Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(7):590-598. doi:10.7150/ijms.11525 This issue Cite
Research Paper
Dysregulated Serum MicroRNA Expression Profile and Potential Biomarkers in Hepatitis C Virus-infected Patients
1. Shenzhen Key Lab of Health Science and Technology, Division of Life Science & Health, Graduate School at Shenzhen, Tsinghua University, Shenzhen 518055, China
2. Zhu Jiang Hospital, Southern Medical University, Guangzhou 510282, China
3. Department of health inspection and quarantine, School of Public Health, Sun Yat-sen University, Guangzhou 510080, China
4. Central Laboratory of Health Quarantine, International Travel Health Care Center, Shenzhen Entry-exit Inspection and Quarantine Bureau, Shenzhen 518033, China
5. The Chinese University of Hong Kong, School of Life Sciences, Biochemistry Programme, The Chinese University of Hong Kong, Room 609, Mong Man Wai Building, Shatin, NT, Hong Kong, China
† Contribute equally.
Received 2015-1-7; Accepted 2015-7-7; Published 2015-7-16
Abstract
Objectives: Circulating microRNAs (miRNAs) play critical roles in pathogen-host interactions. Aberrant miRNA expression profiles might have specific characteristics for virus strains, and could serve as noninvasive biomarkers for screening and diagnosing infectious diseases. In this study, we aimed to find new potential miRNA biomarkers of hepatitis C virus (HCV) infection.
Methods: Expression levels of broad-spectrum miRNAs in serum samples from 10 patients with HCV viremia and 10 healthy volunteers were analyzed using miRNA PCR arrays. Subsequently, the differential expression of four selected miRNAs (miR-122, miR-134, miR-424-3p, and miR-629-5p) was verified by qRT-PCR in the serum of 39 patients compared with that in 29 healthy controls. Receiver operating characteristic (ROC) curve analysis was performed to evaluate their potential for the diagnosis of HCV infection.
Results: miRNA PCR array assays revealed differential expression of 106 miRNAs in sera of HCV patients compared with that in healthy controls. Serum hsa-miR-122, miR-134, miR-424-3p, and miR-629-5p were well identified. The ROC curves showed that miR-122, miR-134, miR-424-3p, and miR-629-5p could distinguish HCV patients with preferable sensitivity and specificity. In addition, Correlation analysis indicated serum miR-122 expression was positive correlation with ALT/AST levels. Functional analysis of target proteins of these miRNAs indicated the involvement of viral replication, inflammation, and cell proliferation.
Conclusion: HCV patients have a broad 'fingerprint' profile with dysregulated serum miRNAs compared with that in healthy controls. Among these, serum hsa-miR-122, miR-134, miR-424-3p, and miR-629-5p are identified as promising indication factors of the serum miRNA profile of HCV infection. Particularly, miR-122 could be one of serum biomarkers for early pathological process of HCV. However, more miRNA biomarkers and biological functions of these miRNAs require further investigation.
Keywords: microRNAs, hepatitis C virus, miR-122, miR-134, miR-424, miR-629
Introduction
Hepatitis C virus (HCV), a type of positive single-stranded RNA virus, is one of the leading causes of viral hepatitis with worldwide pandemic. According to previous reports, the average global prevalence of hepatitis C is approximately 3.0%, and 3.0-4.0 million individuals are subjected to HCV infection every year, of which 75%-80% develop chronic infection and more than 20% have cirrhosis and hepatocellular carcinoma (HCC) [1, 2]. In spite of similar pathologic and transmission characteristics, HCV can be divided into six genotypes with high variability worldwide; thus, effective measures for the prevention and treatment of HCV are difficult to find [3, 4]. New biomarkers for the diagnosis, treatment, and prognosis of HCV infection are urgently needed.
MicroRNAs (miRNAs) are a class of small non-coding single-stranded RNA of about 22 nucleotides (nt). They regulate the post-transcriptional expression of target genes in a classic way of perfect or imperfect complementation to target mRNAs, and cause corresponding mRNA degradation or translation inhibition [5-7]. However, in infectious diseases, miRNAs can also directly target the genome of viruses to regulate their replication. MiR-122, a specific highly expressed miRNA in liver tissues, promotes HCV replication through direct interaction with the 5' end of the HCV RNA genome [8]. By contrast, miR-199a, let-7b, miR-448, and miR-196 have been identified to suppress HCV infection by connecting with their own targets on the genome of HCV [9-11].
Recently, circulating miRNAs have attracted much attention for their potential as noninvasive biomarkers for screening and diagnosing various diseases, including infectious diseases. On one hand, circulating miRNAs are stored in exosomes with sufficient stability [12-15]. On the other hand, they have great specificity for discriminating specific diseases. For example, a combination of let-7c, miR-23b, miR-122, and miR-150 can clearly separate patients with occult hepatitis B virus (HBV) infection from healthy controls [16]. For HCV infection, a very recent report showed that serum miR-134, miR-198, miR-320c, and miR-483-5p are significantly up-regulated in different genotypes, and may serve as biomarkers for the diagnosis of HCV infection [17].
In the present study, 768 miRNAs in sera of HCV patients and healthy controls were screened for different expression profiles to explore the potential biomarkers for the detection of HCV infection or its complications. The results were further verified by qRT-PCR, and potential biological functions were also analyzed by bioinformatics.
Materials and Methods
Sample collection
A total of 68 serum samples (39 patients with active HCV replication and 29 healthy volunteers) were obtained from Zhujiang Hospital of Guangzhou, Guangdong Province. Healthy controls were recruited randomly from individuals who had no clinical symptoms of infectious diseases after regular physical examination, and HCV patients enrolled in this study were confirmed to have no other infectious diseases, such as HBV, HIV, and HSV, and have no drug treatment, and also have no obvious hepatic steatosis, hepatic fibrosis, and hepatic tumors. Serum samples were isolated within 1 h after receiving whole blood and then immediately stored at -80 °C for standby use. This study was approved by the Ethics Committee of Zhujiang Hospital of Guangzhou, Guangdong Province, and written informed consent was obtained from all participants.
RNA extraction
Total RNA was extracted from serum samples by using Trizol LS reagent Invitrogen, USA) following the manufacturer's instructions. For miRNA PCR assay, about 1-2 ml of serum was used to extract total RNA. For RT-PCR validation assays, about 250-500 μl of serum was used to extract mRNA. Here, we take 250 μl of serum for example. Briefly, 250 μl of serum and 750 μl of Trizol LS reagent were efficiently mixed in Eppendorf tubes and incubate at room temperature for 5 minutes. Then, 0.2 ml of chloroform was added into the mixture. The Eppendorf tubes contained the mixture were shaken vigorously by hand for 15 seconds and incubated at room temperature for 2 to 3 minutes. Then, the samples were centrifuged at 13,000 × g for 15 minutes at 4°C. Following centrifugation, the mixture separated into a lower red, phenol-chloroform phase, an interphase, and a colorless upper aqueous phase. RNA remained exclusively in the aqueous phase. After transferring about 0.5 ml of the aqueous phase into a new Eppendorf tube, about 0.5 ml of isopropyl alcohol and 5 μl of RNase-free glycogen per 1 ml of TRIZOL-LS Reagent were further added for the initial homogenization. After incubating at 4 °C for 30 minutes, the samples were centrifuged at 13,000 × g for 15 minutes at 4°C. Then RNA pellets were washed once with 1 ml of 75% ethanol. The RNA pellets were air-dried for 5-10 minutes and dissolved in 20 μl of RNase-free water. The purity and concentration of isolated RNA were evaluated through a NanoDrop® ND-1000 spectrophotometer (Thermo Scientific, USA). Extracted RNA concentration from 1 ml of serum was about 40-50 ng/μl in 20 μl of RNase-free water. OD260/280 and OD260/230 ratios were about 1.8 and 1.6, respectively. For miRNA PCR assay, denaturing agarose gel electrophoresis was carried out to further confirm the quality. No smearing of ribosomal RNA bands were observed. This suggests that RNA was not dissolved or degraded.
MiRNA expression profiles using miRNA PCR arrays
Serum pools produced by mixing 10 of 39 patients' samples and 10 of 29 healthy control samples (mix with identical volume of serum from each sample) were used for miRNA PCR arrays (Human panel I+II, V3.M, KangChen Bio-tech, Shanghai, China). In brief, relative expressions of 768 miRNAs in sera from the HCV positive group and healthy control were screened by using miRNA PCR arrays. The total RNA sample was diluted to 1.5-1.8 ng/µl (20-25 ng, 14 μl) in nuclease-free water. Reverse transcription (RT) was carried out in a RT reaction mix (Exiqon, Denmark) containing 4 μl of fivefold reaction buffer, 2 μl of enzyme mix, and 14 μl of diluted total RNA. cDNA was diluted by 110-fold in nuclease-free water, and amplified using SYBR™ Green master mix (Exiqon, Denmark) with an ABI PRISM 7900 Real-time PCR System (Applied Biosystems, USA) according to the instructions. A Ct detection threshold of more than 38 was defined as beyond the detection limit (undetected), and U6 snRNA was used as the internal reference for normalization because U6 expression was relatively stable in this case.
Verifying miRNA array data by quantitative real-time PCR (qRT-PCR)
For further validation, total RNAs of sera in 39 HCV patients and 29 healthy controls were subjected to further miRNA validation assay via qRT-PCR. An miRNA assay kit (GenePharma, Shanghai, China) was used for miRNA detection and quantification. In brief, the RT reaction was performed using a PrimeScriptTM First Strand cDNA Synthesis Kit (Takara, Dalian, China) with an AlphaTM Unit Block Assembly for DNA EngineH systems (Bio-Rad, USA) under the following reaction conditions: 30 min at 25 °C, 30 min at 42 °C, 5 min at 85 °C, and maintained at 4 °C. The final reaction volume was 10 μl containing 2 μl of miR-RT buffer, 0.375 μl of dNTP, 0.6 μl of miRNA-specific RT primer, 0.125 μl of RNase inhibitor, 0.1 μl of MultiScribe reverse transcriptase, 5.8 μl of nuclease-free water, and 1 μl of total RNA. cDNA was then amplified and quantified using SYBR Green I dye (Takara, Dalian, China) with an ABI PRISM 7300 Real-time PCR System (Applied Biosystems, USA) under 95 °C for 3 min, followed by 40 cycles of 95 °C for 12 s and 62 °C for 40 s. The reaction volume was 20 µl containing 10 μl of SYBR master mix, 0.4 μl of miRNA primer set, 7.6 μl of nuclease-free water, and 2 μl of cDNA. U6 snRNA was used as the internal reference for normalization.
Data analysis
Initial data analysis was performed using the software supplied with the real-time PCR instrument to obtain raw Ct values (Cp or Cq). The relative expression of miRNA was calculated by the 2-ddCt formula, in which dCt = Ct miRNA - Ct U6 snRNA, ddCt = dCt HCV patients - dCt Healthy controls. Subsequently, the relative quantification value underwent log2 transformation to compare the expression levels of candidate miRNAs between healthy controls and patients. The data were expressed as the mean ± SD. Statistical significance of the data was evaluated using one-way ANOVA via SPSS software. Post-hoc comparisons were used to determine the source of significant differences. P < 0.05 was considered statistically significant. Receiver operating characteristic (ROC) curve analysis was performed for selected miRNAs. In addition, the area under the curve (AUC) values and 95% confidence intervals (CIs) were calculated to evaluate the specificity and sensitivity for detecting HCV infection. Correlation and significance analysis were conducted by the website (http://vassarstats.net/); P < 0.05 was considered statistically significant.
Target prediction and functional analysis
To conduct a pilot investigation for the functions of these verified miRNAs whose roles during HCV infection have not been clearly identified, miRNA target prediction and functional analysis were performed through miRecords software (http://mirecords.biolead.org/) and previous reports (http://www.ncbi.nlm.nih.gov/pubmed/). Functional analysis of target proteins was conducted based on the website (http://www.uniprot.org/).
Results
Sample characteristics
For miRNA arrays, the mixed sera of the control group were composed of sera from 10 healthy volunteers, whereas the mixed sera of the positive group were derived from 10 patients with HCV viremia. For q-PCR verification, 68 serum samples (29 controls and 39 patients) were enrolled in this study. The age and sex distribution of the two groups showed no statistically significant differences (P > 0.05, Table 1). No other infectious diseases were involved. Also, these patients belonged to newly diagnosed cases and did not subject to any drug treatment and also did not show any obvious syndromes or complications (e.g. hepatic steatosis, fibrosis, and tumors for HCV) by regular physical examination. Furthermore, 7, 2 and 1 out of 10 patients for miRNA arrays were identified as HCV subtype 1b, 2a and 3a, respectively, by using the method of PCR florescence probe (diagnostic kit for HCV genotyping, Triplex International Biosciences (China) Co. LTD). For q-PCR verification, 30, 4 and 2 out of 39 patients were identified as HCV subtype 1b, 2a and 3a, respectively. Other 3 patients could not be identified as any HCV subtype. Despite this, most of HCV patients (≥70%) belonged to subtype 1b.
Basic characteristics of healthy controls and patients enrolled in the study
| miRNA PCR Array | PCR Validation | |||
|---|---|---|---|---|
| Sample Characteristics | Controls | Patients | Controls | Patients |
| Number | 10 | 10 | 29 | 39 |
| Genotype (1b/2a/3a/others) | No | 7/2/1/0 | No | 30/4/2/3 |
| Sex (male/female) | 4/6 | 6/4 | 16/13 | 24/15 |
| Age (Mean±SD) | 41.7±11.0 | 42.7±7.1 | 45.0±16.1 | 49.0±14.3 |
| Viral load (IU/ml) | RNA(-) | ≥1.0×105 | RNA(-) | ≥5.0×102 |
| Infectious Diseases | No | HCV Only | No | HCV Only |
| Anti-virus treatment | No | No | No | No |
Global analysis of serum miRNA expression profiles by miRNA PCR array
To analyze the possible miRNA changes in sera during HCV infection, a global investigation of relative miRNA expression levels, including 768 miRNAs between patients with HCV viremia and healthy controls, was carried out using miRNA PCR panels. During active virus infection, 367 out of 768 miRNAs were found to be detectable in the serum pool of HCV patients, whereas only 358 out of 768 miRNAs were detectable in the serum pool of healthy controls (Figure 1).
Number and percent composition of miRNAs with different threshold cycle ranges (Ct values) in HCV patients and healthy controls.
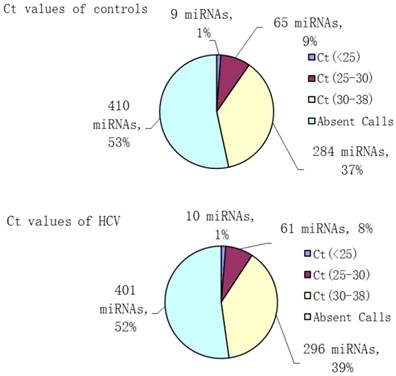
Aberrantly expressed miRNAs associated with HCV infection were defined to meet the following requirements: 1) Ct values <35 either in patients or controls to ensure stable detection; 2) relative fold change ≥2 between the patient and control groups. After screening, 106 miRNAs met the aforementioned conditions, including 51 up- and 55 down-regulated miRNAs, which might be associated with HCV infection (Table 2).
Aberrantly expressed miRNAs in HCV patients compared with healthy controls
| miRNAs | Fold Change (2-ddCt) | miRNAs | Fold Change (2-ddCt) |
|---|---|---|---|
| hsa-miR-629-5p | 20.88 | hsa-miR-26b-3p | 0.46 |
| hsa-miR-424-3p | 18.15 | hsa-let-7d-5p | 0.46 |
| hsa-miR-582-5p | 17.57 | hsa-miR-26b-5p | 0.45 |
| hsa-miR-571 | 15.17 | hsa-miR-1913 | 0.44 |
| hsa-miR-634 | 14.76 | hsa-miR-454-3p | 0.44 |
| hsa-miR-601 | 13.82 | hsa-miR-665 | 0.43 |
| hsa-miR-922 | 13.08 | hsa-miR-329 | 0.42 |
| hsa-miR-647 | 13.03 | hsa-miR-107 | 0.42 |
| hsa-miR-193b-5p | 12.66 | hsa-let-7g-5p | 0.42 |
| hsa-miR-302e | 11.37 | hsa-miR-152 | 0.38 |
| hsa-miR-23a-5p | 9.77 | hsa-miR-339-5p | 0.38 |
| hsa-miR-636 | 9.61 | hsa-miR-495-3p | 0.38 |
| hsa-miR-30a-3p | 9.27 | hsa-miR-122-3p | 0.37 |
| hsa-miR-625-3p | 9.19 | hsa-let-7c | 0.35 |
| hsa-miR-146b-3p | 8.83 | hsa-miR-511 | 0.35 |
| hsa-miR-365b-5p | 8.34 | hsa-miR-382-5p | 0.33 |
| hsa-miR-92a-1-5p | 8.16 | hsa-miR-1249 | 0.33 |
| hsa-miR-105-3p | 8.02 | hsa-miR-136-5p | 0.32 |
| hsa-miR-1245a | 7.97 | hsa-miR-204-5p | 0.31 |
| hsa-miR-635 | 7.70 | hsa-miR-199a-5p | 0.30 |
| hsa-miR-577 | 7.27 | hsa-miR-96-5p | 0.30 |
| hsa-miR-502-3p | 5.53 | hsa-miR-374a-5p | 0.28 |
| hsa-let-7f-1-3p | 5.41 | hsa-miR-300 | 0.27 |
| hsa-miR-1269a | 5.04 | hsa-miR-296-5p | 0.27 |
| hsa-miR-632 | 4.78 | hsa-miR-133b | 0.26 |
| hsa-miR-502-5p | 4.65 | hsa-miR-23b-5p | 0.25 |
| hsa-miR-941 | 4.25 | hsa-miR-566 | 0.23 |
| hsa-miR-598 | 4.24 | hsa-miR-548k | 0.20 |
| hsa-miR-551a | 4.08 | hsa-miR-28-5p | 0.20 |
| hsa-miR-132-5p | 4.01 | hsa-miR-483-3p | 0.18 |
| hsa-miR-24-2-5p | 3.79 | hsa-miR-761 | 0.17 |
| hsa-miR-106b-3p | 3.53 | hsa-miR-141-3p | 0.14 |
| hsa-miR-128 | 3.40 | hsa-miR-373-3p | 0.12 |
| hsa-miR-1207-5p | 3.32 | hsa-miR-570-3p | 0.12 |
| hsa-miR-139-3p | 3.19 | hsa-miR-382-3p | 0.12 |
| hsa-miR-431-3p | 3.13 | hsa-miR-497-5p | 0.11 |
| hsa-miR-409-3p | 3.07 | hsa-miR-374b-3p | 0.11 |
| hsa-miR-206 | 2.88 | hsa-miR-503-5p | 0.09 |
| hsa-miR-942 | 2.68 | hsa-miR-192-3p | 0.09 |
| hsa-miR-134 | 2.61 | hsa-miR-1181 | 0.09 |
| hsa-miR-509-3p | 2.54 | hsa-miR-616-3p | 0.09 |
| hsa-miR-486-5p | 2.48 | hsa-miR-548j | 0.08 |
| hsa-miR-200c-3p | 2.42 | hsa-miR-127-3p | 0.08 |
| hsa-miR-181c-3p | 2.34 | hsa-miR-450b-5p | 0.08 |
| hsa-miR-324-5p | 2.33 | hsa-miR-618 | 0.08 |
| hsa-miR-624-5p | 2.31 | hsa-miR-1271-5p | 0.06 |
| hsa-miR-1238-3p | 2.24 | hsa-miR-519d | 0.06 |
| hsa-miR-874 | 2.22 | hsa-miR-136-3p | 0.06 |
| hsa-let-7b-3p | 2.20 | hsa-miR-937-3p | 0.05 |
| hsa-miR-130b-5p | 2.15 | hsa-miR-543 | 0.03 |
| hsa-miR-25-3p | 2.09 | hsa-miR-301a-3p | 0.03 |
| hsa-miR-551b-3p | 0.48 | hsa-miR-29b-2-5p | 0.03 |
| hsa-miR-33a-5p | 0.47 | hsa-miR-335-3p | 0.01 |
Confirmation of miRNA array data by qRT-PCR
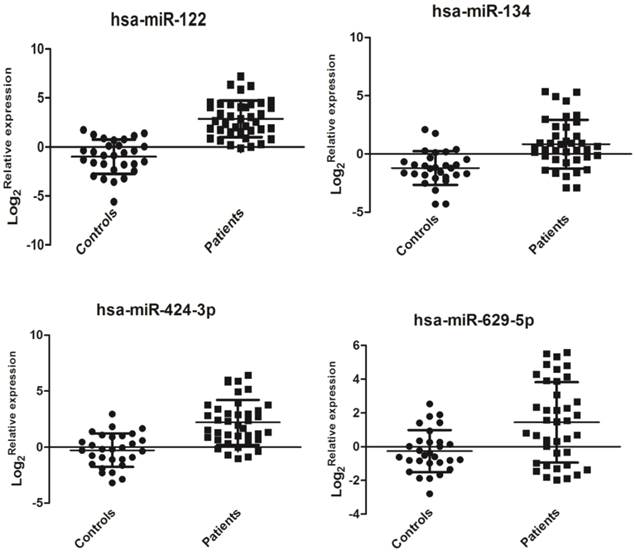
Hsa-miR-122 (miR-122 is usually in a form of miR-122-5p instead of miR-122-3p. Here, actually, the tested has-miR-122 is specific implication for miR-122-5p) plays an important role in HCV replication [8], but it did not show a significant change (data not shown) in the miRNA PCR array. Clinical samples may have huge individual differences that require further validation in future large-scale investigations. Hsa-miR-134 in HCV-infected sera has already been confirmed to be up-regulated in the previous study [17]. In the miRNA array data, we observed that hsa-miR-134 expression was also up-regulated in HCV sera compared with that in healthy controls. Has-629-5p and has-424-3p were the top two highly expressed miRNAs. Also, in the functional analysis by bioinformatics software (see the section of functional analysis), we found these miRNAs may be associated with HCV replication or pathological responses in human. Thus, we selected these four miRNAs for further validation. In this study, the relative expression levels of four selected miRNAs (hsa-miR-122, hsa-miR-134, hsa-miR-424-3p, and hsa-miR-629-5p) were detected by qRT-PCR as previously described. Interestingly, the expression of these four miRNAs was significantly up-regulated in sera of patients with HCV viremia compared with that of healthy controls (P < 0.01, Figure 2). The expression levels of miR-122, miR-134, miR-424-3p, and miR-629-5p may show a finger-print profile in the sera of HCV-infected patient samples compared with healthy controls.
Diagnostic potential of miRNAs
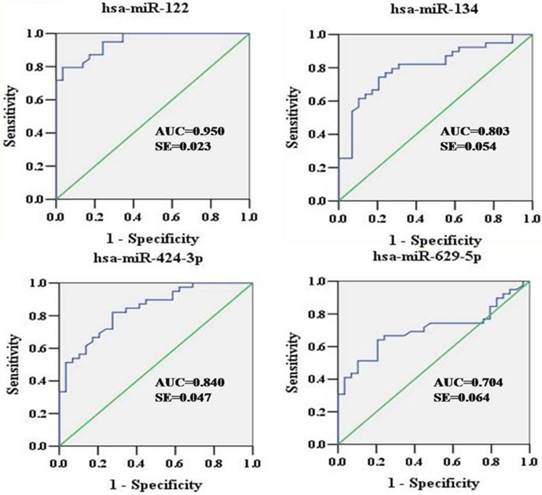
To explore the diagnostic potential of verified miRNAs for HCV infection, ROC curves were constructed (Figure 3). The AUCs for hsa-miR-122, hsa-miR-134, hsa-miR-424-3p, and hsa-miR-629-5p were as follows: 0.950 (95% CI: 0.905-0.994), 0.803 (95% CI: 0.698-0.909), 0.840 (95% CI: 0.748-0.932), and 0.704 (95% CI: 0.580-0.829), respectively. In the sera, hsa-miR-122, hsa-miR-134, and hsa-miR-424-3p showed relatively higher accuracy in indicating HCV viremia than hsa-miR-629-5p.
Relative expressions of serum miRNAs in HCV-infected patients (n=39) and healthy controls (n=29). The relative expression of miRNAs was calculated by the 2-ddCt method, and U6 snRNA was used as the internal control since sera U6 expression was stable in this case. A significant difference was assessed in log 2(Relative expression) between the positive samples and controls. Data were expressed as Mean±SD and statistical analysis was conducted by one-way ANOVA. P<0.05 indicated statistical significance.
ROC curves were constructed to evaluate the diagnostic potential of serum miRNAs for HCV infection. ROC, Receiver operating characteristic; AUC, area under curve; SE, standard error.
Serum ALT and AST levels and correlation analysis
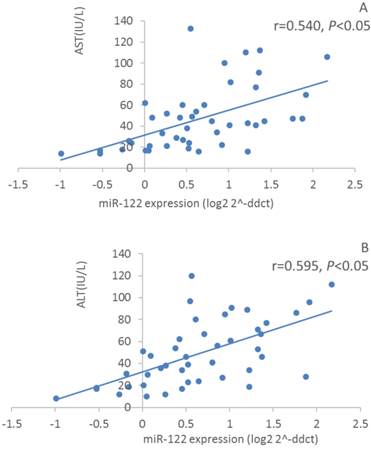
Furthermore, we assayed serum Alanine Transaminase (ALT) and Aspartate Aminotransferase (AST) activities. However, only 31 out of 39 HCV patients and 13 out of 29 healthy controls subject to this assay because part of serum samples were limited to be supplied. Nevertheless, serum ALT and AST activities in HCV patients were significantly increased compared with those in healthy controls (59.0±28.4 IU/L vs 24.8±16.8 IU/L, P<0.01; 57.2±30.8 IU/L vs 22.8±9.5 IU/L, P<0.01, respectively). These results suggested that HCV patients might have a slight hepatitis although no obvious pathological syndromes were observed by regular physical examination. Furthermore, we conducted correlation analysis between selected miRNAs and ALT/AST levels. Surprisingly, we found relative expression of miR-122 was moderately positive correlation with serum ALT and AST activities (r=0.595, P<0.01, n=44; r=0.540, P<0.01, n=44, respectively) (Figure 4). However, relative expression of miR-424 showed a weak positive correlation with serum AST activity but not in a significant manner (r=0.210, P=0.08, n=44). Relative expressions of miR-134 and miR-629 did not show any significant correlation with serum ALT and AST activities. In addition, no correlation was observed between viral load and serum ALT/AST activities, or between viral load and miRNAs levels.
Correlation analysis between relative expression of serum miR-122 expressions and atitivitis of serum ALT (A) and AST (B) in HCV patients and healthy controls (n=44).
Functional analysis
To gain more insights into the possible roles of these miRNAs during HCV infection, previously validated targets of these miRNAs are shown in Table 3. The results indicated that the functions of these target proteins were involved in HCV replication, immune response, cell proliferation, and hepatocarcinogenesis.
Verified targets that might be involved in the pathogenesis of HCV infection
| miRNA | Validated target genes | Functions | References |
|---|---|---|---|
| Has-miR-122 | 5' end of the 5' untranslated region (UTR) of the HCV genomic RNA OCLN IGF1R ADAM17 | HCV replication, cell proliferation, tumorigenesis, and metastasis | [8] [18] [19] [20] |
| Hsa-miR-134 | KRAS STAT5B ITGB1 | Cell proliferation, fiber formation and cell adhesion | [21] [22] |
| Hsa-miR-424 | NFIA PLGA1 SIAH1 SOCS6 FASN c-Myb ICAT | DNA replication and RNA transcription, cell proliferation, monocyte/macrophage differentiation | [23] [24] [25] [26] [27] [28] |
| Hsa-miR-629 | TRIM33 | Regulator of the TGF-beta/Smad signaling pathway, hepatocarcinogenesis, inflammation | [29] |
Discussion
MiRNAs play vital roles in virus-host interactions, including pathogenesis and host resistance, via regulating post-transcriptional translation or gene expression of relevant mRNAs. MiRNA expression levels of patients are dysregulated and distinct from healthy controls, thereby making miRNAs possible as biomarkers of infectious diseases. As of this writing, much data of miRNA expression profiles in in vitro cells and circulating body fluid have been reported. Circulating miRNAs have great potential to facilitate the diagnosis of virus infection, although the discrepancy between the expression levels of intracellular and extracellular miRNAs has been observed in certain situations [30].
In the present study, the expression profiles of serum miRNAs in patients with HCV viremia compared with those in the healthy controls were analyzed. Moreover, further evaluation for their potentials in detecting HCV infection was conducted. Among 768 miRNAs, a number of miRNAs in sera were differentially regulated as a response to HCV infection (51 up-regulated and 55 down-regulated) by a method of miRNA PCR array. PCR array can supply a lot of information and conduct preliminary evaluation but require further validation. Here, qRT-PCR was conducted to verify the expression levels of four selected miRNAs, namely, hsa-miR-122, hsa-miR-134, hsa-miR-424-3p, and hsa-miR-629-5p.
MiR-122 is a vital factor and therapeutic target in liver diseases. Many previous studies have demonstrated that miR-122 can increase the abundance of HCV RNA through direct binding to the 5' UTR of viral genome, and is associated with lipid and cholesterol metabolism during infection [8, 31, 32]. However, miR-122 was also recently reported to show an anti-viral effect by targeting the 3' UTR of OCLN mRNA, an HCV entry molecule, and decreasing the entry of HCV into hepatocytes [18]. MiR-122 is significantly up-regulated in acute or chronic HCV-infected sera, and many studies reported that it can serve as a candidate biomarker of HCV infection [33, 34]. Similarly, we verified that the expression of hsa-miR-122 in sera of patients with HCV viremia was up-regulated. In this study, we selected miR-122-5p as our study objective since miR-122-5p had higher abundance (Ct values, 26~27) in sera than miR-122-3p (Ct values, 33~35) and likely act as one main functional form of miR-122. However, no significant change of miR-122-5p was observed in our miRNA PCR array (data not shown). This discrepancy may suggest pooled sera have some limits and mask part of differences between groups. However, if we can find some differences from pooled serum between different groups, it is reasonable for us to have some clues for further validation of those differences. Furthermore, miR-122-3p was down-regulated in our miRNA arrays. We did not validate serum miR-122-3p level since miR-122-3p was in a very low abundance and neither was one main form of miR-122 in sera compared with miR-122-5p. To ensure the accuracy of the results in the miRNA arrays, hsa-miR-134 was detected and found to be significantly up-regulated in the patients' sera, which was similar to the result in previous research carried out by Shwetha et al [17]. The functions of hsa-miR-134 involve the regulation of cell proliferation, fiber formation, and cell adhesion [21, 22]. We also evaluated the expression levels of two new miRNAs derived from miRNA array by qRT-PCR. In our study, hsa-miR-424-3p was found to be up-regulated in sera of patients with HCV viremia. Hsa-miR-424 was associated with the promotion of viral and cellular DNA replication and RNA transcription. Moreover, hsa-miR-424 is critical in regulating the tumorigenesis and metastatic process of HCC by directly targeting the 3' UTR of c-Myb and ICAT [27, 28]. In addition, hsa-miR-424 mediates monocyte/macrophage differentiation and regulates the response of inflammation [23]. Hsa-miR-629 is considered a key modulator of liver tumorigenesis and inflammation by participating in the feedback loop circuit, which can cause hepatocyte nuclear factor-4α transient inhibition [35, 36]. Thus, over-expressed miR-629-5p may regulate the inflammation response to resist HCV infection. Taken together, hsa-miR-122, has-miR-134, has-424-3p, and miR-629-5p seemed to regulate viral replication and hepatic pathogenesis associated with HCV-infected patients.
Based on the ROC curves, hsa-miR-122 had the highest efficiency in distinguishing between patients with HCV viremia and healthy controls. Furthermore, hsa-miR-122 was highly positive correlation with serum ALT/AST levels. Increased serum ALT/AST levels indicated most of HCV patients had suffered from hepatitis although we did not observe obvious pathological changes (e.g. hepatic steatosis, fibrosis, cancer) by common physical examination. Although hsa-miR-629-5p showed low sensitivity and specificity (AUC = 0.704), hsa-miR-134 (AUC = 0.803) and hsa-miR-424-3p (AUC = 0.840) may serve as good biomarkers for detecting HCV infection. In this study, we proved that miR-122 and miR-134 were good biomarkers for HCV patients, and provided new evidence for serum miRNA diagnosis of HCV or its complications. The results suggested that hsa-miR-424-3p and has-miR-629 were promising biomarkers of serum miRNAs for HCV-infected patients. Future studies should further explore the relationships between these miRNAs and indicators associated with HCV.
In this study, although we have obtained some promising serum miRNA biomarkers for HCV patients, we have to indicate that there are some common limits for this study and also for other similar studies. Firstly, there is no standard reference miRNA for normalization of serum miRNA expression. In different cases, there are different reference miRNAs for serum miRNAs. There is a dispute about whether U6 can be served as reference miRNA control for normalization of cell-free miRNA or not. Some researchers insisted that U6 can be served as reference control for normalization of serum miRNAs [37] while others claimed U6 cannot be served in specific case [38]. In our opinion, if any selected miRNA is stable in specific case, it can be served as reference control miRNA for normalization. In this case, U6 was stable so that we selected U6 as reference control. In the coming study, we should identify and add two or three reference control miRNAs for normalization in sera of HCV patients. On the other hand, we should unify the genotype of HCV patients although we identified most of patients as 1b subtype of HCV. Different HCV genotypes might have different serum miRNA biomarkers, which may be interesting topic for identifying serum miRNA biomarkers in specific genotype of HCV in the future study.
Conclusion
In conclusion, miRNA PCR array assays revealed differential expression of 106 miRNAs in sera of HCV patients compared with that in healthy controls. Serum hsa-miR-122, miR-134, miR-424-3p, and miR-629-5p were identified as potential non-invasive molecular markers for the detection of HCV infection. Particularly, serum miR-122 may serve as an interesting biomarker for early hepatic inflammation responses or other pathological process in HCV patients. However, the biological functions of these miRNAs require further investigation.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81373460 and 81072680), the Shenzhen Science and Technology R&D Foundation (Grant Nos. SGLH20121008144756945, JCYJ20120618172144495 and ZYC201105170341A), the Natural Science Foundation of Guangdong Province (2014A030313744 and 2012B031800126), the China Scholarship Council (201308440130), the ITF Grant (GHX/002/12SZ) and the State Quality Inspection Administration (No. 201310087).
Abbreviations
HCV: hepatitis C virus; HBV: hepatitis B virus; HIV: human immunodeficiency virus; HSV: herpes simplex virus; HCC: hepatocellular carcinoma; RT-PCR: reverse transcription polymerase chain reaction; qRT-PCR: quantitative real-time polymerase chain reaction; UTR: untranslated region; Ct: cycle threshold; ROC: receiver operating characteristic curve; AUC: the area under the curve; CI: confidence interval.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lee MH, Yang HI, Yuan Y. et al. Epidemiology and natural history of hepatitis C virus infection. World J Gastroenterol. 2014;20:9270-9280
2. Razavi H, Elkhoury AC, Elbasha E. et al. Chronic hepatitis C virus (HCV) disease burden and cost in the United States. Hepatology. 2013;57:2164-2170
3. Zhao L, Feng Y, Xia XS. The different epidemic and evolution of HCV genotypes. Yi Chuan. 2014;34:666-672
4. Khaliq S, Jahan S, Pervaiz A. Sequence variability of HCV Core region: important predictors of HCV induced pathogenesis and viral production. Infect Genet Evol. 2011;11:543-556
5. Zhang JF, Fu WM, He ML. et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011;8:829-838
6. Wan G, Xie W, Liu Z. et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10:70-79
7. Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452-460
8. Jopling CL, Schütz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77-85
9. Murakami Y, Aly HH, Tajima A. et al. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453-460
10. Cheng JC, Yeh YJ, Tseng CP. et al. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci. 2012;69:2621-2633
11. Pedersen IM, Cheng G, Wieland S. et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919-922
12. Valadi H, Ekström K, Bossios A. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659
13. Mitchell PS, Parkin RK, Kroh EM. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad Sci USA. 2008;105:10513-10518
14. Mall C, Rocke DM, Durbin-Johnson B. et al. Stability of miRNA ihuman urine supports its biomarker potential. Biomark Med. 2013;7:623-631
15. Zhou Q, Li M, Wang X. et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118-123
16. Chen Y, Li L, Zhou Z. et al. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med Microbiol Immunol. 2012;201:389-395
17. Shwetha S, Gouthamchandra K, Chandra M. et al. Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep. 2013;3:1555
18. Chen Y, Li L, Zhou Z. et al. miR-122 decreases HCV entry into hepatocytes through binding to the 3' UTR of OCLN Mrna. Liver Int. Vision Res. 2015;35:1315-1323
19. Wang B, Wang H, Yang Z. MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PLoS One. 2012;7:e47053
20. Chen YJ, Chang LS. Hydroquinone-induced miR-122 down-regulation elicits ADAM17 up-regulation, leading to increased soluble TNF-alpha production in human leukemia cells with expressed Bcr/Abl. Biochem Pharmacol. 2013;86:620-631
21. Zhang Y, Kim J, Mueller AC. et al. Multiple receptor tyrosine kinases converge on microRNA-134 to control KRAS, STAT5B, and glioblastoma. Cell Death Differ. 2014;21:720-734
22. Zha R, Guo W, Zhang Z. et al. Genome-wide screening identified that miR-134 acts as a metastasis suppressor by targeting integrin beta1 in hepatocellular carcinoma. PLoS One. 2014;9:e87665
23. Rosa A, Ballarino M, Sorrentino A. et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA. 2007;104:19849-19854
24. Imig J, Motsch N, Zhu JY. et al. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 2011;39:1880-1893
25. Wu K, Hu G, He X. et al. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19:739-748
26. Long XH, Mao JH, Peng AF. et al. Tumor suppressive microRNA-424 inhibits osteosarcoma cell migration and invasion via targeting fatty acid synthase. Exp Ther Med. 2013;5:1048-1052
27. Yu L, Ding GF, He C. et al. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One. 2014;9:e91661
28. Zhang Y, Li T, Guo P. et al. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep. 2014;4:6248
29. Jingushi K, Ueda Y, Kitae K. et al. miRNA-629 Targets TRIM33 to Promote TGF-beta/Smad Signaling and Metastatic Phenotypes in ccRCC. Mol Cancer Res. 2015;13:565-574
30. Elhelw DS, Mekky RY, El-Ekiaby N. et al. Predictive prognostic role of with discrepancy in the liver and serum of genotype 4 hepatitis C virus patients. Biomed Rep. 2014;2:843-848
31. Bandiera S, Pfeffer S, Baumert TF. et al. miR-122 - a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448-457
32. Esau C, Davis S, Murray SF. et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87-98
33. Choi Y, Dienes HP, Krawczynski K. Kinetics of miR-122 expression in the liver during acute HCV infection. PLoS One. 2013;8:e76501
34. Bala S, Tilahun Y, Taha O. et al. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151
35. Hatziapostolou M, Polytarchou C, Aggelidou E. et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233-1247
36. Gougelet A, Colnot S. MicroRNA-feedback loop as a key modulator of liver tumorigenesis and inflammation. World J Gastroenterol. 2013;19:440-444
37. Liu B, Xiang Y, Zhang HS. Circulating microRNA-196a as a candidate diagnostic biomarker for chronic hepatitis C. Mol Med Rep. 2015;12:105-110
38. Xiang M, Zeng Y, Yang R. et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454:210-214
Author contact
![]() Corresponding author: E-Mail: xiewdtsinghua.edu.cn (W.X.); Tel: +86-755-26036086; Fax: +86-755-26036086
Corresponding author: E-Mail: xiewdtsinghua.edu.cn (W.X.); Tel: +86-755-26036086; Fax: +86-755-26036086

 Global reach, higher impact
Global reach, higher impact