3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(11):1524-1529. doi:10.7150/ijms.6376 This issue Cite
Research Paper
The -159C/T Polymorphism in the CD14 Gene and Tuberculosis Risk: A Meta-Analysis
Division of Pulmonary Diseases, State Key Laboratory of Biotherapy of China and Department of Respiratory and Critical Care Medicine, West China Hospital of Sichuan University, Chengdu 610041, China.
* These authors are equal contributors and co-first authors to this paper.
Received 2013-4-1; Accepted 2013-8-12; Published 2013-8-31
Abstract
Background: The -159C/T polymorphism in the CD14 gene has been implicated in susceptibility to tuberculosis, but the results were inconclusive. The present meta-analysis aimed to perform a comprehensive assessment of the literature on the possible association between the -159C/T polymorphism and tuberculosis risk.
Methods: We searched in Pubmed and Embase for studies evaluating the association between the -159C/T gene polymorphism and tuberculosis risk. Data were extracted and statistical analysis was performed using Revman 5.1 and STATA 12.0 software.
Results: A total of seven case-control studies involving 3253 subjects (1,574 tuberculosis cases and 1,679 controls) were included. Combined analysis revealed an obvious association between this polymorphism and tuberculosis risk (odds ratio=1.66 and 95% confidence interval: 1.23-2.25, P<0.05 for TT vs. TC+ CC). Sub-group analysis by ethnicity suggested that the risk of tuberculosis associated with the -159C/T polymorphism was significantly elevated among Asians (odds ratio=1.87 and 95% confidence interval: 1.58-2.21, P<0.05 for TT vs. TC+ CC).
Conclusion: This meta-analysis suggests that the -159C/T polymorphism in the CD14 gene contributes to tuberculosis susceptibility. To further investigate gene-gene and gene-environment interactions between this polymorphism and tuberculosis risk, more studies are needed.
Keywords: Tuberculosis, CD14, Polymorphism, Meta-analysis.
1. Introduction
Tuberculosis (TB) is still a public health challenge worldwide [1,2].As reported by the latest 2012 global tuberculosis report, in 2011, it is estimated that there were 8.7 million new cases of TB and 1.4 million people died from TB, including almost 1 million deaths among HIV-negative individuals and 430, 000 among people who were HIV-positive [3]. Recent studies suggest that host genetic factors play critical roles in the pathogenesis and outcome of TB, especially genetic polymorphisms in the immune response to Mycobacterium tuberculosis (M. tuberculosis) may explain individual differences in both susceptibility to disease and the course of infection [4-6]. Up to now, quite a lot studies have investigated the association between TB susceptibility and genetic variants among which the CD 14 gene has been highlighted.
The CD14 gene is localized on chromosome 5q31.1 and CD14 is a membrane recognition factor involved in the binding of bacterial components or inhaled endotoxin, which can induce significant lung inflammation and activate immune pathways [7]. CD14 contributes to receptor-mediated ingestion of non-opsonised M. tuberculosis by mononuclear cells such as macrophages through recognizing components of the cell wall [8]. As previously shown in genetic studies, a polymorphism in the CD14 proximal promoter, -159C/T (rs2569190), might interact with environmental factors in the development of TB. Several studies have investigated whether the -159C/T polymorphism in the CD14 gene is associated with TB risk, and, the results have been inconsistent and inconclusive. Since pooled estimates using meta-analysis have proven to be useful in determining the overall risk of certain polymorphisms [9], the present meta-analysis aimed to clarify the association between the -159C/T polymorphism in the CD14 gene and TB risk.
2. Method
2.1 Literature search
A systematic literature search in Pubmed and Embase databases was performed by two independent reviewers to identify studies examining the possible association between -159C/T polymorphism in the CD14 gene and TB risk on Jan 15, 2013. Search terms were as follows: “TB or tuberculosis” in combination with “CD14” in combination with “polymorphism or variant or mutation”. The reference lists of identified studies and review articles were manually searched to find additional relevant publications.
2.2 Study selection
Studies were included in the meta-analysis if they satisfied the following inclusion criteria: (1) they evaluated the potential association between the -159C/T polymorphism in the CD14 gene and TB risk, (2) they were case-control studies, (3) genotype distributions were available for cases and controls in order to estimate an odds ratio (OR) with 95% confidence interval (CI); and (4) the distribution of genotypes in the control group was consistent with Hardy-Weinberg equilibrium (HWE). Accordingly, abstracts, reviews, and studies in which genotype frequencies were not reported were excluded. When publications involved the same or overlapping data sets, only the study with the largest number of participants was included.
2.3 Data extraction
Two reviewers independently extracted data from the final set of included studies (T Wang and Y Shen), and the differences between them were solved by consensus. The following items were extracted from each study if available: first author, year of publication, country of origin, ethnicity, sample size, TB definition, genotyping method, and genotype number frequencies in TB cases and controls.
2.4 Statistical analysis
The strength of the association between the -159C/T polymorphism in the CD14 gene and risk of TB was assessed using OR and 95% CI. The significance of the pooled OR was determined using the Z-test and P< 0.05 was considered statistically significant. Firstly, we evaluated with the dominant model (TT+TC vs. CC) and recessive model (TT vs. TC+CC) and then evaluated variant genotype TT and compared with the wild-type CC homozygote (TT vs. CC). In addition, we also estimated the risks of TC vs. CC and T vs. C.
Heterogeneity was evaluated using a χ2-based Q statistic and I2 statistic, with P < 0.10 considered statistically significant. When P ≥ 0.10, the pooled OR of each study was calculated using a fixed-effects model; otherwise, a random-effects model was used. Publication bias was assessed using Begg's funnel plots and Egger's test [10,11]. Sensitivity analysis was performed by sequentially excluding individual studies and recalculating the results. HWE was tested using a web-based program (http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl). All statistical tests were performed using Revman 5.1 and STATA 12.0 software.
3. Results
3.1 Characteristics of included studies
After independent review, a total of seven studies evaluating the association between the -159C/T polymorphism in the CD14 gene and TB risk were included in the meta-analysis, involving 3253 subjects (1,574 TB cases and 1,679 controls) [12-18]. There were four case-controls of Asians [12,14,17,18], two of Latinos [15,16], and one of Caucasian [13]. Of the seven included studies, genotype frequencies for control group were all consistent with HWE. The characteristics of each case-control study are summarized in Table 1, and, genotype and allele distributions for each case-control studies are listed in Table 2.
3.2 Quantitative data synthesis
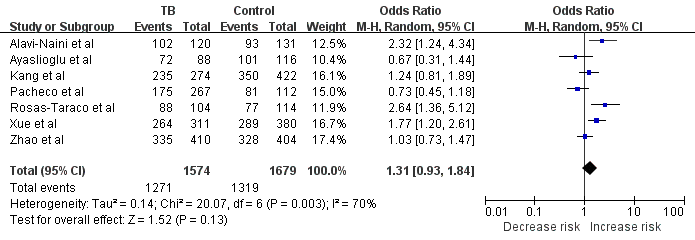
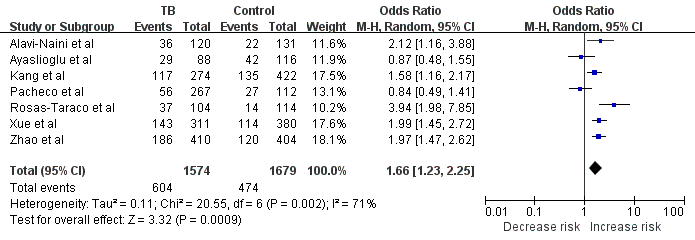
First, we analyzed the heterogeneity of TT+TC vs. CC in order to choose the best calculation model. For included seven studies, the value of χ2 was 20.07 with P=0.003 in a random-effects model, and I-square value, another index of heterogeneity, was 70%, both suggesting a significant of heterogeneity. Thus, we chose the random-effects model to synthesize the data. Overall, the pooled OR was 1.31 (95% CI: 0.93-1.84) and the test for overall effect Z value was 1.52 (P=0.13) (Fig.1). These results suggested that the TT homozygote and TC heterozygote carriers have an increased risk of TB compared with those individuals with the CC homozygote; however, there was no statistical significance. As suggested by Thakkinstian et al [19], the recessive model is the best genetic model for evaluating the association between the -159C/T polymorphism in the CD14 gene and TB risk. We noticed that the TT homozygote carriers have an increased risk of TB compared with those individuals with the TC and CC homozygote with an OR of 1.66 (95% CI: 1.23-2.25) (P<0.05, Fig. 2), indicating the -159C/T polymorphism is a risk factor for TB. Summary of the results of other genetic comparisons are listed in Table 3.
In the subgroup analysis by ethnicity, no associations were found in Latinos (OR= 1.79, 95% CI: 0.39-8.18, P=0.46) in recessive model (TT vs. TC+CC). While we found an obvious association between -159C/T polymorphism in the CD14 gene and the TB susceptibility in dominant model (TT+TC vs. CC), recessive model (TT vs. TC+CC), and allelic genetic model (T vs. C), as shown in Table 3, suggesting -159C/T polymorphism is a genetic risk factor for TB in Asians.
3.3 Sensitivity analysis and Publication bias
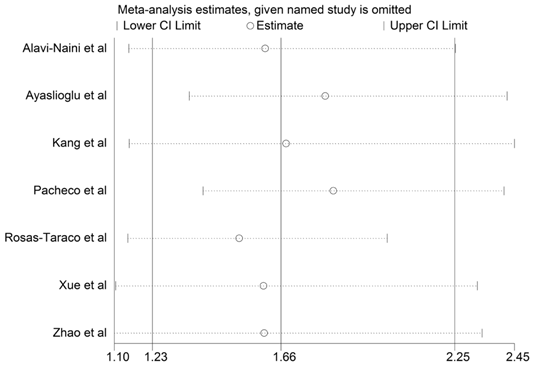
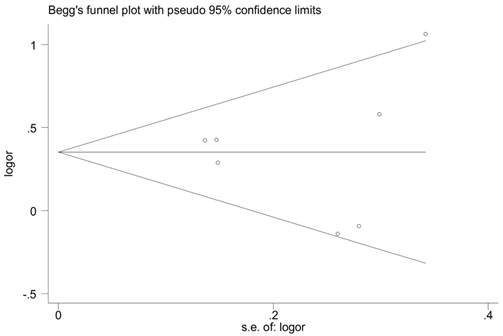
To estimate the stability of our findings, sensitivity analysis was performed by sequentially excluding each study. Statistically similar results were obtained after sequentially excluding each case-control study, showing the stability of the results (Fig.3). Begg's funnel plot and Egger's test were used to assess publication bias. The shape of the funnel plots seemed symmetrical in the TT vs. TC+CC comparison genetic model, suggesting the absence of publication bias (Fig. 4). Then, the Egger's test was performed to provide statistical evidence of funnel plots symmetry. The results indicated a lack of publication bias of the current meta-analysis (P=0.315).
Characteristics of included studies.
| Author | Year | Country | Ethnicity | Tuberculosis | Mean age | Control | Mean age | Genotyping method | TB definition | HWE |
|---|---|---|---|---|---|---|---|---|---|---|
| Alavi-Naini et al | 2012 | Iran | Asian | 120 | 51.5 | 131 | 48.1 | PCR | B | Y |
| Ayaslioglu et al | 2012 | Turkey | Caucasian | 88 | 49.93 | 116 | 32.96 | PCR | B, HP, CD | Y |
| Kang et al | 2009 | Korea | Asian | 274 | 38.0 | 422 | 24.0 | PCR | B, HP, CD | Y |
| Pacheco et al | 2004 | Colombia | Latino | 267 | NA | 112 | NA | PCR-RFLP | B, HP, CD | Y |
| Rosas-Taraco et al | 2007 | México | Latino | 104 | 39.5 | 114 | 26.3 | PCR-RFLP | B, CD | Y |
| Xue et al | 2012 | China | Asian | 311 | 38.0 | 380 | 37.7 | PCR | B, CD | Y |
| Zhao et al | 2012 | China | Asian | 410 | 38.5 | 404 | 36.7 | PCR | B, CD | Y |
NA: Not available; PCR: Polymerase chain reaction; RFLP: Restriction fragment length polymorphism; B: Bacteriology; HP: Histopathology; CD: Clinical diagnosis; HWE: Hardy-Weinberg equilibrium; Y: Yes.
Distribution of CD14 genotype and allele among TB patients and controls
| Author | TB | Control | TB | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC | TC | TT | CC | TC | TT | C | T | C | T | |
| Alavi-Naini et al | 18 | 66 | 36 | 38 | 71 | 22 | 102 | 138 | 147 | 115 |
| Ayaslioglu et al | 16 | 43 | 29 | 15 | 59 | 42 | 75 | 101 | 89 | 143 |
| Kang et al | 39 | 118 | 117 | 72 | 215 | 135 | 196 | 352 | 359 | 485 |
| Pacheco et al | 92 | 119 | 56 | 31 | 54 | 27 | 303 | 231 | 116 | 108 |
| Rosas-Taraco et al | 16 | 51 | 37 | 37 | 63 | 14 | 83 | 125 | 137 | 91 |
| Xue et al | 47 | 121 | 143 | 91 | 175 | 114 | 215 | 407 | 357 | 413 |
| Zhao et al | 75 | 149 | 186 | 76 | 208 | 120 | 299 | 521 | 360 | 448 |
TB: Tuberculosis
Summary of different comparative results.
| TT+TC vs. CC | TT vs. TC+CC | TT vs.CC | TC vs. CC | T vs.C | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | P* | OR (95%CI) | P* | OR (95%CI) | P* | OR (95%CI) | P* | OR (95%CI) | P* | |
| Total | 1.31 (0.93-1.84) | 0.13 | 1.66 (1.23-2.25) | 0.0009 | 1.76 (1.10-2.81) | 0.02 | 1.07 (0.79-1.44) | 0.68 | 1.36 (1.08-1.70) | 0.008 |
| Subgroup by Ethnicity | ||||||||||
| Latino | 1.36 (0.38-4.81) | 0.63 | 1.79 (0.39-8.18) | 0.46 | 2.02 (0.24-16.96) | 0.52 | 1.14 (0.46-2.83) | 0.77 | 1.35 (0.50-3.67) | 0.55 |
| Asian | 1.39 (1.13-1.71) | 0.002 | 1.87 (1.58-2.21) | <0.00001 | 1.93 (1.53-2.44) | <0.00001 | 1.12 (0.76-1.65) | 0.56 | 1.48 (1.31-1.66) | <0.00001 |
The bold values mean that their association is significant, * P value for Z test.
Meta-analysis with a random-effects model for the association between TB risk and the CD14 -159C/T polymorphism (TT+TC vs. CC). The size of the square is proportional to the percent weight of each study; horizontal lines represent the 95% CI.
Meta-analysis with a random-effects model for the association between TB risk and the CD14 -159C/T polymorphism (TT vs. TC+CC). The size of the square is proportional to the percent weight of each study; horizontal lines represent the 95% CI.
Sensitivity analysis of included studies.
Begg's funnel plot for publication bias in selection of studies on the CD14 -159C/T polymorphism (TT vs. TC+ CC).
4. Discussion
TB remains a leading cause of mortality worldwide, while the pathogenesis of TB has not been well understood [1,2]. Gene variants may alter protein production, function and individual's susceptibility to disease, thus, may play a role in pathogenesis of TB [6]. CD14 is one of the extracellular adaptor proteins required for recognition of M. tuberculosis, and it is important for eliciting lung inflammatory response during M. tuberculosis infection which during progressive disease can result enhanced mortality [20]. The -159C/T polymorphism in the gene CD14 might interact with environmental factors in the development of TB. Recently, it has been demonstrated that individuals with CD14 -159 TT genotype had increased serum CD14 levels [12]. It is proposed that the up-regulation of CD14 expression can ease immune interactions with mannosylated lipoarabinomannan, which subsequently enhances the production of transforming growth factor -β and suppresses the immune response, thus, make subjects at a high risk of TB [21]. The correlation of CD 14 polymorphism and TB risk has been studied, but the results remain controversial. Therefore, we performed this meta-analysis to clarify the relationship between this polymorphism and susceptibility to TB. To our best knowledge, it is the most comprehensive meta-analysis regarding the -159C/T polymorphism and TB risk.
A total of seven case-control studies in Asians, Latinos, and Caucasians were included in the meta-analysis. The strength of the present analysis was based on the accumulation of published data giving information to detect more precise conclusion. In this study, the effect of dominant/recessive models and the effect of allele frequency were all estimated to investigate the precise role of -159C/T polymorphism in TB. Moreover, the consistency of genetic effects across populations from different ethnicities was investigated. For the present meta-analysis, research findings do support a role for the CD14-159C/T polymorphism in the development of TB in the total combined analysis (TT vs. TC+CC), the TT homozygote carriers have an increased risk of TB compared with those individuals with the TC and CC homozygote. Rosas-Taraco et al also found that CD14-159TT appear to be a risk factor for the development of pulmonary TB in Latinos [16]. Sub-group analysis suggested that the CD14-159C/T polymorphism is an increased risk factor for TB in Asians, while not in Latinos. Considering the ethnic difference, there may be differences in the genes involved in different populations, thus, to take of a further investigation of population differences will be particularly informative, and susceptibility genes identified in TB patients with different ethnicities may provide an opportunity to explore new mechanisms of disease that are specific in different population.
The findings in this meta-analysis should be interpreted with caution because of several limitations. First, a relatively small number of studies and subjects were included, which may reduce the statistical power for identifying possible association between the -159C/T polymorphism and TB risk. Second, the seven included studies were performed majorly in Asian or Latino populations, only one in Caucasian [13], thus more studies are needed in other ethnic population such as Caucasians because of possible ethnic differences of the -159C/T polymorphism. Third, although we didn't set any restrictions during our literature searching, we included only English-language publications in the present meta-analysis. It is possible that including relevant published studies in other languages or unpublished studies which had null outcomes may alter our results.
5. Conclusions
To our knowledge, this is the first meta-analysis to assess the relationship between the -159C/T polymorphism in CD14 gene and TB risk. Our results suggest that the -159C/T polymorphism was significantly associated with TB risk, especially in Asian population. In the future, large well-designed and multi-center epidemiological studies in different ethnic population should be performed to assess these associations.
Acknowledgements
This work is supported by grants 81230001, 81300032, 31000513 and 31171103 from the National Natural Science Foundation of China. We are indebted to the authors of the primary studies included in this meta-analysis; without their contributions, this work would not have been possible.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lawn SD, Zumla AI. Tuberculosis. Lancet. 2011;378(9785):57-72
2. Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368(8):745-55
3. World Health Organization. Global tuberculosis report 2012. World Health Organization. 2012
4. Yim JJ, Selvaraj P. Genetic susceptibility in tuberculosis. Respirology. 2010;15(2):241-256
5. Qu HQ, Fisher-Hoch SP, McCormick JB. Knowledge gaining by human genetic studies on tuberculosis susceptibility. J Hum Genet. 2011;56:177-182
6. Azad AK, Sadee W, Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun. 2012;80(10):3343-3359
7. Anas A, van der Poll T, de Vos AF. Role of CD14 in lung inflammation and infection. Crit Care. 2010;14(2):209
8. Peterson PK, Gekker G, Hu S. et al. CD14 receptor-mediated uptake of nonopsonized Mycobacterium tuberculosis by human microglia. Infect Immun. 1995;63(4):1598-1602
9. Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med. 2009;6:e28
10. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088-1101
11. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-634
12. Alavi-Naini R, Salimi S, Sharifi-Mood B. et al. Association between the CD14 gene C-159T polymorphism and serum soluble CD14 with pulmonary tuberculosis. Int J Tuberc Lung Dis. 2012;16(10):1383-1387
13. Ayaslioglu E, Kalpaklioglu F, Kavut AB. et al. The role of CD14 gene promoter polymorphism in tuberculosis susceptibility. J Microbiol Immunol Infect. 2012 doi: 10.1016/j.jmii.2012.05.008
14. Kang YA, Lee HW, Kim YW. et al. Association between the -159C/T CD14 gene polymorphism and tuberculosis in a Korean population. FEMS Immunol Med Microbiol. 2009;57:229-235
15. Pacheco E, Fonseca C, Montes C. et al. CD14 gene promoter polymorphism in different clinical forms of tuberculosis. FEMS Immunol Med Microbiol. 2004;40(3):207-213
16. Rosas-Taraco AG, Revol A, Salinas-Carmona MC. et al. CD14 C(-159)T polymorphism is a risk factor for development of pulmonary tuberculosis. J Infect Dis. 2007;196(11):1698-1706
17. Zhao MY, Xue Y, Zhao ZQ. et al. Association of CD14 G(-1145)A and C(-159)T polymorphisms with reduced risk for tuberculosis in a Chinese Han population. Genet Mol Res. 2012;11:3425-3431
18. Xue Y, Zhao ZQ, Chen F. et al. Polymorphisms in the promoter of the CD14 gene and their associations with susceptibility to pulmonary tuberculosis. Tissue Antigens. 2012;80:437-443
19. Thakkinstian A, McEldu P, D'Este C. et al. A method for meta-analysis of molecular association studies. Stat Med. 2005;24:1291-1306
20. Wieland CW, van der Windt GJ, Wiersinga WJ. et al. CD14 contributes to pulmonary inflammation and mortality during murine tuberculosis. Immunology. 2008;125(8):272-279
21. Shams H, Wizel B, Lakey DL. et al. The CD14 receptor does not mediate entry of Mycobacterium tuberculosis into human mononuclear phagocytes. FEMS Immunol Med Microbiol. 2003;36(1-2):63-9
Author contact
![]() Corresponding author: Fuqiang Wen M.D., Ph.D. Division of Pulmonary Diseases, State Key Laboratory of Biotherapy of China and Department of Respiratory and Critical Care Medicine, West China Hospital of Sichuan University, Chengdu 610041, China. E-mail: wenfuqiangcom Phone: +86-28-85422380 Fax: +86-28-85582944.
Corresponding author: Fuqiang Wen M.D., Ph.D. Division of Pulmonary Diseases, State Key Laboratory of Biotherapy of China and Department of Respiratory and Critical Care Medicine, West China Hospital of Sichuan University, Chengdu 610041, China. E-mail: wenfuqiangcom Phone: +86-28-85422380 Fax: +86-28-85582944.

 Global reach, higher impact
Global reach, higher impact