3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(11):1510-1517. doi:10.7150/ijms.5342 This issue Cite
Research Paper
Analysis of Immunoglobulin and T Cell Receptor Gene Rearrangement in the Bone Marrow of Lymphoid Neoplasia Using BIOMED-2 Multiplex Polymerase Chain Reaction
1. Department of Laboratory Medicine, The Catholic University of Korea, College of Medicine, Seoul, Korea;
2. Blood Transfusion Research Institute, Korean Red Cross, Seoul, Korea;
3. Department of Hematology, Catholic Blood and Marrow Transplantation Center, The Catholic University of Korea, College of Medicine, Seoul, Korea.
Received 2012-10-5; Accepted 2013-8-12; Published 2013-8-31
Abstract
The evaluation of bone marrow (BM) involvement is important for diagnosis and staging in patients with lymphoid neoplasia. We evaluated of immunoglobulin (Ig) and/or T-cell receptor (TCR) gene rearrangements in the BM using the standardized BIOMED-2 multiplex PCR clonality assays and compared the results with microscopic findings such as histology and CD10, CD20, CD79a, CD3 and CD5 immunohistochemistry. A total of 151 samples were enrolled; 119 B cell neoplasia, 29 T cell neoplasia, and 3 Hodgkin's lymphoma. The molecular clonality assay and microscopic diagnosis were concordant in 66.9% (n=101) and discordant in 33.1 % (n=50). Ig/TCR gene clonality assay detected 43 cases of BM involvement which was not presented in the morphology. Two cases among them turned into microscopic BM involvement during a close follow up. Clonal TCR gene rearrangements were detected in 12.6% of B cell neoplasia and Ig gene rearrangement were found in 3.4% of T cell neoplasia. This molecular clonality assay is valuable particularly in diagnosing BM involvement of lymphoid neoplasia if it is morphologically uncertain. But it should be carefully interpreted because molecular clonality may be present in the reactive lymphoproliferation. Therefore, comprehensive analysis with morphologic analysis should be important to reach a final diagnosis.
Keywords: immunoglobulin (Ig) /T-cell receptor (TCR) gene rearrangements, BIOMED-2 multiplex PCR.
Introduction
The evaluation of bone marrow (BM) samples in patients with lymphoid neoplasia is an important aspect for the diagnosis, staging and assessing the treatment response [1]. Histology and morphology supplemented with immunohistochemistry (IHC) and/or flow cytometric immunophenotyping can be helpful to accurate diagnosis [2].
However, it is more complicated to discriminate BM involvement of malignant lymphoma accurately with only microscopic findings. Molecular studies of immunoglobulin (Ig) and/or T-cell receptor (TCR) gene rearrangements can be a useful additional tool to support the clonality of the involved cells [3,4].
Although Southern blot analysis has proved to be a gold standard method in the detection of clonal rearrangement, it has several technical disadvantages, such as time necessary to obtain results, the use of radioactivity, and the susceptibility of the method to various artifacts [5].
The BIOMED-2 multiplex PCR approach is a rapid and reliable procedure that is more sensitive than that of the Southern blot analysis in detecting clonality. It seems that the Southern blot analysis can now be reliably replaced in a routine laboratory setting [6].
In this study, we evaluated Ig and TCR gene rearrangements in the BM of patients who were diagnosed as lymphoid neoplasia, using the standardized BIOMED-2 multiplex PCR clonality assays (InVivoScribe Technologies, San Diego, CA, USA), and compared the results with microscopic findings of the BM.
Materials and Methods
Materials
A total of 151 BM aspirates from 131 patients with lymphoid neoplasia were submitted for clonality analysis of Ig and/or TCR gene rearrangements as a part of diagnostic work-up, from July 2009 to February 2010, at Seoul St Mary's Hospital, Seoul, Korea. There were 72 males and 59 females enrolled. The median age was 50 (17- 77). This study was approved by the institutional review board (IRB) of Seoul St. Mary's Hospital (Approval number: KC11RISI0759). The diagnoses were performed, according to the joint World Health Organization-European Organisation for Research and Treatment of Cancer classification, on the bases of clinical, histologic, and IHC criteria [7]. These included 119 B cell neoplasia, 29 T cell neoplasia, and 3 Hodgkin's lymphoma. BM study was done using BM aspirates, clot section, and biopsy to detect the involvement of neoplastic cells. IHC, including CD10, CD20, CD79a and CD3, and CD5 were performed to identify involvement status.
Ig and TCR gene clonality
Genomic DNA was extracted from 200µl of 151 BM aspirates using a commercial genomic DNA purification kit (Qiagen Gmbh, Germany). Each DNA was quantified using the NanoDrop® ND-1000 spectrophotometer. The resulting purity (A260/A280) demonstrated 1.8 to 2.0, and it means to warrant DNA purity.
PCR amplifications were performed to identify the clonal Ig and TCR gene rearrangements, according to the manufacturer's instruction using a commercial BIOMED-2 multiplex PCR, IdentiClone Ig/TCR gene clonality assay (InVivo-Scribe Technologies, San Diego, CA, USA). All samples were analysed in duplicate. The full set of reactions for Ig gene arrangements included five reactions targeting IGH (IGHA: VHFR1-JH; IGHB: VHFR2-JH; IGHC: VHFR3-JH; IGHD: DH1-6-JH; IGHE: DH7-JH), two reactions targeting IGK (IGKA: Vκ-Jκ; IGKB: Vκ-Kde and JκCκ intron-Kde) and one reaction targeting IGL (Vλ-Jλ). The full set of reactions for TCR gene arrangements included two reactions targeting TCRG (TCRGA: VγI/γ10-Jγ; TCRGB: Vγ9/γ11-Jγ), three reactions targeting TCRB (TCRBA:Vβ-Jβ1/2.2/2.6/2.7; TCRBB: Vβ-Jβ2.1/2.3/2.4/2.5; TCRBC: Dβ-Jβ) and one reaction targeting TCRD (Vδ-Jδ). A standard 35- cycle PCR protocol was used for each primer set. After initial denaturation at 95°C for 7 minutes, each cycle was composed of denaturation at 95°C for 45 seconds, annealing at 60°C for 45 seconds and extension at 72°C for 90 seconds. Final extension was at 72°C for 10 minutes. The PCR products were analyzed on an automated capillary electrophoresis system (ABI 3100, Applied Biosystems, Foster City, Calif) with GeneScan software (Applied Biosystems).
All electropherograms were examined by at least two experts experienced in clonality analysis. The criteria for defining a positive peak is that products generated from diagnostic samples are discrete bands appeared within expected size range and at least three times the amplitude of the third largest peak in the polyclonal background. [8]
Results
We consecutively investigated 151 BM samples from 131 patients that were submitted for routine diagnostics. The initial diagnosis, BM findings including IHC, and molecular results, are summarized in Table 1.
The microscopic BM involvement was demonstrated in 40 out of 151 (26.5%) BM cases. Among them, 15 cases were determined after IHC was performed. Among the 119 B cell neoplasias, microscopic BM involvement was positive in 31 cases and Ig gene rearrangement was detected in 25 of 31. In 6 cases, Ig/TCR gene rearrangement test did not detect BM involvement, which was presented by microscopy. The diagnosis of 6 cases were as follows; three diffuse large B cell lymphomas, two mucosa-associated lymphoid tissue lymphomas and one of other B cell neoplasias. Among the 29 T cell neoplasias, microscopic BM involvement was positive in 6 cases and TCR gene rearrangement was detected in all of them. Three Hodgkin's diseases were enrolled whose BM involvement was revealed by microscopy and 2 of them revealed TCR gene rearrangement.
Ig and TCR gene clonality assay detected 43 cases (29 B cell neoplasia and 14 T cell neoplasia) of BM involvement of lymphoid neoplasia which was not presented in the morphology. In 2 cases with positive clonal Ig gene rearrangement and negative microscopic findings turned into positive BM involvement in a microscopic level, during a close follow up BM evaluation. The Ig rearrangement profiles and TCR rearrangement profiles are detailed in Table 2 and Table 3. In one case of MALT lymphoma with microscopic BM involvement, there was clonality only for Ig light chain rearrangement (IGKA). In one follicular lymphoma and one lymphoplasmacytic lymphoma without microscopic bone marrow involvement, there was clonality only for Ig light chain rearrangement(IGKA). In one case of Burkitt lymphoma without microscopic bone marrow involvement, there was clonality only for Ig light chain rearrangement (IGKB). Clonal TCR gene rearrangements were detected in 15 of 119 cases of B cell neoplasias, with or without clonal Ig gene rearrangement. Ig gene rearrangements were found in 1 of 29 cases of T cell neoplasias combined with TCR gene rearrangement. The BM involvements were not detected neither by microscopy nor molecular clonality assay in 68 cases. In total, the result of Ig and TCR clonality assay and the microscopic diagnosis were concordant in 101 cases (66.9%) and discordant in 50 cases (33.1%) (Table 4).
We compared BM Ig/TCR gene rearrangement results with the molecular results of the original lymphoma tissues in some cases and showed the profiles in Fig.1.
Morphologic involvement results and Ig/TCR rearrangement results according to the diagnosis.
| Diagonosis (n) | Microscopic involvement | Total number | Total- | Total+ | Ig- | Ig+ | Ig not done | TCR- | TCR+ | TCR not done |
|---|---|---|---|---|---|---|---|---|---|---|
| DLBCL(62) | Negative | 48 | 36 | 12 | 40 | 7 | 1 | 23 | 5 | 20 |
| Positive | 14 | 3 | 11 | 3 | 11 | 0 | 4 | 2 | 8 | |
| MALT (29) | Negative | 26 | 19 | 7 | 21 | 5 | 0 | 9 | 4 | 13 |
| Positive | 3 | 2 | 1 | 2 | 1 | 0 | 0 | 0 | 3 | |
| FL(6) | Negative | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| Positive | 4 | 0 | 4 | 0 | 4 | 0 | 1 | 0 | 3 | |
| MCL(6) | Negative | 2 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 2 |
| Positive | 4 | 0 | 4 | 1 | 3 | 0 | 1 | 1 | 2 | |
| BL(4) | Negative | 3 | 1 | 2 | 2 | 1 | 0 | 1 | 1 | 1 |
| Positive | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | |
| CLL(3) | Negative | 2 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 1 |
| Positive | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | |
| LPL(2) | Negative | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| Positive | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | |
| Other B(7) | Negative | 4 | 2 | 2 | 3 | 1 | 0 | 1 | 1 | 2 |
| Positive | 3 | 1 | 2 | 1 | 2 | 0 | 2 | 0 | 1 | |
| HD(3) | Negative | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Positive | 3 | 1 | 2 | 3 | 0 | 0 | 1 | 2 | 0 | |
| PTCL(9) | Negative | 6 | 2 | 4 | 3 | 0 | 3 | 2 | 4 | 0 |
| Positive | 3 | 0 | 3 | 3 | 0 | 0 | 0 | 3 | 0 | |
| TLL(8) | Negative | 7 | 4 | 3 | 5 | 0 | 2 | 4 | 3 | 0 |
| Positive | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | |
| ALCL(4) | Negative | 2 | 0 | 2 | 1 | 0 | 1 | 0 | 2 | 0 |
| Positive | 2 | 0 | 2 | 1 | 1 | 0 | 0 | 2 | 0 | |
| ENK/TL(6) | Negative | 6 | 3 | 3 | 1 | 0 | 5 | 3 | 3 | 0 |
| Positive | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| NK/TL(2) | Negative | 2 | 0 | 2 | 1 | 0 | 1 | 0 | 2 | 0 |
| Positive | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | 151 | 75 | 76 | 92 | 45 | 14 | 55 | 37 | 59 |
Abbreviations: DLBCL, diffuse large B cell lymphoma; MALT, MALT lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; BL, Burkitt lymphoma; CLL,chronic lymphocytic lymphoma; LPL,lymphoplasmacytic lymphoma; Other B, Other B cell neoplasia; HD,Hodgkin's disease; PTCL, peripheral T cell lymphoma; TLL, T lymphoblastic lymphoma; ALCL, anaplastic large cell lymphoma; ENK/TL, extranodal NK/T cell lymphoma; NK/TL,NK/T cell lymphoma. Ig/TCR rearrangement negative means: no specific product, polyclonal or pseudoclonal. Ig/TCR rearrangement positive means: clonal or clonal with a polyclonal background.
Morphologic involvement results and Ig rearrangement profiles.
| Dx | M/I | IGHA ( VHFR1-JH) | IGHB (VHFR2-JH) | IGHC ( VHFR3-JH) | IGHD ( DH1-6-JH) | IGHE (DH7-JH) | IGKA ( Vκ-Jκ) | IGKB (Vκ-Kde and JκCκ intron-Kde) | IGL (Vλ-Jλ) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | ||
| DLBCL | Neg | 47 | 47 | 47 | 44 | 3 | 46 | 1 | 45 | 2 | 46 | 1 | 47 | ||||
| Pos | 11 | 3 | 10 | 4 | 11 | 3 | 12 | 2 | 13 | 1 | 5 | 9 | 10 | 4 | 12 | 2 | |
| MALT | Neg | 25 | 1 | 25 | 1 | 26 | 25 | 1 | 26 | 22 | 4 | 26 | 25 | 1 | |||
| Pos | 3 | 3 | 3 | 3 | 3 | 2 | 1 | 3 | 3 | ||||||||
| FL | Neg | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | |||||||
| Pos | 4 | 4 | 3 | 1 | 4 | 4 | 4 | 2 | 2 | 3 | 1 | ||||||
| MCL | Neg | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | ||||
| Pos | 1 | 3 | 1 | 3 | 1 | 3 | 4 | 4 | 1 | 3 | 2 | 2 | 2 | 2 | |||
| BL | Neg | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 1 | 3 | |||||||
| Pos | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| CLL | Neg | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | |||
| Pos | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| LPL | Neg | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| Pos | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| Other B | Neg | 4 | 3 | 1 | 4 | 3 | 1 | 4 | 4 | 3 | 1 | 4 | |||||
| Pos | 1 | 2 | 1 | 2 | 2 | 1 | 2 | 1 | 3 | 1 | 2 | 2 | 1 | 2 | 1 | ||
| HD | Neg | ||||||||||||||||
| Pos | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | |||||||||
| PTCL | Neg | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | ||||||||
| Pos | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | |||||||||
| TLL | Neg | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||||||||
| Pos | |||||||||||||||||
| ALCL | Neg | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| Pos | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 2 | ||||||
| ENK/TL | Neg | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| Pos | |||||||||||||||||
| NK/TL | Neg | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| Pos | |||||||||||||||||
| Total | 119 | 18 | 119 | 18 | 128 | 9 | 127 | 10 | 134 | 3 | 105 | 32 | 119 | 18 | 129 | 8 | |
Abbreviations: DLBCL, diffuse large B cell lymphoma; MALT, MALT lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; BL, Burkitt lymphoma; CLL,chronic lymphocytic lymphoma; LPL,lymphoplasmacytic lymphoma; Other B, Other B cell neoplasia; HD,Hodgkin's disease; PTCL, peripheral T cell lymphoma; TLL, T lymphoblastic lymphoma; ALCL, anaplastic large cell lymphoma; ENK/TL, extranodal NK/T cell lymphoma; NK/TL,NK/T cell lymphoma; M/I, microscopic involvement. Ig/TCR rearrangement negative means: no specific product, polyclonal or pseudoclonal. Ig/TCR rearrangement positive means: clonal or clonal with a polyclonal background.
Morphologic involvement results and TCR rearrangement profiles.
| Dx | M/I | TCRGB(Vγ9/γ11-Jγ) | TCRBA(Vβ-Jβ1/2.2/2.6/2.7) | TCRBB ( Vβ-Jβ2.1/2.3/2.4/2.5) | TCRBC( Dβ-Jβ) | TCRD (Vδ-Jδ) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | ||
| DLBCL | Neg | 26 | 2 | 27 | 1 | 28 | 28 | 27 | 1 | ||
| Pos | 6 | 6 | 6 | 5 | 1 | 6 | |||||
| MALT | Neg | 10 | 3 | 13 | 13 | 12 | 1 | 12 | 1 | ||
| Pos | |||||||||||
| FL | Neg | 1 | 1 | 1 | 1 | 1 | |||||
| Pos | 1 | 1 | 1 | 1 | 1 | ||||||
| MCL | Neg | ||||||||||
| Pos | 2 | 2 | 2 | 2 | 1 | 1 | |||||
| BL | Neg | 2 | 1 | 1 | 2 | 2 | 2 | ||||
| Pos | 1 | 1 | 1 | 1 | 1 | ||||||
| CLL | Neg | 1 | 1 | 1 | 1 | 1 | |||||
| Pos | |||||||||||
| LPL | Neg | ||||||||||
| Pos | 1 | 1 | 1 | 1 | 1 | ||||||
| Other B | Neg | 2 | 1 | 1 | 2 | 2 | 2 | ||||
| Pos | 2 | 2 | 2 | 2 | 2 | ||||||
| HD | Neg | ||||||||||
| Pos | 1 | 2 | 3 | 3 | 3 | 2 | 1 | ||||
| PTCL | Neg | 5 | 1 | 5 | 1 | 6 | 6 | 6 | |||
| Pos | 1 | 2 | 3 | 3 | 3 | 3 | |||||
| LBL | Neg | 5 | 2 | 7 | 7 | 7 | 7 | ||||
| Pos | 1 | 1 | 1 | 1 | 1 | ||||||
| ALCL | Neg | 2 | 2 | 2 | 2 | 2 | |||||
| Pos | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | ||
| ENK/TL | Neg | 5 | 1 | 6 | 6 | 5 | 1 | 6 | |||
| Pos | |||||||||||
| NK/TL | Neg | 2 | 1 | 1 | 2 | 2 | 2 | ||||
| Pos | |||||||||||
| Total | 74 | 18 | 85 | 7 | 92 | 0 | 88 | 4 | 87 | 5 | |
Abbreviations: DLBCL, diffuse large B cell lymphoma; MALT, MALT lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; BL, Burkitt lymphoma; CLL,chronic lymphocytic lymphoma; LPL,lymphoplasmacytic lymphoma; Other B, Other B cell neoplasia; HD,Hodgkin's disease; PTCL, peripheral T cell lymphoma; TLL, T lymphoblastic lymphoma; ALCL, anaplastic large cell lymphoma; ENK/TL, extranodal NK/T cell lymphoma; NK/TL,NK/T cell lymphoma; M/I, microscopic involvement. Ig/TCR rearrangement negative means: no specific product, polyclonal or pseudoclonal. Ig/TCR rearrangement positive means: clonal or clonal with a polyclonal background.
Concordance between molecular results and microscopic BM involvement of B cell neoplasia (n=119) and T cell neoplasia (n=29).
| BM involvement of lymphoid neoplasm | Ig rearrangement(B cell neoplasia) | TCR rearrangement(T cell neoplasia) | ||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Positive | 25(21.0%) | 6(5%) | 6(20.7%) | 0(0%) |
| Negative | 29(24.4%) | 59(49.6%) | 14(48.3%) | 9(31%) |
Ig/TCR rearrangement negative means: no specific product, polyclonal or pseudoclonal. Ig/TCR rearrangement positive means: clonal or clonal with a polyclonal background.
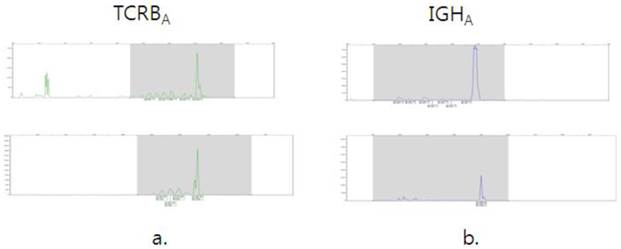
Examples of cases with identical clones in the bone marrow (top) and lymphoma tissue(bottom). Clonal analysis of TCRBA of T lymphoblastic lymphoma (a) and IGHA of mantle cell lymphoma(b).
Discussion
Morphologic examination is the generally accepted method for determining BM involvement of lymphoma in patients evaluated for extent of disease at diagnosis and after the therapy. Molecular methods for determining clonality provide alternative techniques for evaluating suspicious or indeterminate cases, and have the potential to increase the detection of minimal degrees of marrow involvement. The detection of identical T- or B-cell clones in a tissue sample and BM is of great importance in staging lymphoid neoplasia and prognostic relevance [9-12].
The BIOMED-2 concerted action, comprising 47 institutes from seven European countries, developed a set of standardized multiplex-PCR protocols for the diagnosis of clonality, based on capillary electrophoresis. This group designed IGH, IGK, and IGL clonality assays for B-cell neoplasias, and TCRB, TCRD and TCRG for T-cell neoplasias (TCRA was not included as a PCR target due to its complexity). A total of 97 new primers were designed, covering the majority of functional gene segments and representing 418 single PCR procedures [13-15].
We evaluated BIOMED-2 multiplex PCR clonality assay in the BM samples of lymphoid neoplasia. Microscopic findings and BIOMED-2 multiplex PCR were concordant in 66.9% (n=101), and discordant in 33.1% (n=50).
In 5% (6/119) of B cell neoplasias, the BM involvements were detected by microscopic evaluation, but Ig gene rearrangements were negative. This discordance may be related to the sample variation between the trephine biopsy and aspirates because of heterogeneous or patchy marrow involvement, a failure to aspirate sufficient lymphoma cells, or insufficient primer homology for amplification. [16].
In 24.4% (29/119) of B cell neoplasia, the microscopic BM involvements were not present and Ig gene rearrangements were detected. Further, in 48.3% (14/29) of T cell neoplasia, the microscopic BM involvements were not present, and TCR gene rearrangements were detected. Two from these cases, with clear monoclonal Ig/TCR products without morphologic evidence, prompted further pathological review. We experienced that the BM involvement was confirmed after additional IHC in a follow up BM. This demonstrates that it is important to interpret the PCR results within the whole diagnostic context.
According to the literature of the evaluation for BIOMED-2 assay in tissue samples, with reactive lymphoproliferations, most cases show Ig/TCR polyclonality, but clear monoclonal Ig/TCR products were observed in 10%, which prompted further pathological review. Clonal cases included two missed lymphomas in national review and nine cases that could be explained as diagnostically difficult cases or probable lymphomas upon additional review [4].
The use of Ig/TCR clonality assessment is a tool for diagnosing malignant lymphoma with high certainty, but monoclonality is not always equal malignancy. Clonality findings could often occur in some reactive inflammatory conditions such as Helicobacter-associated gastritis or lymphoid hyperplasia of the skin. [17,18,19]
In one case of MALT lymphoma with microscopic BM involvement, there was clonality only for Ig light chain rearrangement (IGKA). The increased detection of clonality by combined analysis of IGK and IGH has been demonstrated in some papers [14,20-22]
Clonal TCR gene rearrangements were detected in 12.6% (15/119) of B cell neoplasia with or without clonal Ig gene rearrangement. Ig gene rearrangements were found in 3.4% (1/29) of T cell neoplasia combined with TCR gene rearrangement. TCR rearrangements occur in 10-20 % of B cell neoplasias and Ig gene rearrangements occur in 5-10 % of T cell neoplasias. These rearrangements may correspond to the presence of a small additional B-or T cell population, therefore, B/T lineage assignment should be based on complete Ig/TCR multiplex pattern [16,17,23].
The biallelic rearrangements in a single clone result in two different PCR products.
When interpreting multiple clonal products, careful consideration of immunobiological and technical explanations is needed. Multiple rearrangements (2-4 clonal rearrangements) occur on IGK loci in 8 cases and 2-4 rearrangements occur on TCRB loci in 7 cases. These are summarized in table 5 and table 6. As reported by Anon W., due to specific configuration of the IGK and TCRB loci, multiple rearrangements can occur on one allele resulting in the possibility that up to four PCR products could be compatible with a single clone. When 2 > clonal Ig/TCR products (> 4 for IGK and TCRB) are detectable, this could reflect biclonality, oligoclonality, or even pseudoclonality [24].
Summary of different IGK rearrangement configurations that are compatible with a single clone.
| IGK configuration | Cases (n) | Number of rearrangements detectable | ||
|---|---|---|---|---|
| Vκ-Jκ (multiplex tube A) | Intron/Vκ-Kde(multiplex tube B) | Total | ||
| V-Ja/V-Ja | 3 | 2 | 0 | 2 |
| V-Kde/V-Kde | 3 | 0 | 2 | 2 |
| V-Ja+intr-Kde/V-Ja+intr-Kde | 2 | 2 | 2 | 4 |
Summary of different TCRB rearrangement configurations that are compatible with a single clone.
| TCRB configuration | Cases (n) | Number of rearrangements detectable | ||
|---|---|---|---|---|
| Vβ-Jβ (multiplex tube A/B) | Dβ-Jβ (multiplex tube C) | Total | ||
| G/D-J1+D-J2 | 1 | 0 | 2 | 2 |
| V-J1 or 2/V-J1 or 2 | 4 | 2 | 0 | 2 |
| D-J1+D-J2/D-J1+D-J2 | 1 | 0 | 4 | 4 |
| V-J1+D-J2/V-J1+D-J2 | 1 | 2 | 2 | 4 |
Two primary difficulties in the interpretation of clonal results are pseudoclonality and oligoclonality, and a careful interpretation of Ig/TCR gene rearrangement is required.
Molecular specialists, pathologists and clinicians should be aware of several technical and immunologic pitfalls that should be considered when interpreting TCR clonality findings. If there is no clonal signal and no polyclonal Gaussian curve in TCR clonality assessment, it may be few T cells in sample, low DNA input and poor DNA quality. In such cases, the checking of the presence of T cells, DNA concentration and DNA quality verification process is needed. Few T cells in a sample may result in the selective amplification and pseudoclonality due to low level of specific template. In such cases, it is needed to repeat PCR in multiplicates and compare patterns for consistency. The exaggerated immune response with dominant specificity can show the oligoclonality or monoclonality in histologically reactive lesion. In such cases, it is needed to repeat PCR in multiplicates and reevaluate histopatholoy for large germinal centers containing dominant T cell clones [25].
Pseudoclonality may be erroneously interpreted as clonal population and generate false-positive results. Specifically, TCR/Ig assays in a reactive tissue, where a few B or T-cells are present, usually show weak clonal products coexisting in one tube that may be incorrectly considered clonal [26]. Duplicate analyses and mixing of PCR products should help to clarify whether the seemingly clonal PCR products are derived from different lymphocytes (polyclonal).
BIOMED-2 multiplex PCR could be a good diagnostic tool in that it offers the evidence of BM involvement of lymphoid neoplasia. If it is molecular positive but not detected microscopically, close follow-up and careful observation with IHC are needed. In addition, comparing BM Ig/TCR gene rearrangement results with the molecular results of the original lymphoma tissues could be helpful. We confirmed that the clone that was detected in the BM was identical to lymphoma clone (5 T cell lymphomas and 5 B cell lymphomas).
In conclusion, Ig/TCR gene rearrangement analysis using BIOMED-2 multiplex PCR is of great value, particularly, in diagnosing BM involvement of lymphoid neoplasia if it is morphologically uncertain. Clonal rearrangements appear to be suspicious of malignant disease, but may be present in the reactive lymphoproliferation, so comprehensive analysis with morphologic analysis should be important to reach a final diagnosis.
Acknowledgements
This study was supported by a grant of the Korea Healthcare technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea. (SN: A092258)
Competing Interests
The authors have declared that no competing interest exists.
References
1. Buhr T, Langer F, Schlue J, von Wsielewski R, Lehmann U, Braumann D, Kreipe H. Reliability of lymphoma classification in bone marrow trephines. Br J Haematol. 2002;118:470-6
2. Sah SP, Matutes E, Wotherspoon AC, Morilla R, Catovsky D. A comparision of flow cytometry, bone marrow biopsy, and bone marrow aspirates in the detection of lymphoid infiltration in B cell disorders. J Clin Pathol. 2003;56:129-32
3. Krober SM, Horney HP, Greschniok A, Kaiserling E. Reactive and neoplastic lymphocytes in human bone marrow: morphological, immunohistological and molecular biological investigations on biopsy specimens. J Clin Pathol. 1999;52:521-6
4. Langerak AW, Molina TJ, Lavender FL, Pearson D, Flohr T, Sambade C. Polymerase chain reaction-based clonality testing in tissue samples with reactive lymphoproliferations: usefulness and pitfalls. A report of the BIOMED-2 Concerted Action BMH4-CT98- 3936. Leukemia. 2007;21:222-9
5. Bourguin A, Tung R, Galili N, Sklar J. Rapid, nonradioactive detection of clonal T-cell receptor gene rearrangements in lymphoid neoplasms. Proc Natl Acad Sci U S A. 1990;87:8536-40
6. Sandberg Y, van Gastel-Mol EJ, Verhaaf B, Lam KH, van Dongen JJ, Langerak AW. BIOMED-2 multiplex immunoglobulin/T-cell receptor polymerase chain reaction protocols can reliably replace Southern blot analysis in routine clonality diagnostics. J Mol Diagn. 2005;7:495-503
7. Jaffe ES, Harris N, Stein H, Vardiman JW. WHO classification of Tumours of Haematopoietic and Lymphoid Tissues. World Health Organization Classification of Tumours. IARC Press. 2008
8. Chute DJ, Cousar JB, Mahadevan MS, Siegrist KA, Silverman LM, Stoler MH. Detection of immunoglobulin heavy chain gene rearrangements in classic hodgkin lymphoma using commercially available BIOMED-2 primers. Diagn Mol Pathol. 2008;17:65-72
9. Mitterbauer-Hohendanner G, Mannhalter C, Winkler K, Mitterbauer M, Skrabs C, Chott A, Simonitsch-Klupp I, Gleiss A, Lechner K, Jaeger U. Prognostic significance of molecular staging by PCR-amplification of immunoglobulin gene rearrangements in diffuse large B-cell lymphoma (DLBCL). Leukemia. 2004;18:1102-7
10. Bruce DC, Beate P, Malik EJ. et al. Revised Response Criteria for Malignant Lymphoma. J Clin Oncol. 2007;25:579-86
11. Lassmann S, Gerlach UV, Technau-Ihling K, Werner M, Fisch P. Application of BIOMED-2 primers in fixed and decalcified bone marrow biopsies. J Mol Diagn. 2005;7:582-91
12. Braunschweig R, Baur AS, Delacretaz F, Bricod C, Benhattar J. Contribution of IgH-PCR to the evaluation of B cell lymphoma involvement in paraffin-embedded bone marrow biopsy specimen. Am J Clin Pathol. 2003;119:634-42
13. Van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257-317
14. Evans PA. et al. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 2007;21:207-14
15. M Bru¨ggemann, H White, P Gaulard, R Garcia-Sanz. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia. 2007;21:215-21
16. Coad JE, Olson DJ, Christensen DR, Lander TA, Chibbar R, McGlennen RC, Brunning RD. Correlation of PCR-detected clonal gene rearrangements with bone marrow morphology in patients with B-lineage lymphomas. Am J Surg Pathol. 1997;21:1047-56
17. Calvert R, Evans PA, Randerson JA, Jack AS, Morgan GJ, Dixon MF. The significance of B-cell clonality in gastric lymphoid infiltrates. J Pathol. 1996;180:26-32
18. Hsi ED, Greenson JK, Singleton TP, Siddiqui J, Schnitzer B, Ross CW. Detection of immunoglobulin heavy gene rearrangement by polymerase chain reaction in chronic active gastritis associated with Helicobacter pylori. Hum Pathol. 1996;27:290-6
19. Dubus P, Vergier B, Beylot-Barry M. et al. Contribution of histopathologic and molecular analyses to the diagnosis of cutaneous B-cell infiltrates. Mod Pathol. 1996;9:1147-55
20. Gong JZ, Zheng S, Chiarle R, De Wolf-Peeters C, Palestro G, Frizzera G, Inghirami G. Detection of immunoglobulin kappa light chain rearrangements by polymerase chain reaction. An improved method for detecting clonal B-cell lymphoproliferative disorders. Am J Pathol. 1999;155:355-63
21. Claudia M, Anna G, Francesco B, Elena S, Carlo S, Fernando R, Maura R. et al. Use of IGK gene rearrangement analysis for clonality assessment of lymphoid malignancies: a single center experience. Am J Blood Res. 2011;1(2):167-74
22. Liu H. et al. A practical strategy for the routine use of BIOMED-2 PCR assays for detection of B- and T-cell clonality in diagnostic Haematopathology. Br J Haematol. 2007;138(1):31-43
23. Garcia-Castillo H, Barros-Nunez P. Detection of clonal immunoglobulin and T-cell receptor gene recombination in hematological malignancies: monitoring minimal residual disease. Cardiovascular & Hematological Disorders-Drug Targets. 2009;9:124-35
24. Langerak AW. et al. Multiple clonal Ig/TCR products: implications for interpretation of clonality findings. J Hematopathol. 2012;5:35-43
25. Patricia JTA. et al. Pitfalls in TCR gene clonality testing: teaching cases. J Hematopathol. 2008;1:97-109
26. Lee SC, Berg KD, Racke FK, Griffin CA, Eshleman JR. Pseudo-Spikes Are Common in Histologically Benign Lymphoid Tussues. J Mol Diagn. 2000;2(3):145-52
Author contact
![]() Corresponding author: Myungshin Kim, M.D., Ph D., Department of Laboratory Medicine, College of Medicine, The Catholic University of Korea, Seoul St. Mary's Hospital, Seoul 137-701, Korea (South). Telephone: +82-2-2258-1645; Fax: +82-2-2258-1719; E-mail: microkimac.kr.
Corresponding author: Myungshin Kim, M.D., Ph D., Department of Laboratory Medicine, College of Medicine, The Catholic University of Korea, Seoul St. Mary's Hospital, Seoul 137-701, Korea (South). Telephone: +82-2-2258-1645; Fax: +82-2-2258-1719; E-mail: microkimac.kr.

 Global reach, higher impact
Global reach, higher impact