target volumes (CTV) _Low | Histology | Histology | Histology |
| 12 | Dose to clinical target volumes (CTV)_High | Number to clinical target volumes (CTV) _High | Radiotherapy (RT) | Dose to clinical target volumes (CTV) _Low | Tumor Size | Tumor Size |
| 13 | Smoking | Radiotherapy (RT) | Differentiation | Radiotherapy (RT) | Differentiation | Differentiation |
| 14 | Radiotherapy (RT) | Surgical | Number to clinical target volumes (CTV) _High | Differentiation | Number to clinical target volumes (CTV) _High | Number to clinical target volumes (CTV) _High |
| 15 | Radiotherapy (RT) surgery | Differentiation | Surgical | Number to clinical target volumes (CTV) _High | Dose to clinical target volumes (CTV) _High | Dose to clinical target volumes (CTV) _High |
| 16 | Behavior Code | Tumor Size | Tumor Size | Surgical | Radiotherapy (RT) | Radiotherapy (RT) |
| 17 | Sequence of Local regional Therapy and Systemic Therapy | Dose to clinical target volumes (CTV) _High | Dose to clinical target volumes (CTV) _High | Dose to clinical target volumes (CTV) _High | Surgical | Surgical |
| 18 | Tumor Size | Behavior Code | Behavior Code | Behavior Code | Behavior Code | Behavior Code |
| 19 | Surgical | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery | Radiotherapy (RT) surgery |
Acknowledgements
This paper is based on results obtained from a project commissioned by the New Energy and Industrial Technology Development Organization (NEDO).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kourou K, Exarchos TP, Exarchos KP. et al. Machine learning applications in cancer prognosis and prediction. Computational and structural biotechnology journal. 2015;13:8-17
2. Ng A K, Travis L B. Subsequent malignant neoplasms in cancer survivors. The Cancer Journal. 2008;14(6):429-434
3. Kaaks R, Lukanova A, Kurzer M S. Obesity, endogenous hormones, and endometrial cancer risk: a synthetic review. Cancer Epidemiology and Prevention Biomarkers. 2002;11(12):1531-1543
4. WHO: Cancer. https://www.who.int/news-room/fact-sheets/detail/cancer
5. Sun CC, Chang CC. Multiple primary malignant neoplasms: Results from a 5-year retrospective analysis in a Metropolitan Hospital. Formosan Journal of Surgery. 2017;50(6):209-214
6. Muneer JA, Hadeer HM, Anas MSU. et al. Risk and survival of chronic myeloid leukemia after breast cancer: A population-based study. Current Problems in Cancer. 2018
7. Lisik-Habib M, Czernek U, Dębska-Szmich S. et al. Secondary cancer in a survivor of Hodgkin's lymphoma: A case report and review of the literature. Oncology letters. 2015;9(2):964-966
8. Fowble B, Hanlon A, Freedman G. et al. Second cancers after conservative surgery and radiation for stages I-II breast cancer: identifying a subset of women at increased risk. International Journal of Radiation Oncology, Biology, Physics. 2001;51(3):679-690
9. Lin CY, Chen SH, Huang CC. et al. Risk of secondary cancers in women with breast cancer and the influence of radiotherapy: A national cohort study in Taiwan. Medicine (United States). 2016;95:1-7
10. Lee K D, Chen SC, Chan C H. et al. Increased risk for second primary malignancies in women with breast cancer diagnosed at young age: a population-based study in Taiwan. Cancer Epidemiology and Prevention Biomarkers. 2008;17(10):2647-2655
11. Mellemkjær L, Friis S, Olsen J H. et al. Risk of second cancer among women with breast cancer. International journal of cancer. 2006;118(9):2285-2292
12. NCBI: Holland-Frei Cancer Medicine. 6th edition. https://www.ncbi.nlm.nih.gov/books/NBK12712/
13. Rubino C, de Vathaire F, Diallo I. et al. Increased risk of second cancers following breast cancer: role of the initial treatment. Breast cancer research and treatment. 2000;61(3):183-195
14. Travis LB. The epidemiology of second primary cancers. Cancer Epidemiology and Prevention Biomarkers. 2006;15(11):2020-2026
15. Suresh S, Alexander VL. Correlation, Causation and Confounding-What Is the True Risk of Lung Cancer following Breast Cancer Radiotherapy? Journal of Thoracic Oncology. 2017;12(5):773-775
16. Travis LB, Rabkin C S, Brown L M. et al. Cancer survivorship genetic susceptibility and second primary cancers: research strategies and recommendations. Journal of the National Cancer Institute. 2006;98(1):15-25
17. Soni J, Ansari U, Sharma D, Soni S. Predictive data mining for medical diagnosis: an overview of heart disease prediction. Int J Comput Appl. 2011;17(8):43-48
18. Tseng CJ, Lu CJ, Chang CC. et al. Integration of data mining classification techniques and ensemble learning to identify risk factors and diagnose ovarian cancer recurrence. Artificial intelligence in medicine. 2017;78:47-54
19. Liang JD, Ping XO, Tseng YJ. et al. Recurrence predictive models for patients with hepatocellular carcinoma after radiofrequency ablation using support vector machines with feature selection methods. Computer methods and programs in biomedicine. 2014:117 (3), 425-434
20. Santos MS, Abreu PH, García-Laencina PJ. et al. A new cluster-based oversampling method for improving survival prediction of hepatocellular carcinoma patients. Journal of biomedical informatics. 2015;58:49-59
21. Lu CJ, Kao LJ. A clustering-based sales forecasting scheme by using extreme learning machine and ensembling linkage methods with applications to computer server. Engineering Applications of Artificial Intelligence. 2016;55:231-238
22. Ye X, Sakurai T. Robust Similarity Measure for Spectral Clustering Based on Shared Neighbors. ETRI Journal. 2016;38(3):540-550
23. Ye X, Sakurai T. Spectral clustering with adaptive similarity measure in Kernel space. Intelligent Data Analysis. 2018;22:751-765
24. Ye X, Li H, Sakurai T. et al. Large Scale Spectral Clustering Using Sparse Representation Based on Hubness. Proceeding of IEEE CBDCom. 2018
25. Chawla NV, Bowyer KW, Hall LO. et al. SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research. 2002;16:321-357
26. k-nearest neighbors algorithm. https://en.wikipedia.org/wiki/K-nearest_neighbors_algorithm
27. Decision tree. https://en.wikipedia.org/wiki/Decision_tree
28. Support vector machine. https://en.wikipedia.org/wiki/Support_vector_machine
29. Classification: ROC and AUC. https://developers.google.com/machine-learning/crash-course/classification/roc-and-auc
30. F1 score. https://en.wikipedia.org/wiki/F1_score
31. Second Cancers. https://www.livestrong.org/we-can-help/healthy-living-after-treatment/second-cancers
Author contact
![]() Corresponding author: Xiucai Ye, PhD, Department of Computer Science, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8577, Japan. E-mail: yexiucaitsukuba.ac.jp.
Corresponding author: Xiucai Ye, PhD, Department of Computer Science, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8577, Japan. E-mail: yexiucaitsukuba.ac.jp.

 Global reach, higher impact
Global reach, higher impact