Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(7):538-543. doi:10.7150/ijms.11700 This issue Cite
Research Paper
Investigation of Somatic NKX2-5 Mutations in Chinese Children with Congenital Heart Disease
1. Institute of Pediatric Translational Medicine, Shanghai Children's Medical Center Affiliated to School of Medicine, Shanghai Jiao Tong University, Shanghai, China.
2. Department of Pediatric Cardiology, Shanghai Children's Medical Center Affiliated to School of Medicine, Shanghai Jiao Tong University, Shanghai, China.
3. Department of Cardiothoracic Surgery, Shanghai Children's Medical Center Affiliated to School of Medicine, Shanghai Jiao Tong University, Shanghai, China.
4. Clinical Laboratory, Shanghai Children's Medical Center Affiliated to School of Medicine, Shanghai Jiao Tong University, Shanghai, China.
5. Molecular Diagnostics Laboratory, Shanghai Children's Medical Center Affiliated to School of Medicine, Shanghai Jiao Tong University, Shanghai, China.
6. Pediatric Cardiology Department, Xinhua Hospital Affiliated to School of Medicine, Shanghai Jiao Tong University, Shanghai, China.
Received 2015-1-27; Accepted 2015-5-18; Published 2015-6-12
Abstract
The purposes of this study are to investigate somatic NKX2-5 mutations in Chinese children with congenital heart disease (CHD) and assess the reliability of somatic mutation detection in formalin-fixed, paraffin-embedded (FFPE) tissues. The study cohort included frozen and FFPE cardiac tissues as well as blood samples from 85 Chinese children with CHD who had the cardiac operations. The right atrial appendage far from the diseased heart was used as normal control. Genomic DNA was isolated from cardiac tissues and blood samples using TIANamp Blood DNA kit. Two exons and exon-intron boundaries of NKX2-5 were amplified by polymerase chain reaction (PCR) and sequenced by dideoxynucleotide chain termination approach. The acquired sequences were aligned with GenBank sequences to identify the sequence variations. No somatic mutation in the NKX2-5 gene was observed in both frozen and FFPE cardiac tissues in 85 Chinese children with CHD. Nonetheless, a common single nucleotide polymorphism (SNP), c.63 A > G (E21E), was identified in all the three kinds of DNA samples with the same allele frequency 82.3%. Moreover, another common SNP c.606 G > C (L202L) was found in 2.3% of our patients. There were no significant differences in the allele frequencies of two SNPs between the cardiac diseased tissues and right atrial appendage (P > 0.05). PCR artefact as mutations was not found in the FFPE tissues stored for one year. Our findings demonstrate that somatic NKX2-5 mutations do not represent an important aetiologic pathway in Chinese children with congenital heart disease.
Keywords: Congenital Heart Disease (CHD), NKX2-5, somatic mutation, single nucleotide polymorphisms (SNP), formalin-fixed, paraffin-embedded (FFPE) tissues
INTRODUCTION
Congenital heart disease (CHD) affects the structure and function of the heart that is present at birth. It is the most common type of birth defect, with a prevalence of approximately 0.8% among live births, and is the most important reason of infant death, especially in the children under 5 years old in China1. The causes of CHD contain external and internal factors. The external factors relate to the environmental factors such as drugs, infections and disease, while the internal factors are associated with the genetic factors. The gene-environment interaction leads to the occurrence of CHD2.
The germline mutations of cardiac specific transcription factors are the major causes of CHD. Nowadays, many studies focus on the transcription factors NKX2-5, GATA-4, dHAND, eHAND, MEF2C, Irx4 and TBX5. It has been reported that NKX2-5, GATA-4 and TBX5 are the key genes associated with the occurrence of simple CHD. Among them, NKX2-5 plays an important role in the heart development3. NKX2-5 is a gene which encodes a homeobox-containing transcription factor. It functions in heart formation and development. NKX2-5 is the earliest known marker of myocardial progenitor cells in all species in which it has been studied4-7.
In 2003, Reamon-Buettner et al first reported that NKX2-5 mutations occurred somatically and these mutations probably resulted from postzygotic errors in cell division, particularly in cardiac progenitors cells8. Then, they studied the pathology and molecular genetics of NKX2-5 in 68 patients with complex CHD and found that many patients had multiple NKX2-5 mutations in diseased heart tissues fixed in formalin over 22 years9. These observed mutations were absent in normal heart tissues from the same CHD patients and lymphocytic DNA of patients with CHD and healthy individuals, indicating somatic origin of mutations. However, contradictory results have been reported in which somatic mutations in NKX2-5 do not represent an important aetiologic pathway in pathologic cardiac development in patients with cardiac septal defects10.
Our study investigated somatic NKX2-5 mutations in Chinese children with congenital heart disease. The cohort included blood and cardiac tissue samples from 85 patients who had the cardiac operations. Moreover, we assessed the reliability of somatic mutation detection in the FFPE tissues stored at room temperature for one year.
MATERIALS AND METHODS
Materials
The study was approved by the Ethics Committee of Xinhua Hospital in Shanghai Jiao Tong University. The heart tissues and blood samples were obtained from the CHD bank at Shanghai Children's Medical Center of Xinhua Hospital. This study included 85 pairs of blood samples and cardiac tissues from children with CHD who had the cardiac operations. They were 36 males and 9 females, with mean age of 25.8 months at the surgery time. Amongst 85 patients, 23 were diagnosed with tetralogy of Fallot (TOF) who had be removed the hypertrophied myocardium on right ventricular outflow tract. Other patients were diagnosed with ventricular septal defect (VSD, n = 48), atrial septal defect (ASD II, n = 5), complete atrioventricular canal (CAVC, n = 1) and other complex CHD with VSD (n = 6) and without VSD (n = 2). All of them do not have family history of CHD. As a normal control, the tissues of right atrial appendage far from the diseased heart were collected from the same cohort of patients during the surgery.
Methods
All the heart tissues and blood samples from the 85 patients were immediately frozen and stored at -80℃ for DNA isolation and NKX2-5 amplification within one month after the operations. For 62 patients, the part of the diseased tissue was immersed in formalin at the time immediately after operation and paraffin-embedded after 24 hours. The FFPE tissues were stored at room temperature over one year for DNA isolation and NKX2-5 amplification.
DNA isolation
Genomic DNA was isolated from frozen cardiac tissues and blood samples using TIANamp Blood DNA kit according to the instruction from the manufacturer (Tiangen, Beijing, China). The digestion of proteinase K was for 24 hours. FFPE cardiac tissues were immersed in dimethylbenzene and then 96% - 100% ethylalcohol to remove the paraffin. Genomic DNA from FFPE tissues was isolated using the same DNA kit with the digestion of proteinase K for 24 - 48 hours.
The quality and quantity of isolated genomic DNA were checked on 1% agarose gel and ultraviolet spectrophotometer. The DNA samples were stored at -20℃.
Amplification of NKX2-5 fragments
For the amplification of NKX2-5 fragments, the sequences of primer sets for DNA samples from blood and frozen tissues were A and B, while the sequences of primer sets for DNA samples from FFPE cardiac tissues were A, C and D (Table 1). A PCR reaction consisted of 50 - 100 ng of genomic DNA, GC Buffer I 12.5μl, dNTP 4μl, 1μl (10μmol/L) of each primer pair, Takara DNA polymeras 0.25μl in a final volume of 25μl. The PCR reactions of first exon of DNA from blood and frozen tissues started with 5 min at 95℃, followed by 35 cycles of 30s at 95℃, 30s at 62℃, and 45s at 72℃, and finished with a 5 min extension period at 72℃. The reactions of DNA from FFPE tissues were the same as mentioned in the blood and frozen tissues, with the exception of a 30s anneal at 58℃. The reactions of the second exon from the two kinds of tissues and blood were the same as the first exon from blood and frozen tissues, with the exception of a 60s extension at 72℃.
Primer sequences for the amplification of NKX2-5 fragments
| Primer | Exon | Forward (5'-3') | Reverse (5'-3') | Length (bp) |
|---|---|---|---|---|
| A | 1 | CTTGTGCTCAGCGCTACCT | CTGAGTTTCTTGGGGACGAA | 550 |
| B | 2 | AGTGCACTTGGCAGAGTGAG | CTAGGTCTCCGCAGGAGTGA | 922 |
| C | 2 | AGTGCACTTGGCAGAGTGAG | ATAGGCGGGGTAGGCGTTAT | 586 |
| D | 2 | AAGCCATGCCTAGGGGACT | CTCATTGCACGCTGCATAAT | 500 |
DNA sequencing
Sequencing the PCR products were conducted by Mastercycler (Eppendorf AG, Hamburg, Germany). The acquired sequences were aligned with GenBank sequences by the aid of BLAST program to identify the sequence variations.
STATISTICAL ANALYSIS
Statistical analysis was carried out with SPSS 11.5 package and two-sided χ2-test was used to evaluate the appearance of sequence variants between the disease located and faraway from the disease. P < 0.05 was considered to indicate a statistically significant difference.
For comparing DNA quantity in the frozen and FFPE tissues, paired t-test was performed on the log2 transformed data to determine the significance of difference between two different types. The concentration of DNA was defined as M, while the volume of DNA solution was named as V. Quantity of DNA from the frozen tissue was M1×V1, and quantity of DNA from the FFPE tissue was M2×V2. The ratio λ of DNA quantity between the frozen and FFPE tissues was calculated using the equation: λ = (M1×V1)/ (M2×V2).
RESULTS
Amplification of NKX2-5 fragments
The exon 1 in the NKX2-5 gene was successfully amplified in all three sample types using the primer set A (Table 1). However, the exon 2 in the NKX2-5 gene could not be amplified in the FFPE tissues using the primer set B. Therefore, the exon 2 was divided into two parts and additional primer sets were designed. Using the primer set C and D (Table 1), the two fragments of exon 2 in the NKX2-5 gene were successfully amplified in the FFPE tissues.
NKX2-5 somatic mutations
Comparing the sequences of NKX2-5 fragments amongst the blood samples and frozen tissues from the diseased heart and normal control from the right atrial appendage, no sequence difference was observed in the NKX2-5 gene. Therefore, no somatic mutation in the NKX2-5 gene was found in the diseased cardiac tissues in 85 Chinese children with CHD.
NKX2-5 sequence variations
Two common single nucleotide polymorphisms (SNPs) were found both in the blood samples and frozen tissues from the same patient. They were rs2277923 c.63 A > G (E21E) located in the exon 1 and rs3729753 c.606 G > C (L202L) located in the exon 2 of the NKX2-5 gene (Figure 1). The frequencies of genotypes and alleles for the two sequence variations are presented in Table 2. For the c.63 A > G (E21E) variation, the genotypes were AA, AG and GG with the frequency of 15, 32 and 38, respectively. The alleles were A and G with the frequency of 62 and 108, respectively. There was no significantly difference in the frequencies of genotypes (P = 0.732) and alleles P = 0.662) between TOF and other CHD (Table 3). For the c.606 G > C (L202L) variation, the genotypes were GG, GC and CC with the frequency of were 82, 3 and 0, respectively. The alleles were G and C with the frequency of 167 and 3, respectively (Table 2). Similarly, there was no significantly difference in the frequencies of genotypes (P = 0.118) and alleles (P = 0.120) between the TOF and other CHD (Table 3).
Frequencies of genotypes and alleles of NKX2-5 sequence variations
| Frequency (%) of genotypes | Frequency (%) of alleles | |||||
|---|---|---|---|---|---|---|
| Genotype of c.63 | ||||||
| AA | AG | GG | A | G | ||
| All CHD | 15 (17.6) | 32 (37.6) | 38 (44.7) | 62 (36.5) | 108 (63.5) | |
| TOF | 5 (21.7) | 8 (34.8) | 10 (43.5) | 18 (39.1) | 28 (60.9) | |
| Other CHD | 10 (16.1) | 24 (38.7) | 28 (45.1) | 44 (35.5) | 80 (64.5) | |
| Z | -0.343 | - 0.438 | ||||
| P | 0.732 | 0.662 | ||||
| Genotype of c.606 | ||||||
| GG | GC | CC | G | C | ||
| All CHD | 82 (96.5) | 3 (3.53) | 0 (0) | 167 (98.2) | 3 (1.8) | |
| TOF | 21 (91.3) | 2 (8.7) | 0 (0) | 44 (95.6) | 2 (4.3) | |
| Other CHD | 61 (98.4) | 1 (1.6) | 0 (0) | 123 (99.2) | 1 (0.8) | |
| Z | -1.563 | -1.553 | ||||
| P | 0.118 | 0.12 | ||||
Somatic mutation detection in FFPE tissues
Of 85 patients, only 38 had enough materials in both frozen and FFPE tissues for the DNA isolation. The concentration (range) of genomic DNA in the FFPE tissues was 100 - 200μg/ml and OD (mean ± SD) was 2.0 ± 0.143. Evidently, the quantity of DNA obtained from the cardiac tissue stored in the paraffin for one year was significantly lower than that in the frozen cardiac tissues ( P = 3.3-36). The quantity of DNA in the frozen tissues was nearly 4 times more than that of the FFPE tissues (Table 4). Nonetheless, the quality of DNA in the FFPE tissues was similar to that in the frozen tissues.
Comparing the sequences of NKX2-5 fragments between the frozen and FFPE tissues, no additional sequence variation was observed in the FFPE tissue. The genotypes of the two common SNPs were identical in both frozen and FFPE tissues from the same patients. PCR artefact as mutations was not found in the cardiac FFPE tissues stored at room temperature over one year.
DISCUSSION
In 1980, researchers proposed that somatic mosaicism in the fetus may manifest as congenital malformations, spontaneous abortions and childhood cancers11. In the past, some studies reported higher frequency of NKX2-5 mutations in the FFPE diseased heart tissues and suggested that somatic mutation might be a mechanism of CHD9. In contrast to earlier reports, other studies questioned the high frequency of somatic mutations in the FFPE tissues and considered that long-time storage in formalin might create PCR artefacts that may appear as mutations12. Do somatic NKX2-5 mutations exist in CHD? To address this issue, our study investigated somatic NKX2-5 mutations in 85 Chinese children with CHD and assessed the reliability of somatic mutation detection in FFPE tissues from the same cohort of patients.
Genotypes of NKX2-5 sequence variations in CHD patients
| CHD | Genotype of c.63 | Genotype of c.606 | ||||
|---|---|---|---|---|---|---|
| AA | AG | GG | GG | GC | ||
| TOF | 5 | 8 | 10 | 21 | 2 | |
| VSD | 8 | 16 | 24 | 47 | 1 | |
| ASD (II) | 1 | 3 | 1 | 5 | 0 | |
| CAVC | 0 | 0 | 1 | 1 | 0 | |
| CoA/VSD | 0 | 0 | 2 | 2 | 0 | |
| IAA/VSD | 0 | 1 | 0 | 1 | 0 | |
| PTA/VSD | 0 | 1 | 0 | 1 | 0 | |
| PA/VSD | 1 | 0 | 0 | 1 | 0 | |
| DORV/VSD | 0 | 1 | 0 | 1 | 0 | |
| TR/PFO | 0 | 1 | 0 | 1 | 0 | |
| Cor triatriatum/TR | 0 | 1 | 0 | 1 | 0 | |
Comparison of DNA quantity in frozen and FFPE tissues
| DNA quantity (ug) | Ratio (λ) | ||
|---|---|---|---|
| Frozen tissue (n = 38) | FFPE tissue (n = 38) | ||
| Median | 7.6 | 1.8 | 4.0 |
| Mean | 7.6 | 1.9 | 4.0 |
| Range | 4.2 - 11.6 | 1.2 - 3.1 | 2.8 - 5.4 |
| SD | 1.9 | 0.5 | 0.6 |
| P value | 3.3E-36 | ||
NKX2-5 sequence variations. (A) A NKX2-5 homozygous sequence variant c.65A/A. (B) A NKX2-5 heterozygous sequence variant c.65A>G (E21E). (C) A NKX2-5 homozygous sequence variant c.65G/G. (D) A NKX2-5 homozygous sequence variant c.606G/G. (E) A NKX2-5 homozygous sequence variant c.606 G > C (L202L).
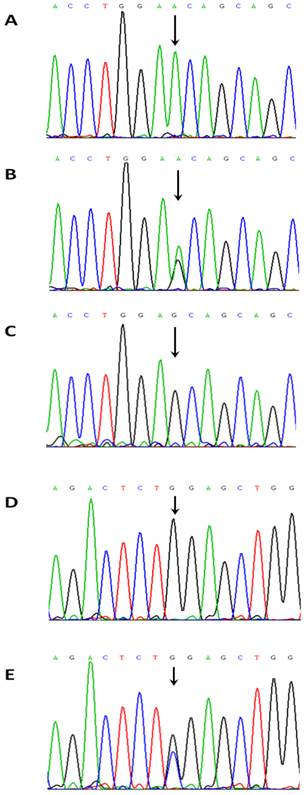
In this study, we collected fresh heart tissues at the location of disease and right atrial appendage far away from the diseased heart to explore whether or not the mosaic exist in the patients with CHD. In the same patients, the sequences from the diseased heart and right atrial appendage were identical. Furthermore, the sequences from the blood and frozen tissues were also identical. We found only two common SNPs and their frequencies are similar to that published in the Genbank database. Our results demonstrate that somatic NKX2-5 mutations do not represent an important aetiologic pathway in Chinese children with CHD.
Large numbers of surgically resected tissues and biopsy specimens are preserved as FFPE blocks and stored at room temperature for decades. Therefore, FFPE tissues have been and still are the most abundant supply of clinical samples, and are often accompanied with sufficient follow-up data. In addition, FFPE tissues also play an irreplaceable role in the fast growing translation medical research through bridging laboratory work and clinical applications. Fully exploration of FFPE tissues is essential for medical research. Unfortunately, DNA is more difficult extracted from fixed tissues and PCR may affect the mutations induced in vitro by the formalin fixation12-14.
The effects of formalin fixation on DNA present in tissues are still uncertain. In fact, as a technique for analyzing DNA evidence, PCR is affected by many factors, such as the quality and quantity of DNA and other unknown inhibitions and facilitors. Some studies proved that formalin would change the physico-chemical properties of DNA, but many steps could remove these changes during the purification of DNA15. Other studies found that the prolonged fixation intervals were associated with extensive DNA degradation and progressive inability to amplify longer DNA templates16. However, none of the studies reported that the tissues stored in formalin could create PCR artefacts that appear as mutations. To investigate this possibility, we compared the sequences of PCR products in the NKX2-5 gene between DNA extracted from the fresh and fixed tissues stored at room temperature for one year. We found that there were no differences of sequences between them, indicating PCR artefact as mutations did not present in the cardiac FFPE tissues for a short term storage. Nonetheless, PCR artifacts as mutations could not be excluded on the reported high frequency of NKX2-5 mutations in the FFPE tissues stored at room temperature for more than 22 years9. The studies should be continued to assess the reliability of somatic mutation analysis in the FFPE tissues for a long-time storage.
A barrier to the analysis of FFPE samples is that DNA extracted from FFPE tissues is often significantly degraded to an average fragment length around 300 - 400 bp 17. In our study, the quantity of genomic DNA from the FFPE tissues was nearly four times less than that from the frozen FFPE tissues. When it came to FFPE tissues, the exon 2 with 922 bp in the NKX2-5 gene could not be amplified with only one pair of primer set. Therefore, we divided the exon 2 into two overlapping fragments and successfully amplified the DNA extracted from the FFPE tissues with two pairs of primer sets.
It has been recognized that NKX2-5 mutations mainly appear in patients with ASD (II), TOF and VSD 18. In our 85 patients with CHD, we only found two common SNPs in the NKX2-5 gene, indicating low mutation frequency of NKX2-5 in Han population. Much more work still needs to be done for the discovery of the causes in Chinese children with CHD.
Acknowledgements
We would like to express our great gratitude for the support of the Joint Research Projects of Shanghai Municipal Health Bureau for health system major diseases (No. 2013ZYJB0016) and 973 Project of National Science and Technology Ministry -Early diagnosis and epidemiological investigation of congenital heart disease (No. 2010CB529501).
We would like to thank all the doctors of cardio-thoracic surgery of Shanghai Children's Medical Center for their CHD heart tissue support, all the doctors of clinical laboratory, all the doctors of pathology department.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
1. Hoffman J I, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890-1900
2. Gong LG, Qiu GR, Jiang H. et al. Analysis of single nucleotide polymorphisms and haplotypes in HOXC gene cluster within susceptible region 12q13 of simple congenital heart disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005;22:497-501
3. Ueyama T, Kasahara H, Ishiwata T. et al. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol Cell Biol. 2003;23:9222-32
4. Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145-9
5. Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203-16
6. Turbay D, Wechsler SB, Blanchard KM. et al. Molecular cloning, chromosomal mapping and characterization of the human cardiacspecific homeobox gene hCsx. Mol Med. 1996;2:86-96
7. Tanaka M, Kasahara H, Bartunkova S. et al. Vertebrate homologs of tinman and bagpipe:roles of the homeobox genes in cardiovascular development. Dev Genet. 1998;22:239-49
8. Reamon-Buettner SM, Borlak J. Somatic NKX2-5 mutations as a novel mechanism of disease in complex congenital heart disease. J Med Genet. 2004;41:684-90
9. Reamon-Buettner SM, Hecker H, Spanel-Borowski K. et al. Novel NKX2-5 mutation in diseased heart tissues of patients with cardiac malformations. Am J Pathol. 2004;164:2117-25
10. Draus JM Jr, Hauck MA, Goetsch M. et al. Investigation of somatic NKX2-5 mutations in congenital heart disease. J Med Genet. 2009;46:115-22
11. Sorsa M. Somatic mutation theory. J Toxicol Environ Health. 1980;6:977-82
12. Quach N, Goodman MF, Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin Pathol. 2004;4:1
13. Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961-1971
14. Greer CE, Peterson SL, Kiviat NB. et al. PCR amplification from paraffin-embedded tissues. Effects of fixative and fixation time. Am J Clin Pathol. 1991;95:117-124
15. Al-soud WA, Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485-93
16. Romer RL, Juston AC, Ballantyne J. et al. The applicability of formalin-fixed and formalin fixed paraffin embedded tissues in forensic DNA analysis. J Forensic Sci. 1997;42:708-14
17. Jacobs S, Thompson ER, Nannya Y. et al. Genome-wide, high-resolution detection of copy number, loss of heterozygosity, and genotypes from formalin-fixed, paraffin-embedded tumor tissue using microarrays. Cancer Res. 2007;67:2544-2551
18. Reamon-Buettner SM, Borlak J. NKX2-5. An update on this hypermutable homeodomain protein and its role in human congenital heart disease (CHD). Hum Mutat. 2010;31:1185-94
Author contact
![]() Corresponding author: Professor Kun Sun, Pediatric Cardiology Department, Xinhua Hospital Affiliated to School of Medicine, Shanghai Jiao Tong University, Shanghai, China. Tel: +86-21-2507899. Fax: +86-21-58393915. E-mail: sunkuncom.cn.
Corresponding author: Professor Kun Sun, Pediatric Cardiology Department, Xinhua Hospital Affiliated to School of Medicine, Shanghai Jiao Tong University, Shanghai, China. Tel: +86-21-2507899. Fax: +86-21-58393915. E-mail: sunkuncom.cn.

 Global reach, higher impact
Global reach, higher impact