Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(7):931-938. doi:10.7150/ijms.32413 This issue Cite
Research Paper
First-line Screening of OXPHOS Deficiencies Using Microscale Oxygraphy in Human Skin Fibroblasts: A Preliminary Study
1. Univ. Lille, Inserm, UMR-S 1172 - JPArc - Centre de Recherche Jean-Pierre AUBERT Neurosciences et Cancer, F-59000 Lille, France
2. CHU Lille, Centre de Biologie-Pathologie Banque de Tissus, F-59000 Lille, France
3. CHU Lille, Centre de Biologie-Pathologie UF Métabolisme général, hormonal et maladies rares, F-59000 Lille, France
4. CHU Lille, Centre de Référence des maladies héréditaires du métabolisme, F-59000 Lille, France
*JK and PM share co-seniorship of this paper
Received 2018-12-19; Accepted 2019-4-11; Published 2019-6-7
Abstract
The diagnosis of mitochondrial diseases is a real challenge because of the vast clinical and genetic heterogeneity. Classically, the clinical examination and genetic analysis must be completed by several biochemical assays to confirm the diagnosis of mitochondrial disease. Here, we tested the validity of microscale XF technology in measuring oxygen consumption in human skin fibroblasts isolated from 5 pediatric patients with heterogeneous mitochondrial disorders. We first set up the protocol conditions to allow the determination of respiratory parameters including respiration associated with ATP production, proton leak, maximal respiration, and spare respiratory capacity with reproducibility and repeatability. Maximum respiration and spare capacity were the only parameters decreased in patients irrespective of the type of OXPHOS deficiency. These results were confirmed by high-resolution oxygraphy, the reference method to measure cellular respiration. Given the fact that microscale XF technology allows fast, automated and standardized measurements, we propose to use microscale oxygraphy among the first-line methods to screen OXPHOS deficiencies.
Keywords: mitochondria, oxidative metabolism, reserve capacity, respiratory chain complex, mitochondrial diseases
1. Introduction
Mitochondrial diseases refer to a heterogeneous group of disorders resulting from primary dysfunctions of the mitochondrial electron transport chain and/or ATP synthase (1). Mitochondrial diseases have different genotypes as well as presenting with highly different clinical, and biochemical phenotypes rendering the diagnostic evaluation very challenging for clinicians. Thus, they present different aspects. Clinical presentations range from Leigh syndrome, a devastating neurodegenerative pathology, occurring in children under 2 years of age and progressing rapidly towards death (2) to MELAS syndrome, associating encephalopathy, lactic acidosis and stroke-type episodes characterized by normal psychomotor development (3,4). Defects in mtDNA account for only around 15% of known mitochondrial pathologies indicating that mitochondrial disorders are frequently related to mutations of nuclear DNA. In recent years, the diagnosis diagram has been greatly disrupted by the appearance of next generation sequencing (NGS) techniques leading to a better understanding of gene-related mitochondrial dysfunction. This is a new way for clinicians to evidence mitochondrial dysfunction and the genetic approach is now widespread (5). However, despite the power of this genetic tool, interpretation is difficult because of the huge number of target genes and the poor correlation between genetic data and clinical/biochemical phenotype (6). Regardless of clinical presentations and the localization of DNA mutations, mitochondrial pathologies have common dysfunctions in the mitochondrial respiratory chain. Consequently, the determination of mitochondrial function by biochemical techniques is useful to help establish a mitochondrial disease diagnosis. Functional in vitro assays in skeletal muscle have been the gold standard for diagnosis of mitochondrial disorders. However, it needs an invasive skeletal muscle biopsy often performed under general anesthesia limiting their practical use in pediatric patients. Alternatively, the easily accessible primary skin fibroblasts from patients can be used to identify mitochondrial dysfunction (7).
Functional investigations usually include spectrophotometric assays of ETC enzyme activity as well as polarographic measurements of oxygen consumption, each assessment contributing to giving clues to the diagnosis of an OXPHOS dysfunction. Determinations of oxygen consumption by polarographic measurements in intact cells are sensitive and close to the “in vivo” situation. It provides a complete study of the bioenergetic mitochondrial state including the determination of ATP renewal speed, mitochondrial coupling and adaptability of mitochondria to react to stress. However, the polarographic assays including high-resolution respirometry are complex and time consuming, limiting their clinical interests in routine diagnosis. As an alternative to polarographic studies, the microscale fluorescent based technology (microscale XF technology) allows the analysis of automated oxygen consumption with oxygen-sensing fluorophores in unpermeabilized cells on 24 or 96 plates. To avoid the drawbacks of polarographic studies, microscale oxygraphy has commonly been used in research and is starting to be used for screening purposes. It has recently been experimented for the diagnosis of Leigh syndrome in combination with enzymatic and genetic approaches (8). Studies indicate that the Microscale XF technology is highly efficient for detecting mitochondrial respiratory defects in genetically proven mitochondrial disease patients (8,9).
Moreover, one major advantage of the microscale XF technology is its ability to determine simultaneously various integrated bioenergetics parameters including the respiration linked to ATP production, proton leak rate, maximum respiratory rate, as well as spare respiratory capacity in the same population of cells (10).
Here we outline a simple protocol, compatible with diagnostic use, and optimized to determine the basic bioenergetics functions of fibroblasts using the microscale XF technology. In this protocol, we assessed seven mitochondrial parameters in fibroblasts isolated from 5 pediatric patients with heterogeneous mitochondrial disorders in order to determine which parameters are the most reliable in detecting mitochondrial dysfunctions.
2. Materials and methods
2.1. Patients
The retrospective analysis included 5 patients (from unrelated families) who had a muscle biopsy at the Lille University Hospital center between 2016 and 2017. Respiratory chain disorders were confirmed either by muscle enzymatic assays and/or molecular-genetic testing but also by clinical, histological and biological markers (Table 1). Written informed consent for research purposes was obtained for all patients. The study was performed in accordance with the Declaration of Helsinki for experiments involving human samples.
2.2. Cell Culture
We analyzed a total of 6 fibroblast cell lines including different genetically proven OXPHOS-related defects. Frozen skin fragments taken from the thigh of patients (n = 5) and pediatric healthy volunteers (n = 1) were used for the preparation of fibroblast cultures. Cells were thawed and cultured at 37°C to 85% confluence according to the established hospital culture protocol in a 2: 1 mixture of Advanced DMEM F12 (Gibco - Thermo Fisher Scientific, Waltham, USA) and reconstituted AmnioMAX (AmnioMAX C-100 complement vial (Gibco) reconstituted in a bottle of AmnioMAX C-100 basal medium (Gibco)) supplemented with 10% fetal bovine serum (Gibco), 1% penicillin and streptomycin (Invitrogen - Thermo Fisher Scientific, Waltham, USA) and 50 μg / ml uridine (Sigma-Aldrich - Merck, Darmstadt, Deutschland). All cells (from patients and control) used in this study were in the 5th and 10th passage.
2.3. Enzyme activities of mitochondrial respiratory chain complexes
Activities of mitochondrial respiratory chain complexes were determined in skeletal muscle suspension according to established method and adjusted to citrate synthase activity, used as indicator of mitochondrial content, as previously described (11). All patient enzymatic muscle activities were calculated as percentage of the control.
2.4. Genetic analysis
Genetic analysis was retrieved from the patient medical file when available. They were conducted in human genetic diagnostic reference centers as indicated (12).
2.5. Microscale oxygraphy
Oxygen consumption rate (OCR) was measured in adherent fibroblasts with a XFe24 Extracellular Flux Analyzer (Seahorse Bioscience - Agilent Technologies, Santa Clara, CA, USA). Each control and mutant fibroblast cell lines were seeded in 12 wells of a XF e24-well cell culture microplate (Seahorse Bioscience) at a density of 25*103 cells/well in 100 μL of standard culture media and incubated for 18 hours at 37°C in 5% CO2 atmosphere. After replacing the growth medium with 500 μL of pre-warmed at 37 °C bicarbonate-free DMEM (DMEM, Sigma -Aldrich - Merck) supplemented with 10 mL of 100mM L-Glutamine (Thermo Fisher Scientific), 5mL of 100mM Sodium Pyruvate (Thermo Fisher Scientific) and 4,5mL of sterile 20% glucose (Invitrogen - Thermo Fisher Scientific). Cells were preincubated for 30min before starting the assay procedure as previously reported (13). Briefly after baseline measurements of OCR (on endogenous substrates), OCR was measured after sequentially adding to each well 1 μM oligomycin (inhibitor of ATP synthase), then maximal OCR was determined with 1 to 3 μM of carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, Sigma-Aldrich - Merck) (uncoupler of oxidative phosphorylation) and 1 μM of rotenone (Sigma-Aldrich - Merck ) plus antimycin (Sigma-Aldrich - Merck) (inhibitors of mitochondrial complex I and III) for determination of rotenone-antimycin insensitive respiration.
Data were expressed as pmol of O2 per minute and normalized by cell number measured by the CyQUANT Cell proliferation kit (Invitrogen - Thermo Fisher Scientific), which is based on a fluorochrome binding to nucleic acids with fluorescence measured in a microplate luminometer (excitation wavelength at 485±10nm, emission detection wavelength at 530±12.5 nm). Seven parameters were evaluated and all determinations were performed in 12 replicates for each sample:
- Mitochondrial Basal OCR (corresponds to baseline OCR minus rotenone/antimycin-insensitive OCR)
- ATP-linked OCR (corresponds to basal OCR minus oligomycin-insensitive OCR).
- Proton leak-linked OCR (corresponds to oligomycin-insensitive OCR minus rotenone/antimycin-insensitive OCR).
- Maximal OCR (corresponds to FCCP-induced OCR minus rotenone/antimycin-insensitive OCR).
- Spare respiratory capacity measured as the difference between Maximal and Basal OCR.
- Non-mitochondrial OCR (corresponds to rotenone/antimycin-insensitive OCR).
Bioenergetic Health Index, a composite index of mitochondrial wellness, determined according to the following formula (14).
Where a, b, c and d exponents modify the relative weight of each respiratory parameter, they are by default equivalent to 1 and can be modulated to maximize the contrast between two experimental conditions.
2.6. High-resolution respirometry
A 1.5 × 106 cells/mL pellet was resuspended in the same warm (37°C) medium. High-resolution respirometry was carried out using an Oxygraph-2k instrument (Oroboros Instruments GmbH, Innsbruck, Austria). Oxygraph sensors were calibrated once a day before the experiment. Experiments were performed, according to the standard protocol described above. Initially, basal cell respiration (on endogenous substrates) was measured, followed by the addition of 1 μM oligomycin, then maximal OCR was determined by titrating with 0.0275 μM of FCCP, followed by the addition of 1 μM rotenone plus antimycin A. Respirometry data were calculated using the DatLab 5.0 software and normalized to the cell count in the chamber.
2.7. Statistical analysis
All statistical analyses were performed using Anova and T-tests on Prism 7.0 (Graphpad Software, La Jolla, USA). For all Seahorse experiments, data referred to patient cell lines are presented as the mean of replicates ± standard deviation between the replicates (SD). For Oroboros experiments data referred to patient cell line are presented as average OCR of a given range of time +/- standard deviation within this range of time (SD).
3. Results
3.1. Skin fibroblasts from patients
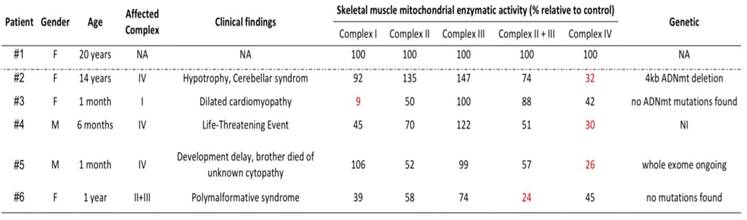
As shown in Table 1, we used skin fibroblast cell lines from 5 pediatric patients with heterogeneous clinical presentation, biochemical results, and genetic data: one specific to complex I; one to complex II+III and three to complex IV. As controls, we used fibroblast cells derived from a healthy young female donor of 20 years, present at each experiment (Table 1).
3.2. Optimization of microscale oxygraphy: adjustment of the number of fibroblasts and FCCP concentration
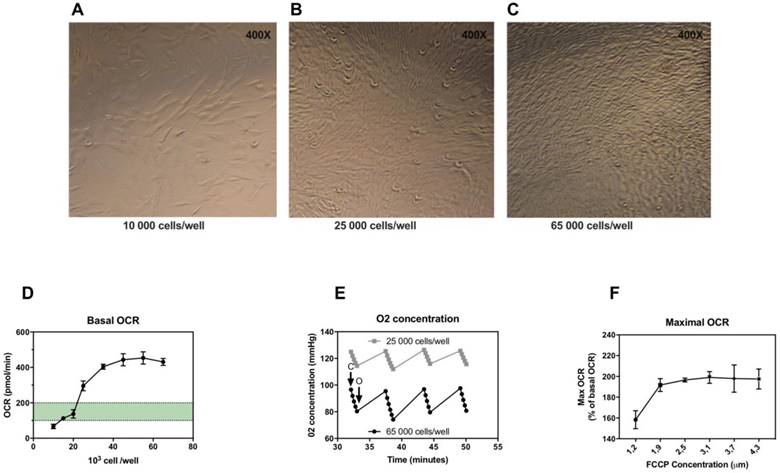
In the first experiments, we determined the optimal number of skin fibroblasts needed to obtain a measurable and reproducible OCR. According to (15), optimal density of cells must be chosen to target OCR values above background values comprised between 100 and 200 pmol/min. We determined the optimal seeding number of skin fibroblasts per well (figure 1A, 1B and 1C) checking cell confluence in each well after 18 hours of incubation and determining oxygen consumption rate (OCR) for each cell concentration. Fibroblast sub-confluence was reached for a concentration of 25,000 cells/well corresponding to an OCR value above background values. OCR increased with increasing cell number from 20,000 to 40,000 per well (Figure 1D), after which OCR signals reached a plateau. Maximal OCR was reached as early as 35,000 cells/well (Figure 1D). We also determined the oxygen concentration during maximal OCR stage, in order to check the good re-oxygenation (back to baseline O2 concentration) of the well at each measurement cycle. As shown in Figure 1E, the oxygenation was sufficient with a return to O2 concentration baseline regardless of cell concentration. Thus, we recommend to use for microscale oxygraphic assays a seeding density in the range of 25,000 to 35,000 skin fibroblasts per well.
Secondly, FCCP, the uncoupling agent injected, should be optimized for the concentration providing the maximal respiratory effect. Optimal concentrations of FCCP were determined by monitoring OCR during FCCP titration (Figure 1F). Maximal stimulation of OCR was achieved for concentrations between 1.85 and 2.50 μM of FCCP.
3.3. Repeatability and reproducibility of microscale oxygraphy
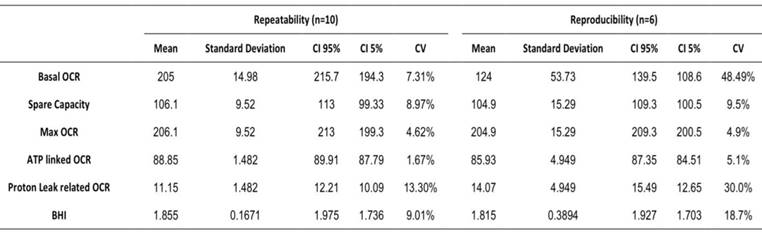
In order to determine if microscale oxygraphy can be used as a diagnostic method we evaluated its reproducibility and repeatability. Repeatability was studied within the same plate during the same assay whereas reproducibility was studied during six different assays at different times. Results are shown in Table 2, and indicate that calculated coefficients of variation were mostly lower than 10 % and thus compatible with a clinical use.
Optimization of microscale oxygraphy for skin fibroblasts. A-C, Seahorse cell culture plates after 18 hours incubation, 10,000, 25,000 and 65,000 cells/well respectively, confluence is reached for 25,000 cells per well. D, Basal oxygen consumption rate (OCR) in relation to cell number per well. E, Oxygen concentration in relation to cell number per well in maximal oxygen consumption state after FCCP injection. C: closing of the chamber, O: opening of the chamber (re-oxygenation). F, Maximal OCR in relation to FCCP concentration (first and second injection combined).
Patient characteristics. Enzymatic activities normalized by Citrate Synthase activity and expressed as a percentage of control enzymatic activities.
Repeatability and reproducibility of OCR analysis on Seahorse XFe24. Mean, standard deviation, confidence interval (CI) and coefficient of variation. Basal OCR in pmol/min and normalized by Cyquant, spare capacity, maximal, ATP linked, proton leak-related OCR are expressed as a percentage of basal OCR. Bioenergetic Health Index is expressed as arbitrary units.
3.4. Microscale oxygraphy in skin fibroblasts from patients
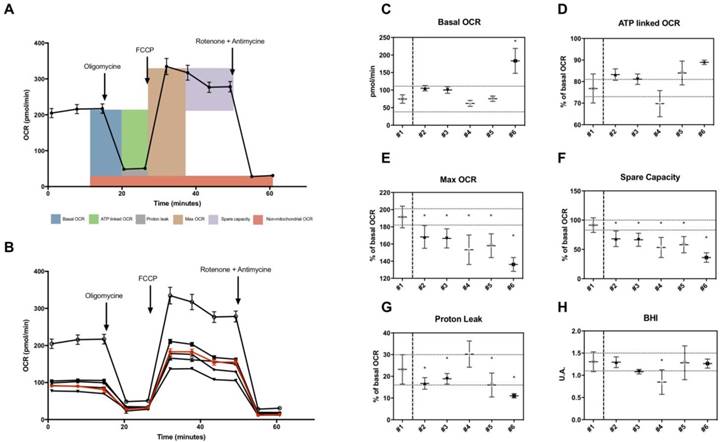
Using the XF Extracellular Flux analyzer, we measured OCR in different conditions following the general scheme of analyses shown in Figure 2A. Values of OCR were used to estimate several parameters including basal OCR, ATP-linked OCR, maximal OCR, spare capacity, proton leak-related OCR in skin fibroblasts (Figure 2B). Basal OCR and ATP-linked OCR were not decreased in fibroblasts from patients (Figure 2C and 2D). Proton leak-linked OCR were decreased in most patients except for patient n°4 (Figure 2G). Among OCR-derived parameters measured, only maximal OCR and spare capacity, representing the mitochondrial reserve to respond to energy demand, were significantly lower in all patients (Figure 2E and 2F). Mean values of spare capacity (% of basal OCR) in patients were 56.5 ± 9,4. There was no significant difference regarding BHI (Figure 2H).
3.5. High-resolution respirometry in skin fibroblasts from patients
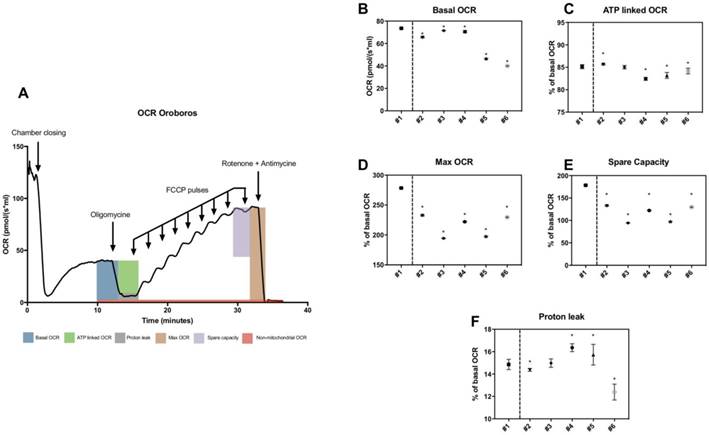
High-resolution respirometry was also used to determine basal OCR, maximal OCR, ATP-linked OCR, spare capacity, proton leak-related OCR in patients (figure 3A). All parameters including maximal OCR and spare capacity were significantly lower in all patients (figure 3D and 3E). Mean values of spare capacity (% of basal OCR) in patients were 66.21 ± 17.93.
Thus, variable but consistent reduction of maximal OCR and spare capacity were observed in all patients by both techniques.
4. Discussion
OXPHOS disorders represent a diagnostic challenge due to their clinical heterogeneity but also their genetic complexity underlining the importance of combining multiple diagnostic methods (8). Although widely recommended for the diagnosis of OXPHOS disorders (4), polarographic measurements of oxygen consumption are not a routine technique used on a large scale in clinical laboratories. The reasons are essentially that the methods proposed were time-consuming, laborious and required technical personnel with substantial experience. Here we propose a procedure readily compatible with clinical use. Monitoring respiration with micrograph oxygraphy allows for faster, more automated and exhibiting higher throughput measurements than classical methods such as polarographic measurement. Micrograph oxygraphy also has the advantage to be easy to use at hospital in first-level screening conditions of patients with suspected mitochondrial disorders. Furthermore, in contrast to polarographic methods, micrograph oxygraphy allows to work on adherent cells, a situation closer to in vivo conditions. For all these reasons, micrograph oxygraphy is a simple, fast and reliable technique that could be useful as a first-line OXPHOS deficiency screening technique.
Microscale oxygraphy in skin fibroblasts from patients. A, General scheme of OCR measurement under basal conditions followed by the sequential addition of oligomycin, FCCP and rotenone plus antimycin A, as indicated. Each data point represents an OCR measurement (mean +/- SD; n> 10). B, OCR profile in studied fibroblasts. All patients are represented in black, control appears in red. Each data point represents an OCR measurement (mean +/- SD; n> 10). C, Basal OCR in pmol/min. D-G, ATP-linked, maximal, spare capacity and proton leak linked OCR as a percentage of basal OCR. Dotted lines represent estimated variation range within the control (coefficient of variation as calculated in reproducibility test). H, Bioenergetic Health index in arbitrary units. Healthy control is #1 and patients #2 to #6. The asterisk indicates the ratios which significantly differ (p < 0.05) between the control patient and fibroblast cultures from patients with mitochondrial cytopathy. Mean +/- SD.
Herein, we established a reliable protocol to measure mitochondrial respiratory function by microscale oxygraphy using the Seahorse XF24 extracellular flux analyzer adapted for skin fibroblasts isolated from patients. Our protocol was developed to provide reliable results while maintaining the simplicity of the procedure, fully compatible with clinical use. We carried out this protocol on skin fibroblasts which, even if they have a metabolic activity often less important than muscle, allow for less invasive samples. In these conditions, we established that this method is both reproducible and repeatable. Among the parameters determined, we observed that basal respiration on its own, even with normalized results, presents a significant variability while calculated OCR parameters, such as maximal OCR and spare capacity, were more reliable with variation coefficients under 10%.
Most importantly, our works identified potential mitochondrial parameters relevant for detection of mitochondrial disorders in human skin fibroblast. Results from microscale oxygraphy demonstrated that maximal OCR and spare capacity were the only parameters decreased in all patients tested, regardless of their OXPHOS disorders. We also confirmed these results using high-resolution oxygraphy, i.e. the gold standard method to detect mitochondrial respiration. Both methods achieved high sensitivity in the measurement of maximum OCR and spare capacity (i.e. representing the difference between maximum and basal OCR). These results are in agreement with previous data (9), showing a significant decreased spare capacity measured with microscale oxygraphy in patients with mtDNA mutations. Moreover, maximum OCR determined with microscale oxygraphy was more sensitive than the spectrometric determination of enzyme activities in fibroblasts from patients with Leigh syndrome (8). The decrease in maximum OCR and spare capacity may indicate a loss of mitochondrial adaptation capacity that logically could be reduced in patients with mitochondrial cytopathy. Interestingly, we observed in our study a correlation between enzymatic activities of Complex II+III and the values of maximal OCR determined by microscale oxygraphy (p: 0.0033, R2 0.91 Pearson) confirming previous data indicating a dependence of spare capacity on complex III activity (16). Altogether, these findings indicate that maximal OCR and spare capacity are reliable to use for objective measurement of mitochondrial function on skin fibroblasts in clinical assessment of OXPHOS disorders. Thus, we suggest that, in addition to genetic screening and enzymatic assays, a maximum OCR/spare capacity determination by microscale oxygraphy to help analyze mitochondrial activity.
High-resolution respirometry in skin fibroblasts from patients. A, General trace of OCR measurement under basal conditions followed by the sequential addition of oligomycin, FCCP and rotenone plus antimycin A, as indicated. B, Basal OCR in pmol/(s*ml). C-F, ATP-linked, maximal, spare capacity and proton leak-linked OCR as a percentage of basal OCR. Healthy control is #1 and patients #2 to #6. The asterisk indicates the ratios which significantly differ (p < 0.05) between the control patient and fibroblast cultures from patients with mitochondrial cytopathy. Mean +/- SD.
5. Conclusion
Our work suggests the promising value of determining maximum OCR/spare capacity by microscale oxygraphy as a first-line screening tool to detect MRC deficits, especially in skin fibroblasts. Further studies enrolling high number of patients are needed to confirm their pertinence in a routine screening setting.
Highlights
- Measurement of respiratory parameters by microscale oxygraphy can be used in human skin fibroblasts of pediatric patients with OXPHOS deficiencies.
- A clear protocol is provided to allow reproduction by others
- Maximum respiration and spare capacity are the best parameters to detect OXPHOS deficiencies
- We propose to include microscale oxygraphy as a first-line method to screen for OXPHOS deficiencies
Competing Interests
The authors have declared that no competing interest exists.
References
1. Murayama K, Shimura M, Liu Z, Okazaki Y, Ohtake A. Recent topics: the diagnosis, molecular genesis, and treatment of mitochondrial diseases. J Hum Genet. 2019Feb;64(2):113-25
2. Lee JS, Kim H, Lim BC. et al. Leigh Syndrome in Childhood: Neurologic Progression and Functional Outcome. J Clin Neurol. 2016;12:181-187
3. Gorman GS, Chinnery PF, DiMauro S. et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080
4. Bauer MF, Gempel K, Hofmann S. et al. Mitochondrial disorders. A diagnostic challenge in clinical chemistry. Clin Chem Lab Med. 1999;37:855-876
5. Wortmann SB, Mayr JA, Nuoffer JM. et al. A Guideline for the Diagnosis of Pediatric Mitochondrial Disease: The Value of Muscle and Skin Biopsies in the Genetics Era. Neuropediatrics. 2017;48:309-314
6. Dimmock DP, Lawlor MW. Presentation and Diagnostic Evaluation of Mitochondrial Disease. Pediatr Clin North Am. 2017;64:161-171
7. Mitochondrial Medicine Society's Committee on Diagnosis, Haas RH, Parikh S, et al. The in-depth evaluation of suspected mitochondrial disease. Mol Genet Metab. 2008;94:16-37
8. Ogawa E, Shimura M, Fushimi T. et al. Clinical validity of biochemical and molecular analysis in diagnosing Leigh syndrome: a study of 106 Japanese patients. J Inherit Metab Dis. 2017;40:685-693
9. Invernizzi F, D'Amato I, Jensen PB. et al. Microscale oxygraphy reveals OXPHOS impairment in MRC mutant cells. Mitochondrion. 2012;12:328-335
10. Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435:297-312
11. Spinazzi M, Casarin A, Pertegato V. et al. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat Protoc. 2012;7:1235-1246
12. Association Nationale des Praticiens de Biologie Moléculaire. Arbres Décisionnels [online]. Fiche 121-Maladies Mitochondriales-ADNmt. available at http://www.anpgm.fr/.
13. Marinangeli C, Kluza J, Marchetti P. et al. Study of AMPK-Regulated Metabolic Fluxes in Neurons Using the Seahorse XFe Analyzer. Methods Mol Biol. 2018;1732:289-305
14. Chacko BK, Kramer PA, Ravi S. et al. The Bioenergetic Health Index: a new concept in mitochondrial translational research. Clin Sci (Lond). 2014;127:367-373
15. Jensen PB. Measuring Mitochondrial Defects. Genetic Engineering & Biotechnology News. 2014;34:19-19
16. Sriskanthadevan S, Jeyaraju DV, Chung TE. et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood. 2015;125:2120-2130
Author contact
![]() Corresponding author: Prof. Philippe MARCHETTI, MD, PhD. INSERM UMR-S 1172 Faculté de Médecine Université de Lille 1, place Verdun F-59045 Lille Cedex France. Tel: 33-3-20 29 88 51 33-3-20 16 92 29 E-mail: philippe.marchettifr
Corresponding author: Prof. Philippe MARCHETTI, MD, PhD. INSERM UMR-S 1172 Faculté de Médecine Université de Lille 1, place Verdun F-59045 Lille Cedex France. Tel: 33-3-20 29 88 51 33-3-20 16 92 29 E-mail: philippe.marchettifr

 Global reach, higher impact
Global reach, higher impact