Impact Factor
ISSN: 1449-1907
Int J Med Sci 2012; 9(2):148-156. doi:10.7150/ijms.3605 This issue Cite
Research Paper
Rapid Detection of rpoB Mutations in Rifampin Resistant M. tuberculosis from Sputum Samples by Denaturing Gradient Gel Electrophoresis
State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, the First Affiliated Hospital, School of Medicine, Zhejiang University. 79 Qingchun Rd., Hangzhou, China. 310003.
Received 2011-10-6; Accepted 2011-12-29; Published 2012-1-11
Abstract
Objective: To establish a rapid detection method for identifying rpoB mutations associated with rifampin (RIF) resistance in sputum specimens.
Methods: We detected rpoB mutations directly in 90 sputum specimens collected from suspected tuberculosis patients using PCR-based denaturing gradient gel electrophoresis (DGGE) and compared these results with those obtained by rpoB sequencing and conventional drug susceptibility testing.
Results: The positive detection rate of Mycobacterium tuberculosis (M. tuberculosis) was 52.2% by Acid-Fast Bacilli staining and 72.2% by conventional mycobacterial culture. In contrast, the positive rate was significantly higher (93.3%) by PCR-based detection of the rpoB gene in the same specimens. Furthermore, 75% of the tested specimens presented abnormal patterns compared with the wild-type pattern (standard H37Rv strain) analysed by DGGE. A total of 12 different patterns, representing 12 different rpoB mutations, were observed in the 63 abnormal patterns. The match rate of rpoB mutations detected by DGGE reached 96.9% when compared to DNA sequencing.
Conclusion: Our findings indicate that PCR-based DGGE is a rapid and reliable bio-technique for direct detection of rpoB mutations associated with RIF resistance in the sputum of suspected tuberculosis patients.
Keywords: Genotyping, Mycobacterium Tuberculosis, Resistance, Microbial Drug Resistance.
Introduction
The most serious threat related to tuberculosis control is the recent emergence of drug-resistant tuberculosis strains. This emergence has been induced by widespread use of the standard short-course drug regimen. Especially concerning is the emergence of multidrug-resistant tuberculosis (MDR-TB), defined as TB that is resistant to at least isoniazid and rifampin (RIF)(1-3). The re-emergence of TB worldwide and the rise in MDR-TB have increased demand for rapid and reliable drug susceptibility testing (DST) to perform drug-resistance surveillance. Improved testing would also aid in the development of an efficient regimen for appropriate treatment of MDR-TB, the frequency of which has gradually increased in many regions of the world(4, 5). Early detection of MDR-TB isolates is essential for efficient treatment and control of TB. Resistance to RIF is almost exclusively associated with mutations in the rpoB gene, which encodes the β-subunit of RNA polymerase in M. tuberculosis(6). More than 70 distinct rpoB mutations have been characterised from RIF-resistant M. tuberculosis isolates worldwide(7-11). Approximately 95% of RIF-resistant isolates harbour mutations in the rifampin resistance-determining region (RRDR), an 81-bp region of rpoB that spans codons 507 to 533. Mutations of the serine 531, histidine 526, and aspartate 516 codons have been identified in approximately 86% of RIF-resistant isolates. Thus, these sites represent “hot spots” within the RRDR(6, 12).
Conventional, 'gold standard' drug susceptibility testing of M. tuberculosis is based on microbial culture and is performed by observation of either growth or metabolic inhibition in a medium containing anti-TB drug(s). This type of assay may take up to 6 weeks to identify a positive culture. Furthermore, it has been reported that tuberculosis can be transmitted by 17% of smear-negative and culture-positive M. tuberculosis patients (5, 13).
Several new techniques for early detection have been established to shorten turnaround time and improve the convenience of case management, including micro-hole, micro-titre well methods, the non-radiometric BACTEC 960/Mycobacteria Growth Indicator Tube method, and others (14, 15). High costs, technical complexity and the absence of appropriately trained human resources make it difficult to perform any of these new techniques in the countries where they are needed most. In addition, these new methods still depend on bacterial culture and suffer from low predictability associated with clinical irrelevance of the results and unacceptably low reliability resulting from poor reproducibility (5).
Over the last two decades, many studies have utilised molecular biological methods to detect rpoB gene mutations related to RIF-resistant M. tuberculosis in various cohorts of patient samples (7, 16, 17). PCR-based denaturing gradient gel electrophoresis (DGGE) is a simple and rapid method that has been widely applied to detection of mutation(s) and polymorphisms of various genes (18-23). This system can also be used to analyse mutations in the rpoB gene within the RRDR of M. tuberculosis (6, 24). In this study, we detected rpoB mutations directly in the sputum of suspected tuberculosis patients using the PCR-DGGE technique and compared these results with those obtained by rpoB sequencing and conventional DST.
Materials and Methods
Sputum specimens and isolates
A total of 90 sputum specimens were collected from 90 suspected tuberculosis patients diagnosed by experienced pulmonologists with strongly positive reaction to tuberculin skin test and specific signs and syndromes consistent with tuberculosis in the First Affiliated Hospital of Zhejiang University from October 2004 to May 2005. All patients hadn't received any anti-TB treatment. The standard M. tuberculosis isolate H37Rv was purchased from the Shanghai Centre for Disease Control and Prevention, China. Informed consent was obtained from all patients to utilise their sputum specimens for this study. All specimens were tested by conventional M. tuberculosis culture. Before culture, the sputum specimens were digested and decontaminated by the N-acetyl-L-cysteine (NALC)-NaOH to prevent overgrowth of the culture by nonmycobacterial microorganisms. Two volumes of NALC-NaOH solution (4% NaOH, 1.45% Na-citrate, 0.5% NALC) were mixed with the each sputum specimen on a sterilized test tube for digestion. The mixture was cultured at room temperature for 15 minutes with a gentle shaking. Ten volumes of 6.7 mM phosphate buffer solution (PBS, pH 7.4) were added and the mixture centrifuged at 3,000 x g for 15 minutes at room temperature. The supernatant was discarded and the pellet washed twice with PBS. Then the pellet was resuspended with 0.5 ml of PBS. A 100 µl aliquot of the suspension was directly processed for M. tuberculosis culture, and the remainder used for Acid-Fast Bacilli (AFB) staining and PCR. M. tuberculosis culture were used by the conventional Lowensein-Jensen (LJ) medium at 37°C in 5% CO 2 for 1 week, at 37°C in air for another 7 weeks and thereafter were observed once a week for M. tuberculosis growth. The fresh growth was then subcultured onto LJ medium for DST. DST for resistance to the first-line drugs RIF (40 µg/ml), INH (0.2 µg/ml), ethambutol (EMB, 2 µg/ ml) and streptomycin (STR, 4 µg/ ml) using the agar proportion method as previously described (25). In brief, growth from the primary isolation medium was subcultured onto LJ medium. Bacterial inoculation was performed by picking fresh colonies from the LJ medium and adjusting cultures to a turbidity matching that of a McFarland 1 standard. A 100 µl aliquot of 100- and 10,000-fold dilution samples were plated onto drug-containing and no-drug sectors of Middlebrook 7H10 quadrant plates. All inoculations were incubated at 37ºC in 5% CO2 for 8 weeks. Resistance to the drugs was defined as >1% growth in drug-containing medium compared to growth in the drug-free control medium (American National Committee for Clinical Laboratory Standards, NCCLS, 1995). Isolates resistant to at least RIF and INH were classified as MDR-TB.
Acid-fast bacilli staining and DNA extraction
All 90 sputum specimens were smeared for AFB staining. A 100 µl aliquot of the decontaminated sputum specimen was spread onto a slide over an area of 1-2 cm, air-dried, and heat fixed. Smeared specimens were inactivated using 5% phenol in ethanol for 5 min before they were moved from the biosafety cabinet to the staining sink. Smear staining was performed as previously described (26). Extraction of M. tuberculosis DNA from all 90 decontaminated sputum specimens was performed using a QIAamp DNA mini kit (Qiagen, CA, USA) as previously described (27). Briefly, A 100 µl aliquot of the decontaminated sputum specimen was mixed with an equal volume of deionised water and centrifuged for 10 min at 14,000 x g. The pellet was resuspended in ATL buffer (Qiagen, CA, USA) containing 1 mg/ml proteinase K and incubated at 56 °C for 60 min. Subsequently, two cycles of freeze-thawing were performed to lyse the mycobacterial cells. DNA was then purified and collected for PCR detection. The standard M. tuberculosis H37Rv isolates and normal human-derived sputum were cultured as positive and negative controls and DNA was extracted from these samples using the same protocol. DNA extraction and PCR amplification were carried out in a specific PCR diagnosis room to prevent cross-contamination of nucleic acids.
PCR amplification
The 126-bp partial sequence of the rpoB gene (GenBank accession no. L27989), including the RRDR (an 81-bp region within the rpoB gene that encodes residues 507 to 533), was amplified by PCR from all sputum specimens with forward (5'-CGCCGCGATCAAGGAGTTCT-3') and reverse (5'- TCACGTGACAGACCGCCGGG-3') primers purchased from Shanghai Sangon Company (Sangon, Shanghai, China). These primers specifically amplified M. tuberculosis sequences and did not cross-react with non-tuberculosis mycobacterial sequences. PCR reactions were performed according to the manufacturer's technical instructions (Invitrogen, NY, USA) and our previously described methods (7). Briefly, PCR reactions were carried out in 50 μl total volume, containing 1×PCR reaction buffer, 200 µM dNTPs, 1U of Taq polymerase, 20 pmoles of each primer, and 2 µl of the DNA sample. The reaction mixtures were then subjected to one cycle of 94°C for 2 min, followed by 35 cycles of 94°C for 45 s, 55°C for 45 s, 72°C for 30 s, and a final cycle of 72°C for 7 min to complete elongation of the intermediate PCR products. PCR products (5 µl) were analysed by electrophoresis through 1.5% agarose gels and ethidium bromide staining. Each sample was tested in triplicate and the standard M. tuberculosis H37Rv isolate was used as a positive control. The presence of a 126-bp band on the agarose gel indicated successful amplification.
PCR-DGGE
PCR-based DGGE analysis was carried out to rapidly detect rpoB gene mutations from the 90 specimens, followed by subsequent confirmation by conventional DST and DNA sequencing. The method employed was modified from Scarpellini, P. et al. (24) and McCammon, M. T. et al. (6). Briefly, 15 µl of the PCR products were loaded onto a 15% acrylamide gel in 1× TAE buffer. The denaturing gradient consisted of 50% to 80% denaturant (100% denaturant is 40% formamide and 7 M urea in 1× TAE buffer) at a constant temperature of 60°C at 120 V for 4 hours. Gels were stained with SYBR GREEN I (Dingguo Biotechnology, Beijing, China) for 15 min and photographed on a UV transilluminator. The standard M. tuberculosis H37Rv isolate was used as a wild-type pattern control. The DGGE gel images were analysed using the BioNumerics software version 5.1. Position tolerance for band matching was set at 1% to help correct for slight migratory variation. Migratory distance for each band was determined by the BioNumerics software. Similarity matrix and dendrogram of the DGGE profiles were generated on the base of Pearson correlation coefficient.
DNA sequencing
PCR products were purified using a QIAquick gel extraction kit (Qiagen, CA, USA) followed by electrophoresis on 1.5% agarose gels. Five hundred nanograms of the purified PCR products were sequenced by the Shanghai Sangon Company (Sangon, Shanghai, China) on an ABI 373 automated sequencer (Applied Biosystems, CA, USA). The sequence data were analysed using the Sequencer program, version 3.0. DNAssist version 1.02 was used to compare the sequences with the H37Rv genome database (GenBank accession no. L27989).
Statistical analysis
Data were evaluated using the chi-squared test for five different detection methods (Table 2) using the SPSS software, version 16.0. The level of significance for all statistical analyses was P < 0.05.
Results
AFB staining and drug susceptibility testing
Results of the AFB staining and DST for the M. tuberculosis samples tested are shown in Table 1. Overall, 52.2% (47/90) of the specimens from potential tuberculosis patients were classified as positive for M. tuberculosis by AFB staining. And the conventional term culture showed 72.2% (65/90) of sputum samples were positive for M. tuberculosis. The Subsequent results of DST showed that 11 strains were single RIF-resistant strains (8 from AFB-positive and 3 from AFB-negative specimens) and 35 were MDR strains (29 from AFB-positive and 6 from AFB-negative specimens). Moreover, we identified 9 strains (6 from AFB-positive and 3 from AFB-negative specimens) that were susceptible to RIF but resistant to any of the other three anti-TB drugs (INH, EMB and STR) and 10 strains that were susceptible to all four anti-TB drugs (4 from AFB-positive and 6 from AFB-negative specimens).
Detection of rpoB mutations from sputum samples by five different techniques.
| Analysis Method | AFB-Positive n1=47 | AFB-Negative n2=43 | Total Percentage (%) | |
|---|---|---|---|---|
| AFB | 47 | 43 | 47/90 (52.2) | |
| CMC | 47 | 18 | 65/90 (72.2) | |
| DST | R-RIF | 8 | 3 | 11 |
| MDR | 29 | 6 | 35 | |
| R-I/E/S | 6 | 3 | 9 | |
| S-R/I/E/S | 4 | 6 | 10 | |
| rpoB-PCR | 47 | 37 | 84/90 (93.3) * | |
| rpoB-DGGE | 37 | 26 | 63/84 (75.0) | |
| rpoB-DNA Sequencing | 38 | 27 | 65/84 (77.4) | |
AFB: AFB smear, CMC: conventional mycobacterial culture, R-RIF: mono-resistance to rifampin, MDR: multidrug-resistant, R-I/E/S: resistance to isoniazid, ethambutol or streptomycin, S-R/I/E/S: susceptible to rifampin, isoniazid, ethambutol and streptomycin.
*: p < 0.05 compared to the AFB smear (52.2%) and CMC (72.2%).
Genetic alterations of the rpoB gene in 90 sputum specimens collected from suspected tuberculosis patients.
| Mutation codon | DAN sequencing | DST | Frequency (%) of mutation n=84* | PCR-DGGE positive (No.) n=84* | |||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||||
| Mutation | Amino acid change | R-RIF | MDR | R-I/E/S | S-R/I/E/S | ||||
| 533 | CTG→TTG | Leu→Leu | 2 | 2(2.4) | 2 | ||||
| 531 | TCG→TTG | Ser→Leu | 2 | 2 | 1Δ | 9(10.7) | 8 | ||
| TCG→TGG | Ser→Trp | 2 | 2 | ||||||
| 526 | CAC→GAC | His→Asp | 5 | 22 | 5 | 43(51.2) | 42 | ||
| CAC→TAC | His→Tyr | 2 | 7 | 1Δ | |||||
| CAC→CTT | His→Leu | 1 | |||||||
| 521 | CTG→TTG | Leu→Leu | 1 | 1 (1.2) | 1 | ||||
| 519 | AAC→GAC | Asn→Asp | 1 | 1 (1.2) | 1 | ||||
| 518 | AAC→GAC | Asn→Asp | 1 | 1 (1.2) | 1 | ||||
| 516 | GAC→GGA | Asp→Gly | 1 | 2 | 5 (6.0) | 5 | |||
| GAC→TAC | Asp→Tyr | 2 | |||||||
| 513 | CAA→GAA | Gln→Glu | 1 | 2 | 3(3.6) | 3 | |||
| Total mutation (%) | 50 | 15 | 65 (77.4) | 63(75.0) | |||||
| No Mutation | 5 | 10 | 4 | ||||||
R-RIF: mono-resistance to rifampin; MDR: multidrug-resistant; R-I/E/S: resistance to isoniazid, ethambutol or streptomycin, S-R/I/E/S: susceptible to rifampin, isoniazid, ethambutol and streptomycin
*: Total of 84 rpoB-PCR positive specimens.
Δ: The specimens harboured rpoB mutations but present wild-type-like DGGE patterns.
PCR and DNA sequencing
The PCR results (Figure 1) showed that 93.3% (84/90) of sputum specimens were positive for the rpoB gene; these samples included 47 AFB-positive and 37 AFB-negative specimens. The results of subsequent DNA sequencing (Tables 1 and 2) of 84 rpoB-positive PCR products, including samples from 38 AFB-positive and 27 AFB-negative specimens, showed that 65 specimens displayed mutations in the rpoB gene. Our analysis detected a total of 12 mutated positions distributed among 8 codons within the RRDR of the rpoB gene. In total, 61 specimens displayed substitutions and 4 specimens displayed double substitutions (3 at codon 516/GAC → GGA, 1 at 526/CAC → CTT). The most frequent position for a mutation was codon 526 (51.2%, 43/84), followed by codon 531 (10.7%, 9/84) and codon 516 (6.0%, 5/84). Eight resistance-associated mutations in the rpoB gene occurred at codons 513 (3 specimens), 518 (1 specimen), 519 (1 specimen), 521 (1 specimen), and 533 (2 specimens). Mutations at codon 526 of CAC → GAC (His → Asp) occurred in 32 specimens and the CAC → TAC (His → Tyr) substitution occurred in 10 specimens, while the CAC → CTT (His → Leu) mutation occurred in only one specimen. Mutations at codon 531 of TCG → TTG (Ser → Leu) occurred in 5 specimens, and a change of TCG → TGG (Ser → Trp) occurred in 4 specimens. The GAC → GGA (Asp → Gly) mutation at codon 516 occurred in 3 specimens and the GAC → TAC (Asp → Tyr) substitution was observed in 2 specimens. Comparison of these data with the results of conventional DST (showed in Table 2) indicated that 11 of the RIF-resistant specimens and 35 of the MDR-TB specimens harboured mutations at codons 513, 526 or 531. Four specimens susceptible to RIF but resistant to any one of the other three anti-TB drugs (isoniazid, ethambutol and streptomycin) harboured mutations at codons 516, 518, 519 and 521. Fifteen specimens that displayed mutations at codons 516 (4 specimens), 526 (6 specimens), 531 (3 specimens), and 533 (2 specimens) were classified as negative by DST.
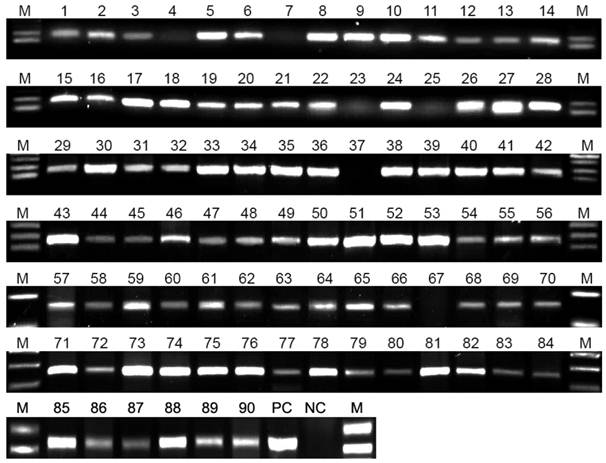
The results of PCR detection. M: DNA marker, 1-90: numbered sputum specimens, PC: positive control (standard H37Rv strain), NC: negative control (normal human sputum). From a total of 90 sputum specimens tested, 84 were positive and 6 (No. 4, 7, 23, 25, 37 and 67) were negative for detection of the rpoB gene. The brightness of the PCR bands was determined by the concentration of M. tuberculosis.
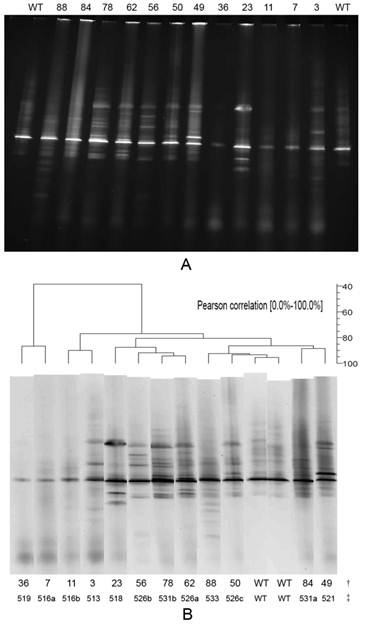
The results of the DGGE analysis. A is the DGGE profile of rpoB RRDR mutations for 12 different mutations. The WT lane is the control wild-type strain H37Rv. The following lanes are various sputum specimens. B is the result of DGGE analysis using BioNumerics software version 5.1. †: the number of specimens (including 2 lanes of wild-type strain H37Rv; each specimen lane is the same as in image A), ‡: rpoB mutation codon. Lane 36, with a mutation at codon 519, represents 1 specimen; lanes 7 and 11, with mutations at codon 516 (a, GAC→GGA, b, GAC→TAC), represent 2 specimens; lanes 3 and 23, with mutations at codons 513 and 518, represent 3 and 1 specimens, respectively; lane 56, with a mutation at codon 526 (b, CAC→TAC), represents 11 specimens; lane 78, with a mutation at codon 531 (b, TCG→TGG), represents 2 specimens; lane 62, with a mutation at codon 526 (a, CAC→GAC), represents 33 specimens; lane 88, with a mutation at codon 533, represents 1 specimen; lane 50, with a mutation at codon 526 (c, CAC→CTT), represents 1 specimen; lane WT, wild-type strain H37Rv, no mutation; lane 84, with a mutation at codon 531 (a, TCG→TTG), represents 5 specimens; lane 49, with a mutation at codon 521, represents 1 specimen.
PCR-DGGE analysis
A total of 84 amplified rpoB-PCR products, generated from 90 sputum specimens, 126 bp in length were analysed by DGGE. The DGGE profile results (Table 2) showed that 25% (21/84) of the specimens displayed the same (wild-type) patterns and 75% (63/84) were abnormal compared to the control wild-type pattern obtained from the fully susceptible standard M. tuberculosis H37Rv isolate. A total of 50 specimens with abnormal DGGE patterns were also DST-positive. These included 11 specimens resistant to RIF, 35 specimens that were MDR and 4 specimens susceptible to RIF but resistant to any one of the other three first-line anti-TB drugs. When we combined these results with the DNA sequencing data, we determined that 42 of the 63 specimens with abnormal patterns harboured mutations at codon 526, 8 displayed mutations at codon 531, 5 samples contained mutations at codon 516, 3 displayed mutations at codon 513 and 2 specimens contained mutations at codon 533; the remaining 3 specimens contained mutations at codons 521, 519 and 518. The results of a BioNumerics analysis (Figure 2B) showed that 12 different DGGE patterns (typical specimens shown in Figure 2A) were observed in the 63 abnormal patterns. Each mutation generated a specific DGGE pattern that was dependent on the nucleotide alterations and/or the position of the mutations within the RRDR of the rpoB gene. The BioNumerics analysis also showed that the Pearson Correlation between 2 lanes of wild-type strain H37Rv (WT) reached 96.4%. The Pearson Correlation was 93.6% between the WT lane and lane 50 (mutation at codon 526); 91.3% between the WT lane and lane 88 (mutation at codon 533); and less than 85% between the WT lane and the remaining lanes. The DGGE method identified 96.9% of the same rpoB mutations as the DNA sequencing method (63 specimens with abnormal DGGE patterns and 65 specimens containing mutations).
Discussion
Resistance to RIF is almost exclusively associated with mutation of the rpoB gene, which encodes the β-subunit of RNA polymerase (28-30). More than 95% of rpoB mutations in RIF-resistant clinical isolates have been found within the RRDR. Over 70 distinct rpoB mutations and four frequent mutations (codons 526, 513, 531 and 516) have been reported for RIF-resistant M. tuberculosis isolates worldwide (31, 32). However, it can take 6 weeks or longer to detect RIF resistance in M. tuberculosis by conventional culture-based DST. Recently, more rapid and accurate biomolecular techniques have received increased attention as alternatives to DST (33, 34) and have been reported to detect rpoB gene mutations linked to RIF-resistant M. tuberculosis (14, 15). Kim et al. (16) used a method of polymerase chain reaction restriction analysis to analyse the rpoB gene for direct characterisation of non-tuberculous mycobacteria in AFB smear-positive respiratory specimens. The results of O'Riordan et al. (35) revealed that rpoB mutation molecular resistance testing has the potential to rapidly identify MDR-TB patients and would enable initiation of appropriate therapy significantly earlier than would be possible using conventional testing methods. Our previous study revealed that PCR-based single strand conformation polymorphism analysis (PCR-SSCP) is a rapid and useful molecular resistance screening approach for detection of MDR-TB (7). DGGE can distinguish mutant amplicons from their wild-type equivalents based on the altered melting temperatures of the mutants as the DNA fragments migrate through a gradient of denaturants (6, 24, 36, 37). McCammon et al. (6) used DGGE combined with DNA sequencing to detect rpoB and pncA mutations in RIF- and pyrazinamide-resistant M. tuberculosis isolates after conventional culture and DST. Their results demonstrated the power and usefulness of DGGE in detecting mutations associated with drug resistance in M. tuberculosis; DGGE is even more sensitive than DNA sequencing in detecting mutations in complex DNA samples. In addition, DGGE can detect point mutations, small insertions, and deletions. Although Scarpellini et al. (24) and Hori et al. (22) ever used DGGE to detect rpoB mutations in DST positive specimens, as far as we are aware, investigators have not yet reported on using DGGE to detect rpoB mutations in the sputum of suspected tuberculosis patients.
In this study, PCR-based DGGE was performed to probe for rpoB mutations associated with RIF resistance in 90 sputum specimens collected from suspected tuberculosis patients in Eastern China. The results of our study indicate that the positive rate of detection of M. tuberculosis is only 52.2% for detection by AFB staining and 72.2% by conventional mycobacterial culture. In contrast, the positive rate of M. tuberculosis detection reached 93.3% for PCR detection of the rpoB gene in the same specimens, which included 47 AFB-positive and 37 AFB-negative sputum samples. The positive rate of M. tuberculosis detection by PCR was significantly higher than the rates obtained with AFB staining or DST (p < 0.05). The PCR-based DGGE results demonstrated that 75% (63/84) of specimens presented a total of 12 DGGE different patterns that differed from the wild-type pattern. The rate of agreement in detection of rpoB mutations between DNA sequencing and DGGE reached 96.9%. Comprehensive analysis also revealed that 50 of the specimens with abnormal DGGE patterns were validated as DST-positive, including 46 RIF-resistant samples (or MDR) and 4 specimens susceptible to RIF but resistant to any one of the other three first-line anti-TB drugs. The other 13 specimens have not been tested by DST but have been analysed by PCR-based DGGE (15 positive by PCR-based DNA sequencing). Of these 13 specimens, 7 harboured mutations at codon 526, 4 contained mutations at codon 516 and 2 displayed alterations at codon 533. Many reports have demonstrated that M. tuberculosis isolates containing rpoB mutations at codons 526 and 516 are always resistant to RIF (31, 32, 38). According to this assumption, at least 11 of the 13 specimens with abnormal DGGE patterns were possibly RIF-resistant. This assumption may also help to explain why 3 specimens that harboured rpoB mutations at codons 521, 519, and 518 presented abnormal DGGE patterns but were susceptible to RIF. However, the mechanism of this phenomenon is unclear. Interestingly, we observed that one strain with a mutation at codon 516 was susceptible to RIF. Together, our results indicate that PCR-based DGGE is more sensitive than AFB and faster than DST (a couple of days vs. 6-8 weeks) in detecting M. tuberculosis isolates. Therefore, the technique of DGGE analysis can reasonably be considered an alternative to other indirect techniques of rpoB mutation in RRDR. Its comparative reproducibility, together with the fact that it is fast and easy to complete, means that it can be used in clinical detection.
In conclusion, as a rapid and valid bio-technique, DGGE has a number of practical advantages compared to other methods. It is less laborious than the manual single-strand conformation polymorphism (SSCP) method used in our previous study (7) because it does not require the casting of ultrathin gels or the use of radioactive isotopes. Furthermore, compared with the conventional culture system, DGGE is performed with a relatively inexpensive machine and lower cost of the molecular commercial kits (BACTEC 960 system, US$ 38,000 plus reagents and drugs cost per test US$ 12; DGGE system, US$ 3,500 plus reagents cost per test US$ 10) used for the rpoB mutation analysis that allows for controlled experimental conditions and optimum reproducibility. Although DGGE is not a novel molecular technique, our results and other studies have demonstrated that it is a rapid and valid bio-technique for early and fast detection of rpoB gene mutations associated with rifampin resistance in the sputum of suspected tuberculosis patients. These findings will be aid in generating a novel and rapid drug resistance screening approach for treatment of MDR-TB.
Acknowledgements
We want to thank Dr. Xutao Deng (Bioinformatics Core Facility, Beckman Research Institute, City of Hope Medical Center) who provided us with many important suggestions and comments during the preparation of this manuscript.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Marin M, Garcia de Viedma D, Ruiz-Serrano MJ, Bouza E. Rapid direct detection of multiple rifampin and isoniazid resistance mutations in Mycobacterium tuberculosis in respiratory samples by real-time PCR. Antimicrob Agents Chemother. 2004;48:4293-4300
2. Viveiros M, Amaral L. Enhancement of antibiotic activity against poly-drug resistant Mycobacterium tuberculosis by phenothiazines. Int J Antimicrob Agents. 2001;17:225-228
3. Zhang SL, Shen JG, Xu PH, Li DX, Sun ZQ, Li L, Yang ZR. et al. A novel genotypic test for rapid detection of multidrug-resistant Mycobacterium tuberculosis isolates by a multiplex probe array. J Appl Microbiol. 2007;103:1262-1271
4. Pfyffer GE, Bonato DA, Ebrahimzadeh A, Gross W, Hotaling J, Kornblum J, Laszlo A. et al. Multicenter laboratory validation of susceptibility testing of Mycobacterium tuberculosis against classical second-line and newer antimicrobial drugs by using the radiometric BACTEC 460 technique and the proportion method with solid media. J Clin Microbiol. 1999;37:3179-3186
5. Kim SJ. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur Respir J. 2005;25:564-569
6. McCammon MT, Gillette JS, Thomas DP, Ramaswamy SV, Graviss EA, Kreiswirth BN, Vijg J. et al. Detection of rpoB mutations associated with rifampin resistance in Mycobacterium tuberculosis using denaturing gradient gel electrophoresis. Antimicrob Agents Chemother. 2005;49:2200-2209
7. Sheng J, Li J, Sheng G, Yu H, Huang H, Cao H, Lu Y. et al. Characterization of rpoB mutations associated with rifampin resistance in Mycobacterium tuberculosis from eastern China. J Appl Microbiol. 2008;105:904-911
8. Ahmad S, Mokaddas E, Fares E. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Kuwait and Dubai. Diagn Microbiol Infect Dis. 2002;44:245-252
9. Spindola de Miranda S, Kritski A, Filliol I, Mabilat C, Panteix G, Drouet E. Mutations in the rpoB gene of rifampicin-resistant Mycobacterium tuberculosis strains isolated in Brazil and France. Mem Inst Oswaldo Cruz. 2001;96:247-250
10. Khan K, Muennig P, Behta M, Zivin JG. Global drug-resistance patterns and the management of latent tuberculosis infection in immigrants to the United States. N Engl J Med. 2002;347:1850-1859
11. Lee AS, Lim IH, Tang LL, Wong SY. High frequency of mutations in the rpoB gene in rifampin-resistant clinical isolates of Mycobacterium tuberculosis from Singapore. J Clin Microbiol. 2005;43:2026-2027
12. Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3-29
13. Patnaik M, Liegmann K, Peter JB. Rapid detection of smear-negative Mycobacterium tuberculosis by PCR and sequencing for rifampin resistance with DNA extracted directly from slides. J Clin Microbiol. 2001;39:51-52
14. Johansen IS, Thomsen VO, Marjamaki M, Sosnovskaja A, Lundgren B. Rapid, automated, nonradiometric susceptibility testing of Mycobacterium tuberculosis complex to four first-line antituberculous drugs used in standard short-course chemotherapy. Diagn Microbiol Infect Dis. 2004;50:103-107
15. Rohner P, Ninet B, Benri AM, Auckenthaler R. Evaluation of the Bactec 960 automated nonradiometric system for isolation of mycobacteria from clinical specimens. Eur J Clin Microbiol Infect Dis. 2000;19:715-717
16. Kim S, Park EM, Kwon OJ, Lee JH, Ki CS, Lee NY, Koh WJ. Direct application of the PCR restriction analysis method for identifying NTM species in AFB smear-positive respiratory specimens. Int J Tuberc Lung Dis. 2008;12:1344-1346
17. Huang HJ, Xiang DR, Sheng JF, Li J, Pan XP, Yu HY, Sheng GP. et al. rpoB nested PCR and sequencing for the early diagnosis of tuberculous meningitis and rifampicin resistance. Int J Tuberc Lung Dis. 2009;13:749-754
18. De Angelis M, Di Cagno R, Gallo G, Curci M, Siragusa S, Crecchio C, Parente E. et al. Molecular and functional characterization of Lactobacillus sanfranciscensis strains isolated from sourdoughs. Int J Food Microbiol. 2007;114:69-82
19. Spano G, Lonvaud-Funel A, Claisse O, Massa S. In Vivo PCR-DGGE analysis of Lactobacillus plantarum and Oenococcus oeni populations in red wine. Curr Microbiol. 2007;54:9-13
20. da Mota FF, Gomes EA, Paiva E, Seldin L. Assessment of the diversity of Paenibacillus species in environmental samples by a novel rpoB-based PCR-DGGE method. FEMS Microbiol Ecol. 2005;53:317-328
21. Taylor MW, Schupp PJ, Dahllof I, Kjelleberg S, Steinberg PD. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ Microbiol. 2004;6:121-130
22. Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y. Direct comparison of single-strand conformation polymorphism (SSCP) and denaturing gradient gel electrophoresis (DGGE) to characterize a microbial community on the basis of 16S rRNA gene fragments. J Microbiol Methods. 2006;66:165-169
23. Strauss J, White A, Ambrose C, McDonald J, Allen-Vercoe E. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe. 2008;14:301-309
24. Scarpellini P, Braglia S, Carrera P, Cedri M, Cichero P, Colombo A, Crucianelli R. et al. Detection of rifampin resistance in Mycobacterium tuberculosis by double gradient-denaturing gradient gel electrophoresis. Antimicrob Agents Chemother. 1999;43:2550-2554
25. Yam WC, Tam CM, Leung CC, Tong HL, Chan KH, Leung ET, Wong KC. et al. Direct detection of rifampin-resistant mycobacterium tuberculosis in respiratory specimens by PCR-DNA sequencing. J Clin Microbiol. 2004;42:4438-4443
26. Di Perri G, Bonora S. Which agents should we use for the treatment of multidrug-resistant Mycobacterium tuberculosis? J Antimicrob Chemother. 2004;54:593-602
27. Wu TL, Chia JH, Kuo AJ, Su LH, Wu TS, Lai HC. Rapid identification of mycobacteria from smear-positive sputum samples by nested PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 2008;46:3591-3594
28. Chaves F, Alonso-Sanz M, Rebollo MJ, Tercero JC, Jimenez MS, Noriega AR. rpoB mutations as an epidemiologic marker in rifampin-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2000;4:765-770
29. Heep M, Brandstatter B, Rieger U, Lehn N, Richter E, Rusch-Gerdes S, Niemann S. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J Clin Microbiol. 2001;39:107-110
30. Klein JL, Brown TJ, French GL. Rifampin resistance in Mycobacterium kansasii is associated with rpoB mutations. Antimicrob Agents Chemother. 2001;45:3056-3058
31. Torres MJ, Criado A, Gonzalez N, Palomares JC, Aznar J. Rifampin and isoniazid resistance associated mutations in Mycobacterium tuberculosis clinical isolates in Seville, Spain. Int J Tuberc Lung Dis. 2002;6:160-163
32. Cavusoglu C, Hilmioglu S, Guneri S, Bilgic A. Characterization of rpoB mutations in rifampin-resistant clinical isolates of Mycobacterium tuberculosis from Turkey by DNA sequencing and line probe assay. J Clin Microbiol. 2002;40:4435-4438
33. Agdamag DM, Kageyama S, Solante R, Espantaleon AS, Sangco JC, Suzuki Y. Characterization of clinical isolates of Mycobacterium tuberculosis resistant to drugs and detection of RpoB mutation in multidrug-resistant tuberculosis in the Philippines. Int J Tuberc Lung Dis. 2003;7:1104-1108
34. Prammananan T, Cheunoy W, Taechamahapun D, Yorsangsukkamol J, Phunpruch S, Phdarat P, Leechawengwong M. et al. Distribution of rpoB mutations among multidrug-resistant Mycobacterium tuberculosis (MDRTB) strains from Thailand and development of a rapid method for mutation detection. Clin Microbiol Infect. 2008;14:446-453
35. O'Riordan P, Schwab U, Logan S, Cooke G, Wilkinson RJ, Davidson RN, Bassett P. et al. Rapid molecular detection of rifampicin resistance facilitates early diagnosis and treatment of multi-drug resistant tuberculosis: case control study. PLoS One. 2008;3:e3173
36. McBain AJ, Bartolo RG, Catrenich CE, Charbonneau D, Ledder RG, Gilbert P. Effects of triclosan-containing rinse on the dynamics and antimicrobial susceptibility of in vitro plaque ecosystems. Antimicrob Agents Chemother. 2003;47:3531-3538
37. McCammon MT, Gillette JS, Thomas DP, Ramaswamy SV, Rosas II, Graviss EA, Vijg J. et al. Detection by denaturing gradient gel electrophoresis of pncA mutations associated with pyrazinamide resistance in Mycobacterium tuberculosis isolates from the United States-Mexico border region. Antimicrob Agents Chemother. 2005;49:2210-2217
38. Tang X, Morris SL, Langone JJ, Bockstahler LE. Microarray and allele specific PCR detection of point mutations in Mycobacterium tuberculosis genes associated with drug resistance. J Microbiol Methods. 2005;63:318-330
Author contact
![]() Corresponding author: Lanjuan Li. Tel: 86-571-87236425; Fax: 86-571-87236456; E-mail: ljliedu.cn
Corresponding author: Lanjuan Li. Tel: 86-571-87236425; Fax: 86-571-87236456; E-mail: ljliedu.cn

 Global reach, higher impact
Global reach, higher impact