Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(7):1612-1629. doi:10.7150/ijms.107773 This issue Cite
Review
Circular RNA in Pancreatic Cancer: Biogenesis, Mechanism, Function and Clinical Application
1. Institute of Hepatopancreatobiliary Surgery, Chongqing General Hospital, Chongqing University, Chongqing, 401147, China.
2. Department of Anesthesiology, Chongqing Seventh People's Hospital, Chongqing University of Technology, Chongqing, 400054, China.
3. Chongqing Medical University, Chongqing, 400016, China.
Hang Chen and Xianxing Wang contributed equally to this work.
Received 2024-11-28; Accepted 2025-2-14; Published 2025-2-28
Abstract
Circular RNAs (circRNAs) are a class of novel RNA molecules featured by single-strand covalently closed circular structure, which not only are extensively found in eukaryotes and are highly conserved, but also conduct paramount roles in the occurrence and progression of pancreatic cancer (PC) through diverse mechanisms. As recent studies have demonstrated, circRNAs typically exhibit tissue-specific and cell specific expression patterns, with strong potential as biomarkers for disease diagnosis and prognosis. On the basis of their localization and specific interactions with DNA, RNA, and proteins, circRNAs are considered to possess specific biological functions by acting as microRNA (miRNA) sponges, RNA binding protein (RBP) sponges, transcriptional regulators, molecular scaffolds and translation templates. On that account, further addressing the technical difficulties in the detection and research of circRNAs and filling gaps in their biological knowledge will definitely push ahead this comparatively young research field and bring circRNAs to the forefront of clinical practice. Thus, this review systematically summarizes the biogenesis, function, molecular mechanisms, biomarkers and therapeutic targets of circRNAs in PC.
Keywords: circRNA, miRNA, pancreatic cancer, biomarker, therapeutic target
1. Introduction
Pancreatic cancer (PC) is a highly lethal malignancy from digestive system. The 5-year survival rate for PC is only 12%, much lower than for other malignancies [1]. In 2024, 66,440 new PC cases and 51,750 deaths are projected to occur in the United States [2]. A cross-sectional study reported that PC was predicted to become the second leading cause of cancer mortality in the United States to the year 2040 [3]. As the detection technology moves ahead speedily, declining trends in various degrees has been observed with regard to the innovation of surgical concepts and techniques, the application of new anti-tumor drugs, and the mortality of most tumors. Nonetheless, the mortality of PC has not been ameliorated and exhibits a slowly upward tendency [2-4]. The leading causes behind it may consist in that the non-specific symptoms of PC, the difficulty of early diagnosis, the speedy progression of the disease and the deficiency of treatments [5]. For these reasons, it's particularly essential to comprehensively reveal the molecular mechanism of PC occurrence and development, identify specific biomarkers that can be used for early diagnosis and prognosis, and discover new therapeutic targets, which are tremendously crucial to better the diagnosis and prognosis of PC patients.
Circular RNAs (circRNAs) are a class of novel single-stranded RNA molecules characterized by covalent closed rings formed by precursor mRNAs (pre-mRNAs) back-splicing or skipping events of thousands of genes and widely existing in eukaryotes. They are featured by stable structure, high conservation, abundant expression and high endogenous level, and have specific expression patterns in dissimilar tissues at various developmental stages [6]. In 1976, Sanger et al. discovered the first circRNA molecule in viroid through electron microscopy [7]. As already indicated by early studies, only a few circRNAs (such as circSRY, circDCC, and circEST1) have biological functions, while most circRNAs are merely "by-products" or "garbage" produced by abnormal splicing [8]. In recent years, as high-throughput RNA sequencing, circRNA-specific microarrays and bioinformatics advance speedily, a multitude of circRNAs have been discovered and identified. Aside from that, their cell/tissue and time-specific expression patterns and functions have been revealed in an all-round manner [9]. On the basis of genomic origin and biogenetic pattern, circRNAs are currently categorized into circular intron RNAs (ciRNAs), exon-intron circRNAs (EIcircRNAs), exon circRNAs (EcircRNAs) and tRNA intronic circular RNAs (tricRNAs) [10]. As suggested by multiplying evidence, circRNAs conduct a key regulatory role in tissue development, aging and disease occurrence (such as tumors, neurological diseases, diabetes, cardiovascular diseases and chronic inflammatory diseases) [11]. In particular, the role of circRNAs in cancer occurrence and progression has captured massive attention. More importantly, they possess the potential to serve as cancer biomarkers and new therapeutic targets [12-15]. As already illustrated by further studies, circRNAs conduct paramount roles in cancer initiation and development through multiple molecular mechanisms, such as regulating gene transcription [9], acting as miRNA sponges [16-18], interacting with RNA binding proteins (RBPs) [19, 20], and translating into proteins/peptides [21]. Consequently, further understanding of the biogenesis, functions and mechanisms of circRNAs in PC may provide brand new references and ideas with respect to the exploration of potential molecular diagnostic markers and therapeutic targets in PC.
2. Biogenesis
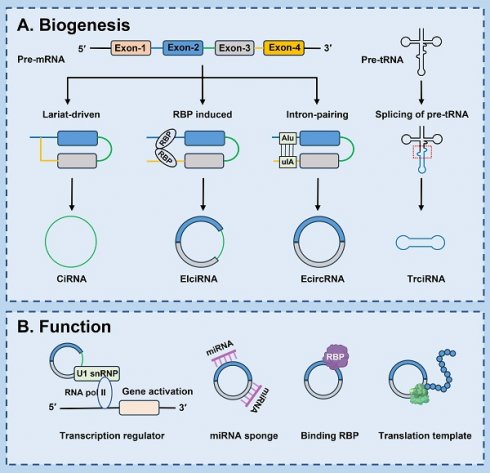
The biogenesis of circRNAs apparently differs from the production mechanism of mRNA. Back-splicing of pre-mRNA is the predominant process for circRNA generation. A splice donor that is downstream of the 5' splice site is joined to a splice acceptor that is upstream of the 3' splice site, producing a circular format with a 3'-5' phosphodiester bond at the back-splicing junction site [22]. Currently, the common models of circRNA circularization include: lariat-driven circularization, RBP induced circularization, intron-pairing-driven circularization and splicing of pre-tRNA-driven circularization [10] (Fig. 1).
2.1 Lariat-driven circularization
Pre-mRNAs are partially folded as a result of exon hopping during transcription. The upstream 3' splicing receptor is covalently linked to the downstream 5' splicing donor to form a lariat intermediate containing exons or introns. Subsequently, the lariat intermediate undergoes reverse splicing, excision or retention of the sequence, and eventually forms a covalently closed ciRNAs [23]. In addition, ciRNA formation depends on conserved elements which are seven nucleotides rich in GU near the 5' splicing site and 11 nucleotides rich in C near the 3' branching point. In the process of reverse splicing, these two elements combine to form a lariat structure, and then the exons in the structure are removed by splicing. These two elements protect ciRNA from intron debranching and degradation, ultimately forming a stable ciRNA [24, 25].
2.2 Intron pairing-driven circularization
In this process, introns on either side of exons are essential elements. The upstream introns and the downstream introns contain reverse complementary sequences (such as Alu repeats and other nonrepetitive elements), after base pairing occurs, the downstream 5-donor and upstream 3-receptor splicing sites are close to each other, ultimately speeding up ecircRNAs formation [26, 27]. Some bioinformatic analyses and experimental studies have also confirmed that reverse complementary Alu repeats in flanking introns are associated with circRNA biogenesis [28]. Li et al. demonstrated that the sequences of flanking introns of cGGNBP2 (hsa_circ_0003930) was highly reversed complementary and necessary for the circRNA biogenesis [29]. What's more, circHIPK3 expression is promoted by Alu elements [30].
2.3 RBP-driven circularization
CircRNA biogenesis is regulated by a variety of proteins, such as RBPs, enzymes, and transcription factors [31-34]. RBPs bind specifically to specific sites in the flanking intron sequence of pre-mRNA to form a bridge, making the splicing sites closer between the receptor and donor. In the process of back splicing, some intron sequences are not removed but remain in the circRNA, known as EIcircRNAs [35]. For instance, mannan binding lectin (MBL) heightens circMBL generation by recognizing and binding highly conserved MBL-binding elements located in the flanking introns of its own pre-mRNA [36]. Fused in sarcoma binds to RHOBTB3 pre-mRNA and then accelerates the back-splicing process of circRHOBTB3 production [32]. Moreover, quaking, acting as an alternative splicing factor [37], induces exon cyclization by binding with the recognition element in the intron and forming a dimer, and subsequently gives rise to the generation of circRNA, which also facilitates the reverse splicing efficiency in the epithelial-mesenchymal transition (EMT) process [38]. Apart from that, adenosine deaminase 1, as an RNA editing enzyme, negatively regulates the production of circRNA by lessening flanking introns and reverse folded RNA pairing structures [33]. Likewise, the RNA/DNA helicase DExH-Box helicase 9 lessens circRNA formation by down-regulating intron pairing [39-41].
2.4 Splicing of pre-tRNA-driven circularization
Additionally, a subset of circRNAs is derived from tRNAs through the splicing of pre-tRNAs [42]. In the process of tRNA maturation, the tRNA splicing endonuclease complex cleaves pre-tRNAs, and an enzyme known as RNA terminal phosphorylase B (RtcB) ligates the exon fragments and introns to generate a tricRNA [43, 44].
To put it simple, the biogenesis of circRNAs and the molecular mechanism that remain inconclusive with regard to how they regulate circularization. It is noteworthy that the origin of circRNA is closely related to parental genes. Currently, the genetic origin of circRNA can be queried through a series of databases, which will drive us to gain in-depth insight into the circularization process of circRNAs (Table 1).
Biosynthesis and functions of circRNAs. (A) Lariat-driven circularization: Pre-mRNA can produce a lariat intermediate by exon-skipping. The intron sequences in the lariat intermediate are retained, and then form ciRNA; RBP induced circularization: RBPs specifically bind to specific sites in the flanking intron sequence of pre-mRNA to form a bridge, thereby increasing the formation of EIciRNA; Intron-pairing-driven circularization: The upstream introns are base-paired with the downstream introns, forming EcircRNA; Splicing of pre-tRNA-driven circularization: Pre-tRNA is cleaved into half of the exon and intron part. TricRNA is produced by joining the termini of the introns. (B) Transcription regulator: CircRNAs can interact with U1 small nuclear ribonucleoproteins (U1 snRNPs) or RNA polymerase II (Pol II) in the promoter region of targeted genes, and enhance the transcription and splicing of genes; microRNA (miRNA) sponge: CircRNAs with complementary binding sites can pairing with miRNAs response elements of mRNAs and sequester miRNAs away from target mRNAs, thereby protecting target mRNAs from miRNA-dependent degradation; Binding RBP: CircRNAs may act as sponge, protein scaffold, or decoy for proteins directly or indirectly regulate their locations and functions; Translation template: CircRNAs may be translated into unique peptide by cap-independent translation initiation mechanisms under specifical conditions.
Databases regarding circRNA
| Database | Description |
|---|---|
| circBank | Search the genomic linear structure, mature sequences, expression patterns across different tissues or developmental stages, miRNA regulatory sites, N6-methyladenosine (m6A) sites, internal ribosome entry site (IRES), coding potential and open reading frames (ORFs). |
| circBase | Search the genomic position, spliced length, gene symbol, best-matching transcript and circRNA study. |
| TransCirc | Search the genomic position, gene symbol, m6A sites, IRES, ORFs, ribosome profiling and mass spectrometry data. |
| circRNADb | Search the genomic position, spliced length, gene symbol, best-matching transcript, sample, IRES and ORFs. |
| Circular RNA Interactome | Search the genomic position, best-matching transcript, mature sequences, RBPs and miRNA regulatory sites, divergent primers and siRNAs. |
| CIRCpedia | Search the genomic position, gene symbol, sample and conservation analysis |
| CircNet | Search the genomic position, gene symbol, RBPs and miRNA regulatory sites, ORFs |
| circAtlas | Search the genomic position, gene symbol, the second RNA structure, sample, conservation analysis, RBPs and miRNA regulatory sites, IRES and ORFs. |
3. Mechanisms of circRNAs in PC
3.1 MiRNA sponges
As ubiquitous and conserved small non-coding RNAs with lengths of 19-25 nucleotides, miRNAs prevent translation or facilitate degradation by binding to specific miRNA responsive element of target mRNA. In such case, it thereby exhibits a wide range of biological functions [45]. On the basis of the available evidence, the researchers define this class of circRNA with miRNA-bound miRNA response element (MRE) as competitive endogenous RNA (ceRNA). By acting as miRNA molecular sponges, these circRNAs bind to corresponding miRNAs and inhibit their functions, and regulate the translation or degradation of target molecules, thus exerting tumor inhibition or carcinogenic effects [46, 47]. As Multiple studies have confirmed that circRNAs can act as miRNA sponges in PC (Supplementary Table 1). For instance, circ001859, circSEC24A, circMBOAT2, circRNF13 and circ000684 affect malignant phenotypes of pancreatic ductal adenocarcinoma (PDAC) cells via sponging dissimilar miRNAs [80, 83, 117, 120, 123]. Likewise, circATG7 and circRHOBTB3 sponges miR-766-5p and miR-600 to regulate the autophagy response of PDAC cells, respectively [19, 32]. Apart from that, hsa_circRNA_001587 and circ_0000284 regulate angiogenesis of PC cells via binding to miR-223 and miR-1179, respectively [124, 125]. As illustrated by other studies, circ_0058058 acted as a molecular sponge of miR-557 to mediate PC immunosuppressive microenvironments [133]. A myriad of evidence confirmed that circRNAs can function as miRNA sponges. Nonetheless, not all circRNAs possess this capability, which is owing to the fundamental fact that some specific circRNAs have little or even no MRE.
3.2 Binding RBPs
RBP is a group of proteins with RNA recognition and binding ability, which conducts an indispensable role in biological processes by binding RNA [135]. As already revealed by recent studies, circRNAs may bind to RBPs to regulate the expressions of core genes in the process of PC (Supplementary Table 1). Meng et al. suggested that circSTX6 interacted with cullin-2 (CUL2) to participate in the ubiquitin-dependent degradation of hypoxia-inducible factor 1-alpha (HIF1A) and subsequently accelerating PDAC progression [79]. Another study demonstrated that circRTN4 interacted with EMT-driver RAB11FIP1 and blocked its ubiquitination site to lower its ubiquitination in PDAC [78]. Furthermore, circPTPN22 attenuates signal transducer and activator of transcription 3 (STAT3)/sirtuin 1 (SIRT1) interaction by binding to STAT3, ultimately facilitating STAT3 acetylation during PC immune resistance [128]. Other investigations have revealed that circMYO1C targets the m6A site of programmed death-ligand 1 (PD-L1) mRNA to reinforce its stability through cooperating with insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), thereby pushing ahead PDAC immune escape [130]. As mentioned above, circRNAs can bind to RBPs to facilitate or inhibit their functions. Nevertheless, rare further studies have been found to probe deep into the interactions between circRNAs and RBPs in PC. For this reason, RBP immunoprecipitation combined with circRNA sequencing can be adopted to discover circRNAs that can interact with RBPs.
3.3 Protein scaffolds
As displayed by new evidence, circRNAs can also serve as protein scaffolds, interacting with proteins to regulate protein structures or functions, thereby participating in pathological and physiological processes (Supplementary Table 1). One study illustrated that circEIF3I was a molecular scaffold that interacted with small mother against decapentaplegic family member 3 and adaptor-related protein complex 2 subunit alpha 1 to form a ternary complex, which immensely facilitated growth and metastasis of PDAC [20]. Furthermore, circATG7 act as a scaffold to increase the interaction between the human antigen R protein and ATG7 mRNA as well as reinforce ATG mRNA stability in PC cells [19]. To go it further, other research has demonstrated that circPTPN22 regulates PC immune microenvironment by facilitating STAT3 acetylation via inhibiting STAT3/SIRT1 interaction [128]. More importantly, circRNAs can also conduct biological functions as the molecular scaffold while functioning as sponges for miRNAs. CircFOXK2 facilitates growth and metastasis of PDAC by complexing with Y-box-binding protein 1 (YBX1) and heterogeneous nuclear ribonucleoprotein K and sponging miR-942 [52]. Likewise, exosomal circPDK1 sponges miR-628-3p to activate the bromodomain plant homeodomain finger transcription factor (BPTF)/c-myc axis and serves as a scaffold to strengthen the interaction between ubiquitin-conjugating enzyme E2 O and bridging integrator 1 (BIN1), which gives rise to PC glycolysis [111]. Altogether, all of these significant findings evidently reveal that circRNAs can act as protein scaffolds to affect the expression or function of certain proteins, which expands our understanding of circRNAs. But it remains essential to look into the specific mechanisms by further exploration.
3.4 Regulators of gene splicing and transcription
In general, the majority of EIciRNAs and ciRNAs are located in the nucleus, thus possessing the ability to exert regulatory effects at the transcriptional or post-transcriptional level [136]. Associated research has illustrated that circ-EIF3J and circ-PAIP2, identified as EIcircRNA, can combine with snRNP to form EIciRNA-U1 snRNP compounds, which further interact with Pol II to facilitate the transcription of EIciRNA parent genes [137]. Additionally, the specific interaction between ci-ANKRD52 and RNA Pol II complex facilitates ankyrin repeat domain 52 transcription [138]. It is particularly noteworthy that alternative splicing is extensively correlated with a variety of biological processes. Furthermore, circRNAs also participate in the regulation of gene expression by affecting pre-mRNA alternative splicing. For instance, circMBL competes with the classical splicing of MBL pre-mRNA, thereby weakening the formation of linear RNA [36]. Furthermore, hsa_circ_0007919 recruits forkhead box protein A1 and ten eleven translocation 1 to lessen the methylation of the ligase I promoter and heighten its transcription, which tremendously facilitate the repair of base excision, mismatch and nucleotide excision [102]. Likewise, Kong et al. uncovered that circ-CDYL cooperated with a novel transcriptional (co-) factor eukaryotic elongation factor 1alpha-2 to initiate collagen, type XIV, alpha1 transcription [139]. Another study investigating breast cancer has proved that antisense circular RNA circSCRIB hindered the splicing of scribble (SCRIB) pre-mRNA, and interacted with SCRIB mRNA sequence, which resulted in the inhibition of SCRIB translation [140]. As jointly revealed by these significant findings, circRNAs affect gene expression at the transcriptional or post-transcriptional level. Nevertheless, the current research on the transcriptional regulation and splicing of circRNA in PC is relatively lagging behind that in other tumors, which apparently necessitates more comprehensive exploration and more conspicuous reinforcement.
3.5 Protein/peptide templates
Early studies have illustrated that circRNAs, as non-coding RNAs, cannot be translated through a cap-dependent mechanism owing to the deficiency of 5' end and 3' Poly A tail [141]. Nonetheless, as the study deepens its depth and width, some circRNAs with initial codon sites, ORF and IRES elements are translated in a cap-independent manner in some cases, producing proteins or peptides with specific functions [142-144]. For instance, circSEMA4B which contains AUG, ORF and IRES encodes a novel protein SEMA4B-211aa. SEMA4B-211aa inhibits the production of phosphatidylinositol 3,4,5-trisphosphate by binding to p85, thus inhibiting the phosphorylation of protein kinase B (AKT) (Thr308), and ultimately inhibiting the progression of breast cancer [145]. As clearly proved by another study with regard to colorectal cancer, a spanning junction ORF with the potential to encode a 121 amino acid protein and an IRES at 207-292 nt are contained in the sequence of circINSIG1. circINSIG1 encods a 121 amino acid protein circINSIG1-121 to accelerate cholesterol metabolism in cancer [146]. Apart from IRES and ORF responsible for circRNA translation, the m6A modification can also initiate or reinforce circRNA translation [147]. The m6A reader YT521-B homology domain 2 drives the hsa_circ_0082002 encoding the protein MET404 in glioblastoma [148]. Nonetheless, no reports have been released to investigate circRNA encoding protein in PC. It is also noteworthy that circRNA translation is inadequate. Worse still, controversial debates are still heating with respect to how its derived proteins or peptides affect PC development. As a consequence, more information is essential to elucidate the mechanisms of circRNA translation and deepen our understanding of the circRNA molecular mechanisms and human proteins.
4. Functions of circRNAs in PC
4.1 Proliferation and tumorigenesis
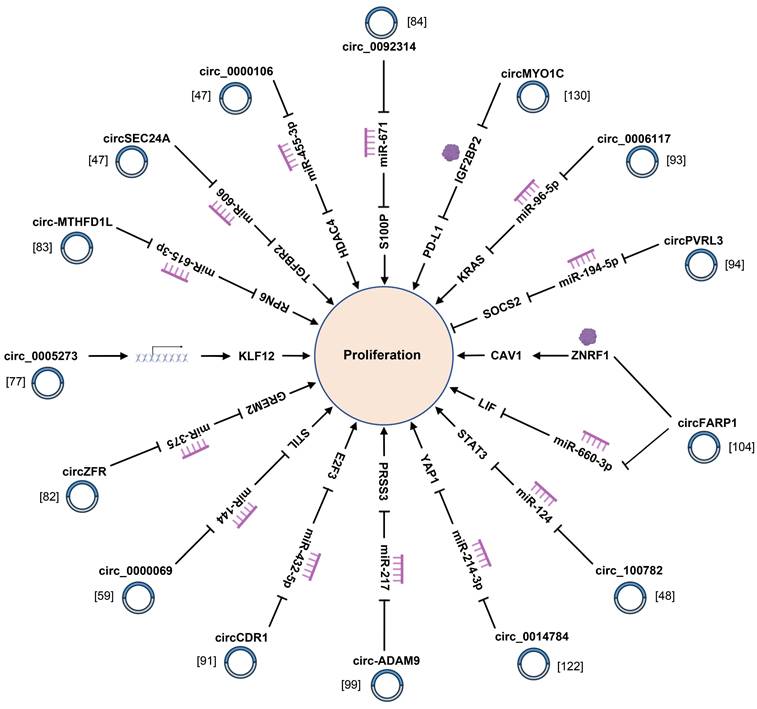
Normally speaking, cancer cells are featured by continuous proliferation and avoidance of apoptosis [149]. These key cellular processes are regulated by multiple factors that have not yet been sufficiently elucidated. As already demonstrated by relevant studies, multiple circRNAs with dysregulated expression in PC tissues and cells are tightly correlated with proliferation and apoptosis of PC cells (Fig. 2, 3) (Supplementary Table 1). For instance, circ-MBOAT2 regulates tumor growth and apoptosis via miR-433-3p/ glutamic-oxaloacetic transaminase 1 signaling axis in PC [117]. Li et al. reported on exosomal circ_0030167 derived from bone marrow mesenchymal stem cells (BM-MSCs) implicated in proliferation, and stemness of PC cells via the miR-338-5p/wingless-type MMTV integration site family (WNT) inhibitory factor-1/Wnt8/β-catenin axis [119]. Moreover, circSFMBT1 affects proliferation and apoptosis of PC cells by augmenting the mRNA level of p21-regulated kinase 1 [54]. As revealed by a recent study, hsa_circ_0006790 is involved in PC cells proliferation and apoptosis by cooperating with chromobox protein homologue 7 [132]. Furthermore, circRNA_102049 interacts with miR-455-3p to accelerate CD80 transcription, thereby inhibiting PC proliferation and attenuating apoptosis [134]. hsa_circ_0000069 facilitates the proliferation and cell cycle progression of PC cells by suppressing the expression of the parent gene stem cell leukemia/T-cell acute lymphoblastic leukemia 1 interrupting locus for hsa_circ_0000069 [59]. To further clarify the indispensable role of circRNAs in proliferation, apoptosis, autophagy and tumorigenesis of PC cells is advantageous for deepening the understanding on the pathogenesis of PC.
CircRNAs affect the proliferation of pancreatic cancer cells via regulating miRNA-mRNA axis.
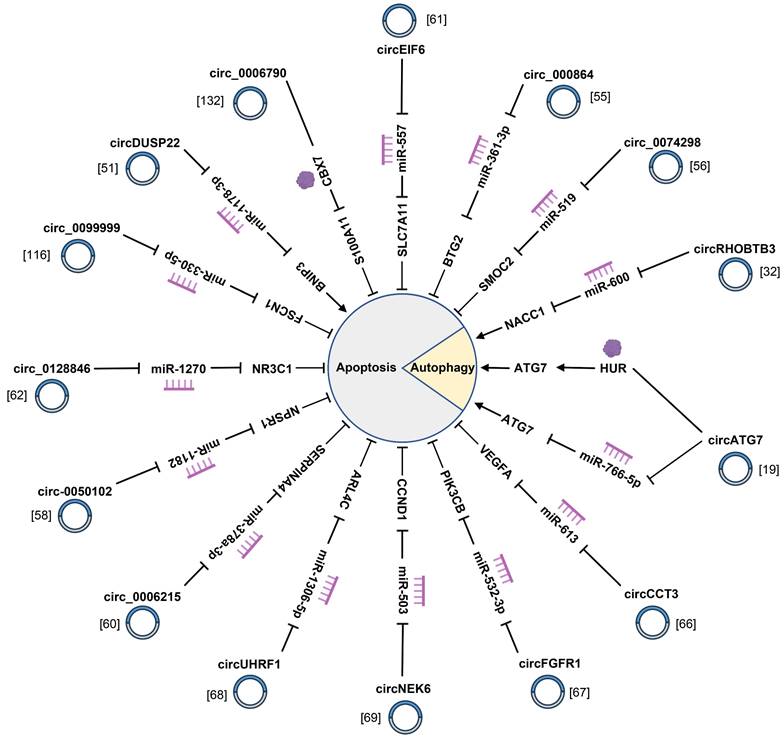
CircRNAs are involved in the apoptosis and autophagy of pancreatic cancer cells by interacting with miRNAs and RNA-binding proteins.
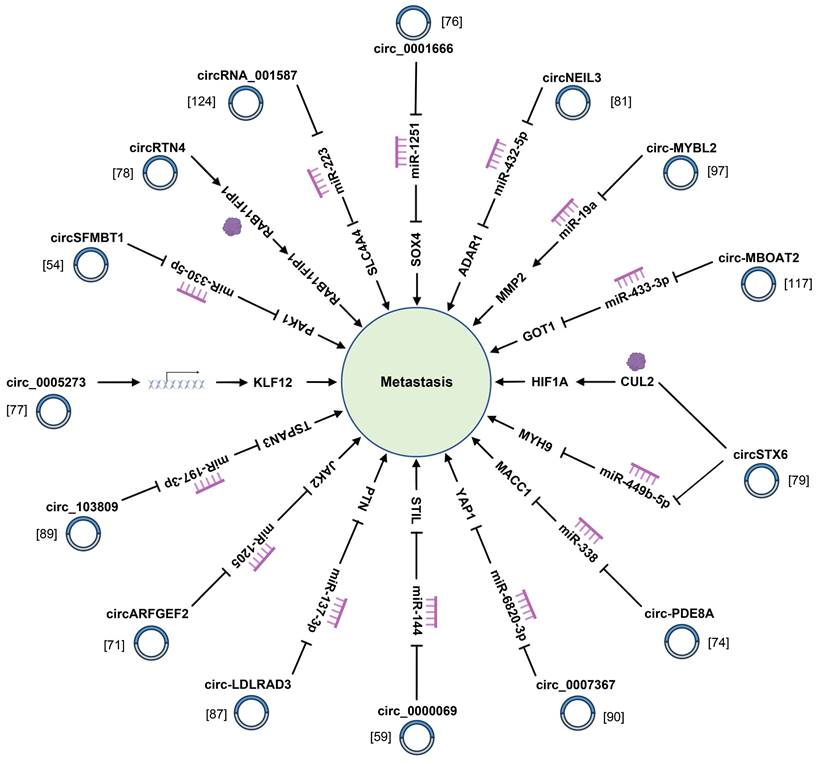
4.2 Metastasis
Aside from the infinite growth of tumor cells, metastasis is another pivotal feature of tumor cells. As revealed by recent studies, some circRNA have close correlations with tumor metastasis (Fig. 4) (Supplementary Table 1). For instance, circ_0000284 is remarkably upregulated in PC tissues and cells, and facilitates PC cell proliferation, migration, and invasion while regulating apoptosis [125]. Besides, hsa_circRNA_001859 acts as a molecular sponge of miR-21-5p and regulates the expression of solute carrier family 38 member A2 to regulate tumor EMT of PC [80]. Additionally, circ-STK39 regulates translocation-associated membrane protein 2-mediated proliferation and the EMT of PC by sponging miR-140-3p [85]. Chen et al. identified that circSEC24A noticeably governed the proliferative, migration and invasive capacity of PC cells through accelerating the expression of TGF-beta receptor 2 (TGFBR2) [83]. Apart from ordinary circRNAs, exosomal circRNAs also conduct a pivotal role in the process of PC cells. For instance, exosomal circPDK1 strikingly facilitates PC cells migration, proliferation, and glycolysis by modulating the miR-628-3p/BPTF axis and degrading BIN1 [111]. Another study suggested that exosomal circ_0030167 inhibited the invasion, migration, proliferation and stemness of PC cells by inactivating the Wnt/β-catenin signal pathway [119]. A great many clinical practices have confirmed that tumor metastasis greatly affects the diagnosis and treatment of PC, so more studies are needed to understand the role of circRNAs in PC metastasis.
CircRNAs modulate the metastasis of pancreatic cancer cells through sponging miRNAs and binding to related proteins.
4.3 Chemotherapy resistance
Chemotherapy is one of the predominant treatments for PC. Nevertheless, the emergence of chemotherapy resistance and multi-drug resistance seriously affects the effectiveness of PC treatment and is one of the crucial factors bringing about undesirable prognosis of PC patients [150]. In recent years, as the study probes deeper into circRNAs, a new perspective has been provided for people to understand the mechanism of drug resistance in PC (Fig. 5) (Supplementary Table 1). For instance, circ_0087502 gives rise to PC cell proliferation, migration, and gemcitabine (GEM) resistance by sponging miR-1179 and facilitating TGFBR2 expression [107]. Moreover, cancer-associated fibroblast (CAF)-specific circFARP1 inhibits caveolin-1 degradation and enhances leukaemia inhibitory factor (LIF) secretion, which ultimately triggers GEM chemoresistance in PDAC [104]. As suggested in the studies conducted by Chen et al., circMTHFD1L, as an endogenous miR-615-3p sponge, upregulated the expression of ribophorin VI, which not only accelerated the repair of DNA damage, but also noticeably weakened the sensitivity of PC to GEM [105]. Another study suggested that circHIPK3 accelerated GEM resistance in PC cells by targeting Ras-association domain family 1 via miR-330-5p [30]. The above circRNAs conduct a catalytic role in GEM resistance, while some circRNAs induce PC cells to be sensitive to GEM. hsa_circ_0007401 and circ_0092367 augment the chemosensitivity of GEM in PC [101, 109]. What's critical to mention is that circRNAs not only regulate GEM resistance of PC cells, but also affect therapeutic efficacy of the small-molecule epidermal growth factor receptor tyrosine kinase inhibitor against PC [108]. On the whole, these studies have persuasively illustrated that circRNAs conduct irreplaceable roles in modulating chemosensitivity in PC. Hence, delving deeply into the potential roles of circRNAs in the mechanism underlying drug resistance in PC is advantageous for the discovery of novel molecular targets and the enhancement of chemotherapy efficacy, which is of paramount value for the prognosis of patients with PC.
CircRNAs play crucial roles in GEM resistance and immune escape of pancreatic cancer.
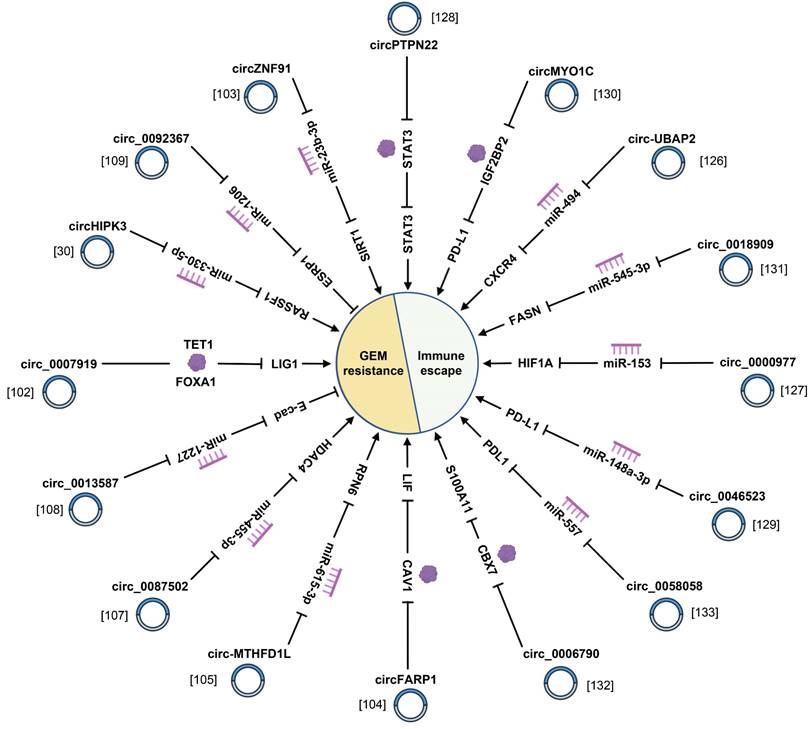
4.4 Immune escape
Tumor-related immunity can exert profound influence in tumor progression and treatment. As reported by recent studies, circRNAs are prevalently involved in tumor immune escape (Fig. 5) (Supplementary Table 1). For instance, hsa_circ_0006790 is highly expressed in PC and is involved in immune escape [132]. Furthermore, circMYO1C mediated by m6A methyltransferase methyltransferase like 3 targets PD-L1 mRNA stability by cooperating with IGF2BP2, thereby accelerating PDAC immune escape [130]. Another study suggested that circPTPN22 could trigger the formation of PC immune microenvironment by facilitating STAT3 acetylation via attenuating STAT3/SIRT1 interaction [128]. Moreover, circ_0058058 exhibits elevated expression in PC tissues and works as a molecular sponge of miR-557 to upregulate PD-L1, thereby resulting in PC progression and immune escape [133]. Aside from cancer cells, other cells can also exert certain influence in the formation of tumor immune microenvironment, such as CAFs, macrophages, and T cells. Fu et al. reported that hsa_circ_0046523 was beneficial for an immunosuppressive tumor microenvironment by propelling the apoptosis and exhaustion of CD8+ T cells, inhibiting CD8+ T cell function, lowering the secretion of immunosuppressive cytokines interleukin-10 and TGF-β, and lessening the secretion of immune effector cytokines interferon-gamma and IL-2 among among peripheral blood mononuclear cells [129]. Additionally, circCUL2 is specifically expressed in CAFs and induces the inflammatory CAF (iCAF) phenotype, subsequently iCAFs facilitates PDAC progression through IL-6 secretion [100]. Other than that, a study on circ_0018909 illustrated that circ_0018909 induced polarization of M0 macrophages to M2 macrophages to regulate the development of PC [131]. With the belief that studies about circRNAs play a pivotal role in PC-related immunity, and more far-reaching research on the mechanism of circRNAs in immune regulation is advantageous to explore a new breakthrough in the treatment of PC.
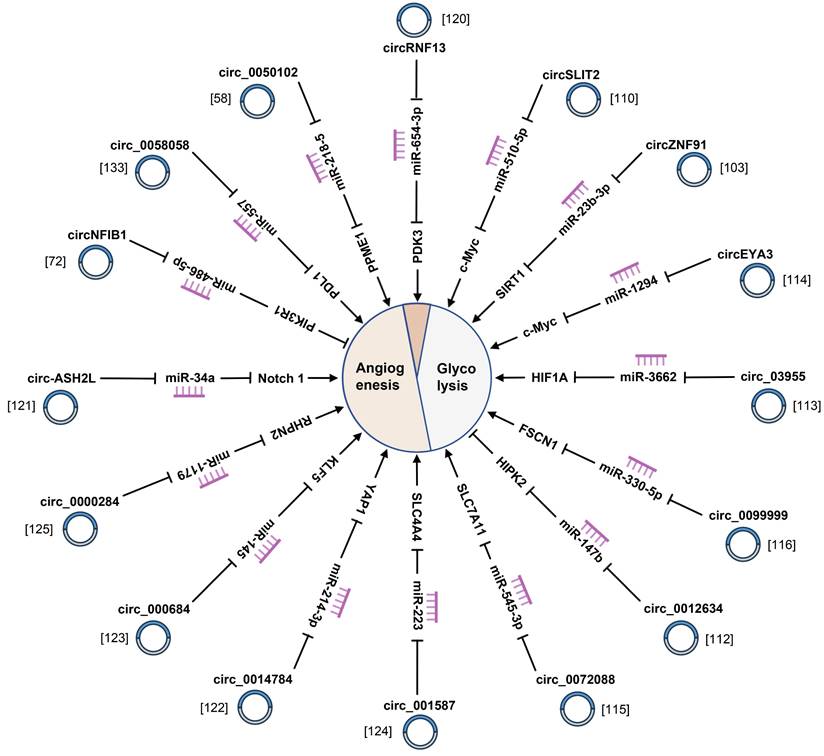
4.5 Glycolysis
Most solid tumors are typically characterized by the hypoxic microenvironment which can be attributed to the speedy proliferation of tumor cells, heteromorphic tumor structure, and aberrant structure and function of tumor vessels. During this process, cancer cells obtain energy through glycolysis under hypoxia [151]. CircRNAs are associated with hypoxia (Fig. 6) (Supplementary Table 1). Zhao et al. reported that circRNF13 induced by hypoxia was upregulated in PC tissues and accelerated tumor glycolysis in PC [120]. Moreover, circ_0072088 facilitates cell extracellular acidification rate, lactate production, glucose uptake, and ATP generation, but suppressed oxygen consumption rate in PDAC cells [115]. Furthermore, circ_03955 regulates Warburg effect of PC by the promotion of HIF-1ɑ [113]. Simultaneously, circSLIT2 can propel the aerobic glycolysis of PDAC via targeting miR-510-5p/c-Myc/ lactate dehydrogenase A axis [110]. These significant findings reveal that to look into how circRNAs affect anaerobic and aerobic glycolysis is likely to provide a novel sight for PC treatment.
4.6 Angiogenesis
Angiogenesis, the formation of new blood vessels from pre-existing ones, is an essential process for growth, development and disorders. Angiogenesis is bound up with tumors, which can be certified by the extensive application of antiangiogenic drug bevacizumab in anti-cancer [152]. Nevertheless, no antiangiogenic drug has been developed to treat PC. As evidently demonstrated by numerous studies, circRNAs can impose remarkable effects on angiogenesis of PC cells (Fig. 6) (Supplementary Table 1). What should be pointed out is that hsa_circRNA_001587 inhibits angiogenesis in PC by impairing miR-223-mediated solute carrier family 4 member 4 inhibition [124]. Furthermore, circ_0000284 facilitates PC angiogenesis resting with the regulation of miR-1179/rhophilin 2 [125]. Meanwhile, hsa_circ_0050102 expedites angiogenesis of PC through growing phosphatase methylesterase 1 abundance [58]. Thus, we provide an all-round overview of recent insights into circRNAs in blood vessel development, which will present brand new opportunities and enlightening references for the design of vascular-directed therapies in PC.
4.7 Other
PC has the marked desmoplastic reaction. As the dominant cell type within the tumor stroma, CAFs critically contribute to biological behaviors in this highly fibrotic solid malignancy [153]. Recently, Rong et al. reported that circBIRC6, a CAF-derived extracellular vesicles-packaged circRNA, induced oxaliplatin resistance by mediating SUMOylation of X-ray repair cross-complementing protein 4 [154]. Moreover, Chen team elucidated two CAF-derived circRNAs. circCUL2 was significantly correlated with the poor survival of PDAC patients. circCUL2 induced normal fibroblasts to transformed into iCAF phenotype, and then secreted IL-6 to activate STAT3 signaling pathway in cancer cells, ultimately inducing PDAC progression [100]. Furthermore, a CAF-specific circFARP1 is positively correlated with poor survival in advanced PDAC patients. circFARP1 interacts with caveolin 1 and miR-660-3p, enhancing LIF secretion to facilitate GEM resistance and tumor cell stemness [104]. Significantly, cancer stem cells are important cells that play a fundamental role in cancer [155]. The roles of PC-associated circRNAs in cancer stem cells has also received attention. circRREB1 is significantly upregulated in PDAC and promotes WNT7B transcription by directly interacting with YBX1 and facilitating its nuclear translocation, consequently activating the Wnt/β-catenin signaling pathway to maintain PDAC stemness [118]. At present, there is still a lack of research on circRNA about CAF and stem cells in PC. To further clarify the biological functions and molecular mechanisms of circRNAs in CAF differentiation and cancer stem cells will help promote the progress of circRNA research.
CircRNAs are responsible for angiogenesis and glycolysis of pancreatic cancer.
5. Clinical application of circRNAs in PC
5.1 Diagnostic and prognostic markers
Early detection, diagnosis and treatment are of vital importance in improving the prognosis of cancer patients. Nevertheless, inappropriate prognostic evaluation will seriously affect the adjustment of treatment strategies and the extension of lifespan for patients. Nowadays, an increasing number of studies have implicated that circRNAs have great potential as biomarkers for PC diagnosis and prognosis (Table 2). To be specific, owing to the special splicing mode of circRNAs, most circRNAs are highly conserved in various species [156]. Moreover, as a consequence of its unique covalent closed-loop structure, circRNA is resistant to RNA exonuclease or RNase R and has a longer half-life in tissues and plasma. On this basis, it is more stable than linear RNA, which not only is instrumental in the circRNA accumulation, but also is more easily to be detected in cells, tissues or body fluids [157]. Apart from that, the expression profiles of circRNAs exhibit cell type specificity, tissue specificity, or developmental stage specificity [158]. Furthermore, circRNAs are widely expressed and can be detected in multiple species (such as yeast, plants, fungi, mice, rats, monkeys, fruit flies, humans, and many other organisms). Last but not least, other than solid tissues, circRNAs are also expressed in exosomes, blood, saliva, and urine, which can be employed for non-invasive diagnosis [159].
CircSTX6 has been demonstrated to be frequently upregulated in PDAC. Moreover, the up-regulation of circSTX6 expression in tumor tissues is positively correlated with tumor size and N stage. On this basis, circSTX6 can serve as a potential biomarker for the management of PDAC [79]. Zhao et al. uncovered that circRNF13 expression was highly abundant in PC tissues and was positively correlated with T stage, N stage, M stage and American Joint Committee on Cancer stage, which might be a potential prognostic indicator [120]. Apart from that, circ-MTHFD1L is a GEM resistance-associated circRNA that is conspicuously heightened and stably expressed in PDAC tissues. And the high circ-MTHFD1L expression level is dramatically bound up with chemotherapy resistance and undesirable prognosis of PDAC patients, which may be a new potential molecular marker for GEM resistance [105]. Circ_001569 is not only highly expressed in tissues and plasma of PC patients, but also is positively associated with lymphatic metastasis, clinical stage, and venous invasion. Moreover, multivariate Cox regression analyses validated that circ_001569 was an independent prognostic indicator for overall survival rates of PC patients. As evidently demonstrated by operating characteristic (ROC) curve analysis, the area under curve of plasma circ_001569 was 0.716 with a sensitivity and specificity of 62.76% and 74.29%, respectively [70]. Likewise, Kaplan-Meier analysis for circNEIL3 revealed that overall survival of PC patients with high expression of circNEIL3 was strikingly shortened. Univariate and multivariate Cox regression analysis collectively revealed that the circNEIL3 expression level was independent prognostic factors for PDAC patients, as were tumor size and TNM stage [81].
On the contrary, some circRNAs are down-regulated in PC, which is associated with a favourable prognosis. For instance, circACTR2 is not only poorly expressed in PC, but also is bound up with overall survival and pathological grade. As further revealed by univariate and multivariate Cox regression analysis, circACTR2 expression levels is independent prognostic factors for PC patients [106]. Kong et al. held a standpoint that circNFIB1 expression was dramatically decreased in PDAC tissues, and was negatively correlated with lymphatic metastasis and TMN stage [72]. Another study exhibited that tumor suppressor circ_0092367 expression was decreased in PC, and attenuated PC progression. Low circ_0092367 expression is tightly connected with shorter survival time [109].
What deserves mentioning is that circRNAs can be detected either in tumor tissues, or in exosomes, blood, saliva, and urine. For instance, circ-PDE8A is upregulated in PDAC tissues and plasma exosomes. Expression level of circ-PDE8A in plasma exosomes is bound up with lymphatic invasion, TNM stage and a disappointing survival rate of PDAC patients. Moreover, exosomal circ-PDE8A may be a useful marker of PDAC diagnosis or progression [74]. In comparison, exosomal circ-IARS secreted by PC cells in plasma can be utilized as an indicator for early diagnosis and prognostic prediction in PDAC. For the critical role of circ-IARS in regulating endothelial monolayer permeability, it can be employed as a therapeutic target [73]. Wang et al. pointed out that exosomal hsa_circ_0012634 was not only isolated from serums and cell culture supernatant, but also was dramatically downregulated in PDAC [112]. What's more, ROC curve analysis about hsa_circ_0012634 showed the Area Under the ROC curve value was 0.868, suggesting that hsa_circ_0012634 could be serve as an indicator for distinguishing the population of PDAC patients and healthy controls. Other than exosomal circRNA secreted by tumor cells, exosomal circRNA can also be produced by other cells in the tumor microenvironment. For instance, exosomal circ_0030167 derived from BM-MSCs is decreased in PC serums and cells [119]. Furthermore, Lin and colleagues first adopted RNA sequencing to screen for hypoxia-induced exosomal circRNAs in PC, and subsequently noticed that circPDK1 was highly expressed in PC tissues and serum exosomes, which may support a novel diagnostic biomarker for early PC patients [111]. These fundamental studies noticeably illustrate that circRNAs have potential as diagnostic and prognostic biomarkers for PC. Nonetheless, it's critical to collect more samples with more typical features to further confirm the reproducibility and specificity of circRNAs detection. Only under such circumstance can they be applied to cancer diagnosis and prognosis evaluation.
5.2 Therapeutic targets
For the time being, a multitude of studies have been carried out to explore molecular drugs that regulate gene level based on technologies such as siRNA, antisense oligonucleotides [160] and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein system [161]. Fortunately, numerous drugs originating from these findings have been applied in clinic. Meanwhile, a mushrooming number of domestic and international studies have implicated that circRNAs have been certified to conduct pivotal roles in the occurrence and development of PC. As a consequence, these significant findings provide adequate evidence to support the conjecture that circRNAs may become therapeutic targets for PC. Guo and colleagues analyzed microarray data in PDAC patients and found therapeutic target of PDAC circBFAR notably upregulated in PDAC. CircBFAR knockdown suppresses the proliferation, migration, and invasion of PDAC cells in vitro and in vivo [98].
CircRNAs act as potential diagnostic and prognostic biomarkers in pancreatic cancer
| circRNA | Expression | Diagnostic | Prognostic | Clinicopathological parameters | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pathological stage | Tumor stage | Lymph node status | Distant metastasis | TNM stage | Other | |||||
| circ-LDLRAD3 | Up | Yes | Yes | / | / | Yes | Yes | / | Vascular invasion | [87] |
| hsa_circ_0007367 | Up | / | Yes | Yes | / | Yes | / | / | / | [90] |
| circSTX6 | Up | / | Yes | Yes | / | Yes | / | / | / | [79] |
| circNEIL3 | Up | / | Yes | / | / | / | / | / | Vascular invasion, Nerve invasion | [81] |
| circ_0087502 | Up | / | Yes | Yes | Yes | Yes | / | / | / | [107] |
| hsa_circ_0046523 | Up | / | Yes | Yes | Yes | Yes | / | Yes | / | [129] |
| circMYO1C | Up | / | / | / | / | Yes | / | Yes | / | [130] |
| circCUL2 | Up | Yes | Yes | / | / | Yes | / | Yes | / | [100] |
| circRNF13 | Up | / | Yes | / | Yes | Yes | Yes | / | / | [120] |
| circ-ASH2L | Up | / | Yes | / | / | Yes | / | Yes | / | [121] |
| circRNA_000684 | Up | / | Yes | Yes | / | Yes | Yes | Yes | / | [123] |
| circSLIT2 | Up | / | Yes | / | / | Yes | / | Yes | / | [110] |
| circ_001569 | Up | Yes | Yes | / | / | Yes | / | Yes | Vascular invasion | [70] |
| circBFAR | Up | / | Yes | / | / | / | / | Yes | / | [98] |
| circ-0005105 | Up | / | Yes | / | Yes | Yes | / | Yes | Vascular invasion | [96] |
| circPTPRA | Up | / | Yes | / | / | Yes | / | / | / | [95] |
| circ_0030235 | Up | / | Yes | / | Yes | Yes | / | / | / | [65] |
| circ_0005273 | Up | Yes | Yes | / | / | Yes | Yes | / | / | [77] |
| Exosomal hsa_circ_0012634 | Down | / | / | / | Yes | / | / | / | / | [112] |
| hsa_circRNA_001587 | Down | / | Yes | Yes | / | Yes | / | / | / | [124] |
| circ_0092367 | Down | / | Yes | Yes | / | Yes | / | / | / | [109] |
| circ_0013587 | Down | / | Yes | Yes | Yes | Yes | / | / | / | [108] |
| circNFIB1 | Down | / | / | / | / | Yes | / | Yes | / | [72] |
| circACTR2 | Down | / | Yes | Yes | / | / | / | / | / | [106] |
| hsa_circ_0001649 | Down | / | Yes | Yes | Yes | / | / | / | / | [50] |
| Exosomal circ-PDE8A | Up | / | Yes | / | Yes | Yes | / | Yes | / | [74] |
| Exosomal circPDK1 | Up | / | Yes | Yes | Yes | Yes | Yes | / | / | [111] |
| Exosomal circ-IARS | Up | / | Yes | / | / | / | Yes | / | / | [73] |
| hsa_circ_0071036 | Up | Yes | Yes | / | / | Yes | / | / | PET-CT SUV max value | [17] |
| circATG7 | Up | Yes | Yes | / | Yes | Yes | / | / | / | [19] |
| circEIF3I | Up | / | Yes | / | Yes | Yes | / | / | / | [20] |
| circRHOBTB3 | Up | / | Yes | / | Yes | / | / | / | Vascular invasion | [32] |
| circRHOT1 | Up | / | / | / | / | Yes | / | / | / | [57] |
| circ_0007534 | Up | / | Yes | / | Yes | Yes | / | / | / | [63] |
| circ_0092314 | Up | / | Yes | / | / | Yes | / | Yes | / | [84] |
| circ-MTHFD1L | Up | / | Yes | Yes | / | / | / | / | CA19-9 | [105] |
| hsa_circ_0000069 | Up | Yes | Yes | / | Yes | / | Yes | / | / | [59] |
| circ-ADAM9 | Up | / | Yes | / | / | Yes | Yes | Yes | / | [99] |
| circEYA3 | Up | / | Yes | / | / | / | / | Yes | CA19-9 | [114] |
| circPCDH10 | Up | / | Yes | Yes | Yes | / | Yes | Yes | / | [49] |
| circEIF6 | Up | Yes | Yes | / | / | / | / | Yes | / | [61] |
| circ_0128846 | Up | / | / | / | / | / | / | Yes | / | [62] |
| circNEK6 | Up | Yes | Yes | / | / | Yes | / | / | / | [69] |
| circCCT3 | Up | / | Yes | / | / | Yes | / | Yes | Peritoneal metastasis, Vascular invasion | [66] |
| hsa_circ_0074298 | Up | Yes | / | Yes | Yes | Yes | / | / | / | [56] |
| ciRS-7 | Up | / | / | / | / | Yes | / | / | Vascular invasion | [88] |
| circ_0075829 | Up | / | / | / | Yes | Yes | / | / | / | [92] |
| circ_0013912 | Up | / | / | / | / | Yes | / | Yes | / | [64] |
| circARFGEF2 | Up | / | Yes | / | / | Yes | / | / | / | [71] |
Apart from that, the aberrantly expression of circ-0005105 in PDAC can modulate the expression of collagen type XI alpha 1, which is conspicuously correlated with undesirable prognosis of PDAC, thereby lowering the tumorigenicity and metastasis of PDAC cells [96]. CircPTPRA, which is positively associated with lymph node invasion and dissatisfactory prognosis, strkingly pushes ahead the migration, invasion, proliferation and EMT of PDAC in vitro and in vivo [95]. Furthermore, circ-ASH2L facilitates PDAC invasion, proliferation and angiogenesis by aberrantly activating Notch signaling pathway, and may be a therapeutic target of PDAC [121]. Intriguingly, circEYA3 regulates energy production via ATP synthesis to accelerate PDAC progression, and circEYA3 may be an efficient molecular therapeutic target in PDAC [114]. It's critical to note that the hypoxic-induced exosomal circZNF91 can heighten deacetylation-dependent stability of HIF-1α protein. knockdown of circZNF91 retarded glycolysis PC cells [103]. Possible therapeutic target circUBAP2 can stabilize the expressions of CXC motif chemokine receptor type 4 and Zinc-finger E-box-binding homeobox 1, hold back antigen presentation, lower the infiltration and function of immune cells, and ultimately facilitate immune escape mechanisms of PC [126]. CircACTR2 overexpression retards GEM resistance in PC through activating the phosphatidylinositol-3-kinase/AKT signaling pathway [106].
In summary, the above findings clearly demonstrate that circRNAs play central roles in PC development, tumor microenvironment composition, metabolic changes, immune escape, and chemotherapy resistance. These studies persuasively prove the conjecture that the potential of circRNAs as therapeutic targets or new drugs for PC, but these viewpoints are mostly based on theoretical assumptions and basic experiments. On this basis, it is extremely significant to conduct more basic research and clinical studies to validate these ideas.
6. Summary and Prospects
As already confirmed by extensive studies, circRNAs play key regulatory roles in the occurrence and development of various tumors, including PC. As described in this review, circRNAs have been revealed to be involved in dissimilar biological processes of PC and have the potential as biomarkers and therapeutic targets for the diagnosis and prognosis of PC. Nonetheless, it is worthy of mentioning that the current research on circRNAs in PC is still in the early stage, and relevant studies have some limitations. First and foremost, there is no unified standard for the naming of circRNA at present, and unifying the naming rules of circRNA is beneficial for sharing research findings and accelerating the research progress of circRNA. Aside from that, the majority of current studies merely concentrate on the downstream molecular mechanism of circRNAs, while their attention is less fixed on how to explore its upstream molecular mechanism. For instance, the mechanisms of biogenesis and degradation of circRNAs as well as how circRNAs are exported from the nucleus to the cytoplasm. In such case, it's crucial to carry out further elucidation of the upstream molecular mechanism of circRNAs, which is evidently advantageous for us to deepen the understanding of circRNAs. Additionally, numerous studies have implicated that circRNAs regulate various biological processes of PC primarily by acting as miRNA sponges, while there are few studies on their biological functions by binding proteins and regulating gene transcription. Confusingly, there is currently no research on PC-specific circRNAs encoded peptides. Apart from that, the expression abundances of most circRNAs are much lower than that of miRNAs. As a result, it is essential to further elucidate other mechanisms by which circRNAs playing roles. To go it further, the detection of circRNAs at present are predominantly conducted in clinical tissue samples, and their expression profiles in various body fluid samples should also be taken seriously. Simultaneously, circRNAs in various clinical samples should be quantified, and more attention should be immersed in the specific expressions of circRNAs in PC. Finally, the strategy based on circRNAs for the treatment of PC is still in its infancy. It is paramount to figure out how to accurately overexpress or knock down circRNAs, how to accurately transport circRNAs to tumor tissues, and how to avoid possible immune rejection during delivery. To cope well with these problems will not only provide a brand-new viewpoint for clarifying the prominent role of circRNAs in PC biology, but also is advantageous for materializing the transformation of circRNAs from basic research to clinical application.
Supplementary Material
Supplementary table.
Acknowledgements
We would like to acknowledge the reviewers for their helpful comments on this paper.
Funding
This work was supported by National Natural Science Foundation of China (No. 82403212), Chongqing Technology Innovation and Application Development Special Key Project (No. CSTB2022TIAD-KPX0170), Chongqing Science and Health Joint Medical Research Major Project (No. 2024DBXM003), Natural Science Foundation of Chongqing Municipal (No. CSTB2024NSCQ-MSX0166) and Medical Research Foundation of Chongqing General Hospital (No. Y2023YXYJMSXM01).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Wagle NS. et al. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48
2. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12-49
3. Rahib L, Wehner MR, Matrisian LM. et al. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA network open. 2021;4:e214708
4. Bray F, Laversanne M, Sung H. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263
5. Halbrook CJ, Lyssiotis CA. et al. Pancreatic cancer: Advances and challenges. Cell. 2023;186:1729-1754
6. Li S, Wang J, Ren G. CircRNA: An emerging star in plant research: A review. Int J Biol Macromol. 2024;272:132800
7. Sanger HL, Klotz G, Riesner D. et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852-3856
8. Long F, Lin Z, Li L. et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. 2021;20:26
9. Kristensen LS, Jakobsen T, Hager H. et al. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188-206
10. Feng XY, Zhu SX, Pu KJ. et al. New insight into circRNAs: characterization, strategies, and biomedical applications. Experimental hematology & oncology. 2023;12:91
11. Pisignano G, Michael DC, Visal TH. et al. Going circular: history, present, and future of circRNAs in cancer. Oncogene. 2023;42:2783-2800
12. Chen H, Yang R, Xing L. et al. Hypoxia-inducible CircPFKFB4 Promotes Breast Cancer Progression by Facilitating the CRL4DDB2 E3 Ubiquitin Ligase-mediated p27 Degradation. Int J Biol Sci. 2022;18:3888-3907
13. Yang R, Chen H, Xing L. et al. Hypoxia-induced circWSB1 promotes breast cancer progression through destabilizing p53 by interacting with USP10. Mol Cancer. 2022;21:88
14. Chen H, Liu S, Li M. et al. circ_0003418 Inhibits Tumorigenesis And Cisplatin Chemoresistance Through Wnt/β-Catenin Pathway In Hepatocellular Carcinoma. Onco Targets Ther. 2019;12:9539-9549
15. Zheng X, Huang M, Xing L. et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19:73
16. Shi X, Yang J, Liu M. et al. Circular RNA ANAPC7 Inhibits Tumor Growth and Muscle Wasting via PHLPP2-AKT-TGF-β Signaling Axis in Pancreatic Cancer. Gastroenterology. 2022;162:2004-2017.e2
17. Han X, Fang Y, Chen P. et al. Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis. Cell Cycle. 2021;20:369-382
18. Liu J, Yuan W, Gong D. Hsa_circ_0000994 Inhibits Pancreatic Cancer Progression by Clearing Immune-Related miR-27a and miR-27b. J Oncol. 2022;2022:7274794
19. He Z, Cai K, Zeng Z. et al. Autophagy-associated circRNA circATG7 facilitates autophagy and promotes pancreatic cancer progression. Cell Death Dis. 2022;13:233
20. Zhao Z, Yang W, Kong R. et al. circEIF3I facilitates the recruitment of SMAD3 to early endosomes to promote TGF-β signalling pathway-mediated activation of MMPs in pancreatic cancer. Mol Cancer. 2023;22:152
21. Song R, Ma S, Xu J. et al. A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of Mtor. Mol Cancer. 2023;22:16
22. Chen LL. The biogenesis and emerging roles of circular RNAs. Nature reviews: molecular cell biology. 2016;17:205-211
23. Guo Y, Yang J, Huang Q. et al. Circular RNAs and their roles in head and neck cancers. Mol Cancer. 2019;18:44
24. Zhang XO, Wang HB, Zhang Y. et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-147
25. Zhang Y, Zhang XO, Chen T. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
26. Salzman J, Gawad C, Wang PL. et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733
27. Jeck WR, Sorrentino JA, Wang K. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157
28. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233-2247
29. Li H, Lan T, Liu H. et al. IL-6-induced cGGNBP2 encodes a protein to promote cell growth and metastasis in intrahepatic cholangiocarcinoma. Hepatology. 2022;75:1402-1419
30. Zheng Q, Bao C, Guo W. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215
31. Ma S, Kong S, Wang F. et al. CircRNAs: biogenesis, functions, and role in drug-resistant Tumours. Mol Cancer. 2020;19:119
32. Yang T, Shen P, Chen Q. et al. FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2021;40:261
33. Ivanov A, Memczak S, Wyler E. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170-177
34. Meng J, Chen S, Han JX. et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res. 2018;78:4150-4162
35. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829-1842
36. Pamudurti NR, Patop IL, Krishnamoorthy A. et al. circMbl functions in cis and in trans to regulate gene expression and physiology in a tissue-specific fashion. Cell Rep. 2022;39:110740
37. Conn SJ, Pillman KA, Toubia J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-1134
38. Xi Y, Shen Y, Wu D. et al. CircBCAR3 accelerates esophageal cancer tumorigenesis and metastasis via sponging miR-27a-3p. Mol Cancer. 2022;21:145
39. Shen Y, Zhang N, Chai J. et al. CircPDIA4 Induces Gastric Cancer Progression by Promoting ERK1/2 Activation and Enhancing Biogenesis of Oncogenic circRNAs. Cancer Res. 2023;83:538-552
40. Wang X, Chen T, Li C. et al. CircRNA-CREIT inhibits stress granule assembly and overcomes doxorubicin resistance in TNBC by destabilizing PKR. J Hematol Oncol. 2022;15:122
41. Wang P, Huang Z, Peng Y. et al. Circular RNA circBNC2 inhibits epithelial cell G2-M arrest to prevent fibrotic maladaptive repair. Nat Commun. 2022;13:6502
42. Lu Z, Filonov GS, Noto JJ. et al. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA (New York, N.Y.). 2015;21:1554-1565
43. Shen H, Liu B, Xu J. et al. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. Journal of hematology & oncology. 2021;14:134
44. Schmidt CA, Giusto JD, Bao A. et al. Molecular determinants of metazoan tricRNA biogenesis. Nucleic acids research. 2019;47:6452-6465
45. Xiong G, Feng M, Yang G. et al. The underlying mechanisms of non-coding RNAs in the chemoresistance of pancreatic cancer. Cancer Lett. 2017;397:94-102
46. Salmena L, Poliseno L, Tay Y. et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358
47. Hao S, Yao Z, Liu Y. Hsa_circ_0000106 Acts as a Tumor Promoter in Pancreatic Cancer by Targeting the MiR-455-3p/HDAC4. Horm Metab Res. 2023;55:722-732
48. Chen G, Shi Y, Zhang Y. et al. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017;10:5783-5794
49. Zhang S, Qiu M, Gao S. et al. Circular RNA PCDH10 regulates the tumorigenesis of pancreatic cancer through the miR-338-3p/hTERT axis. Am J Transl Res. 2021;13:2181-2197
50. Jiang Y, Wang T, Yan L. et al. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88-93
51. Shi Y, Shen M, Yang Y. et al. CircDUSP22 Overexpression Restrains Pancreatic Cancer Development via Modulating miR-1178-3p and Downstream BNIP3. Biochem Genet. 2023;61:651-668
52. Wong CH, Lou UK, Li Y. et al. CircFOXK2 Promotes Growth and Metastasis of Pancreatic Ductal Adenocarcinoma by Complexing with RNA-Binding Proteins and Sponging MiR-942. Cancer Res. 2020;80:2138-2149
53. Yuan H, Huang X, Li Q. et al. SiRNA-circFARSA-loaded porous silicon nanomaterials for pancreatic cancer treatment via inhibition of CircFARSA expression. Biomed Pharmacother. 2022;147:112672
54. Xu S, Lei SL, Liu KJ. et al. circSFMBT1 promotes pancreatic cancer growth and metastasis via targeting miR-330-5p/PAK1 axis. Cancer Gene Ther. 2021;28:234-249
55. Huang L, Han J, Yu H. et al. CircRNA_000864 Upregulates B-cell Translocation Gene 2 Expression and Represses Migration and Invasion in Pancreatic Cancer Cells by Binding to miR-361-3p. Front Oncol. 2020;10:547942
56. Chen H, Wang LS, Xie P. et al. Hsa_circ_0074298 promotes pancreatic cancer progression and resistance to gemcitabine by sponging miR-519 to target SMOC. J Cancer. 2022;13:34-50
57. Ling S, He Y, Li X. et al. CircRHOT1 mediated cell proliferation, apoptosis and invasion of pancreatic cancer cells by sponging miR-125a-3p. J Cell Mol Med. 2020;24:9881-9889
58. Hua S, Gao J, Li T. et al. The promoting effects of hsa_circ_0050102 in pancreatic cancer and the molecular mechanism by targeting miR-1182/NPSR1. Carcinogenesis. 2021;42:471-480
59. Ye Z, Zhu Z, Xie J. et al. Hsa_circ_0000069 Knockdown Inhibits Tumorigenesis and Exosomes with Downregulated hsa_circ_0000069 Suppress Malignant Transformation via Inhibition of STIL in Pancreatic Cancer. Int J Nanomedicine. 2020;15:9859-9873
60. Zhu P, Ge N, Liu D. et al. Preliminary investigation of the function of hsa_circ_0006215 in pancreatic cancer. Oncol Lett. 2018;16:603-611
61. Zhang T, Li M, Lu H. et al. Up-Regulation of circEIF6 Contributes to Pancreatic Cancer Development Through Targeting miR-557/SLC7A11/PI3K/AKT Signaling. Cancer Manag Res. 2021;13:247-258
62. Wang M, Li M, Liu Z. et al. Hsa_circ_0128846 knockdown attenuates the progression of pancreatic cancer by targeting miR-1270/NR3C1 axis. Sci Rep. 2023;13:2792
63. Hao L, Rong W, Bai L. et al. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J Cell Biochem. 2019;120:3780-3789
64. Guo W, Zhao L, Wei G. et al. Blocking circ_0013912 Suppressed Cell Growth, Migration and Invasion of Pancreatic Ductal Adenocarcinoma Cells in vitro and in vivo Partially Through Sponging miR-7-5p. Cancer Manag Res. 2020;12:7291-7303
65. Xu Y, Yao Y, Gao P. et al. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509:138-142
66. Hou JP, Men XB, Yang LY. et al. CircCCT3 Acts as a Sponge of miR-613 to Promote Tumor Growth of Pancreatic Cancer Through Regulating VEGFA/VEGFR2 Signaling. Balkan Med J. 2021;38:229-238
67. Wang KQ, Ye ML, Qiao X. et al. Circular RNA Fibroblast Growth Factor Receptor 1 Promotes Pancreatic Cancer Progression by Targeting MicroRNA-532-3p/PIK3CB Axis. Pancreas. 2022;51:930-942
68. Liu W, Deng L, Xu A. et al. Identifying a novel IRF3/circUHRF1/miR-1306-5p/ARL4C axis in pancreatic ductal adenocarcinoma progression. Cell Cycle. 2022;21:392-405
69. Shao Z, Chen X, Qiu H. et al. CircNEK6 promotes the progression of pancreatic ductal adenocarcinoma through targeting miR-503/CCND1 axis. Transl Oncol. 2024;39:101810
70. Shen X, Chen Y, Li J. et al. Identification of Circ_001569 as a Potential Biomarker in the Diagnosis and Prognosis of Pancreatic Cancer. Technol Cancer Res Treat. 2021;20:1533033820983302
71. Kong Y, Luo Y, Zheng S. et al. Mutant KRAS Mediates circARFGEF2 Biogenesis to Promote Lymphatic Metastasis of Pancreatic Ductal Adenocarcinoma. Cancer Res. 2023;83:3077-3094
72. Kong Y, Li Y, Luo Y. et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol Cancer. 2020;19:82
73. Li J, Li Z, Jiang P. et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177
74. Li Z, Wu YF, Li J. et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237-250
75. Xie H, Zhao Q, Yu L. et al. Circular RNA circ_0047744 suppresses the metastasis of pancreatic ductal adenocarcinoma by regulating the miR-21/SOCS5 axis. Biochem Biophys Res Commun. 2022;605:154-161
76. Zhang R, Zhu W, Ma C. et al. Silencing of circRNA circ_0001666 Represses EMT in Pancreatic Cancer Through Upregulating miR-1251 and Downregulating SOX4. Front Mol Biosci. 2021;8:684866
77. Hou YS, Li X. Circ_0005273 induces the aggravation of pancreatic cancer by targeting KLF12. Eur Rev Med Pharmacol Sci. 2020;24:11578-11586
78. Wong CH, Lou UK, Fung FK. et al. CircRTN4 promotes pancreatic cancer progression through a novel CircRNA-miRNA-lncRNA pathway and stabilizing epithelial-mesenchymal transition protein. Mol Cancer. 2022;21:10
79. Meng L, Zhang Y, Wu P. et al. CircSTX6 promotes pancreatic ductal adenocarcinoma progression by sponging miR-449b-5p and interacting with CUL2. Mol Cancer. 2022;21:121
80. Li L, Wang N, Wang J. et al. Hsa_circRNA_001859 regulates pancreatic cancer progression and epithelial-mesenchymal transition through the miR-21-5p/SLC38A2 pathway. Cancer Biomark. 2023;37:39-52
81. Shen P, Yang T, Chen Q. et al. CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing. Mol Cancer. 2021;20:51
82. Wang J, Zheng L, Hu C. et al. CircZFR promotes pancreatic cancer progression through a novel circRNA-miRNA-mRNA pathway and stabilizing epithelial-mesenchymal transition protein. Cell Signal. 2023;107:110661
83. Chen Y, Xu S, Liu X. et al. CircSEC24A upregulates TGFBR2 expression to accelerate pancreatic cancer proliferation and migration via sponging to miR-606. Cancer Cell Int. 2021;21:671
84. Shen Q, Zheng G, Zhou Y. et al. CircRNA circ_0092314 Induces Epithelial-Mesenchymal Transition of Pancreatic Cancer Cells via Elevating the Expression of S100P by Sponging miR-671. Front Oncol. 2021;11:675442
85. Li C, Cai J, Liu W. et al. Downregulation of circ-STK39 suppresses pancreatic cancer progression by sponging mir-140-3p and regulating TRAM2-mediated epithelial-mesenchymal transition. Apoptosis. 2023;28:1024-1034
86. Wang Y, Zhang J, Jia J. et al. Circ_0008768 Suppresses the Pancreatic Cancer Progression via miR-330- 3p/PTEN Axis. Protein Pept Lett. 2022;29:796-805
87. Yao J, Zhang C, Chen Y. et al. Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic cancer progression through miR-137-3p/PTN axis. Life Sci. 2019;239:116871
88. Liu L, Liu F.B, Huang M. et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;18:580-586
89. Wu X, Zhou S, Wang L. et al. Circ_103809 Aggravates the Malignant Phenotype of Pancreatic Cancer Through Modulating miR-197-3p/TSPAN3 Axis. Mol Biotechnol. 2024;66:2455-2466
90. Zhang H, Ma X, Wang L. et al. Circular RNA hsa_circ_0007367 promotes the progression of pancreatic ductal adenocarcinoma by sponging miR-6820-3p and upregulating YAP1 expression. Cell Death Dis. 2022;13:736
91. Xiong X, Feng J, Yang X. et al. Circular RNA CDR1as promotes tumor progression by regulating miR-432-5p/E2F3 axis in pancreatic cancer. Cancer Cell Int. 2021;21:112
92. Zhang X, Xue C, Cui X. et al. Circ_0075829 facilitates the progression of pancreatic carcinoma by sponging miR-1287-5p and activating LAMTOR3 signalling. J Cell Mol Med. 2020;24:14596-14607
93. Liu T, Zhou L, He Z. et al. Circular RNA hsa_circ_0006117 Facilitates Pancreatic Cancer Progression by Regulating the miR-96-5p/KRAS/MAPK Signaling Pathway. J Oncol. 2021;2021:9213205
94. Chi B, Zheng Y, Xie F. et al. Increased expression of miR-194-5p through the circPVRL3/miR-194-5p/SOCS2 axis promotes proliferation and metastasis in pancreatic ductal adenocarcinoma by activating the PI3K/AKT signaling pathway. Cancer Cell Int. 2022;22:415
95. Fu W, Wang X, Xiang J. et al. CircPTPRA promotes the progression of pancreatic ductal adenocarcinoma via the miR-140-5p/LMNB1 axis. Cancer Med. 2023;12:11651-11671
96. Ma G, Li G, Fan W. et al. Circ-0005105 activates COL11A1 by targeting miR-20a-3p to promote pancreatic ductal adenocarcinoma progression. Cell Death Dis. 2021;12:656
97. Qian X, Zong W, Ma L. et al. MM-associated circular RNA downregulates microRNA-19a through methylation to suppress proliferation of pancreatic adenocarcinoma cells. Bioengineered. 2022;13:9294-9300
98. Guo X, Zhou Q, Su D. et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol Cancer. 2020;19:83
99. Xing C, Ye H, Wang W. et al. Circular RNA ADAM9 facilitates the malignant behaviours of pancreatic cancer by sponging miR-217 and upregulating PRSS3 expression. Artif Cells Nanomed Biotechnol. 2019;47:3920-3928
100. Zheng S, Hu C, Lin H. et al. circCUL2 induces an inflammatory CAF phenotype in pancreatic ductal adenocarcinoma via the activation of the MyD88-dependent NF-κB signaling pathway. J Exp Clin Cancer Res. 2022;41:71
101. Han C, Zheng H, Hu D. et al. Hsa_circ_0007401 regulates gemcitabine resistance of pancreatic cancer through the hsa-miR-6509-3p/fli1 axis. Medicine (Baltimore). 2023;102:e33775
102. Xu L, Ma X, Zhang X. et al. hsa_circ_0007919 induces LIG1 transcription by binding to FOXA1/TET1 to enhance the DNA damage response and promote gemcitabine resistance in pancreatic ductal adenocarcinoma. Mol Cancer. 2023;22:195
103. Zeng Z, Zhao Y, Chen Q. et al. Hypoxic exosomal HIF-1α-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene. 2021;40:5505-5517
104. Hu C, Xia R, Zhang X. et al. circFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol Cancer. 2022;21:24
105. Chen ZW, Hu JF, Wang ZW. et al. Circular RNA circ-MTHFD1L induces HR repair to promote gemcitabine resistance via the miR-615-3p/RPN6 axis in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2022;41:153
106. Xu C, Ye Q, Ye C. et al. circACTR2 attenuates gemcitabine chemoresiatance in pancreatic cancer through PTEN mediated PI3K/AKT signaling pathway. Biol Direct. 2023;18:14
107. Chen M, Liu X, Lu J. et al. Dysregulation of the circ_0087502/miR-1179/TGFBR2 pathway supports gemcitabine resistance in pancreatic cancer. Cancer Biol Ther. 2023;24:2258566
108. Xu H, Chen R, Shen Q. et al. Overexpression of Circular RNA circ_0013587 Reverses Erlotinib Resistance in Pancreatic Cancer Cells Through Regulating the miR-1227/E-Cadherin Pathway. Front Oncol. 2021;11:754146
109. Yu S, Wang M, Zhang H. et al. Circ_0092367 Inhibits EMT and Gemcitabine Resistance in Pancreatic Cancer via Regulating the miR-1206/ESRP1 Axis. Genes (Basel). 2021;12:1701
110. Guan H, Luo W, Liu Y. et al. Novel circular RNA circSLIT2 facilitates the aerobic glycolysis of pancreatic ductal adenocarcinoma via miR-510-5p/c-Myc/LDHA axis. Cell Death Dis. 2021;12:645
111. Lin J, Wang X, Zhai S. et al. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J Hematol Oncol. 2022;15:128
112. Wang L, Wu X, Ruan Y. et al. Exosome-transmitted hsa_circ_0012634 suppresses pancreatic ductal adenocarcinoma progression through regulating miR-147b/HIPK2 axis. Cancer Biol Ther. 2023;24:2218514
113. Liu A, Xu J. Circ_03955 promotes pancreatic cancer tumorigenesis and Warburg effect by targeting the miR-3662/HIF-1α axis. Clin Transl Oncol. 2021;23:1905-1914
114. Rong Z, Shi S, Tan Z. et al. Circular RNA CircEYA3 induces energy production to promote pancreatic ductal adenocarcinoma progression through the miR-1294/c-Myc axis. Mol Cancer. 2021;20:106
115. Sun H, Liu F, Zhang H. Circ_0072008, an oncogene in pancreatic ductal adenocarcinoma, contributes to tumour cell malignant progression and glycolysis by regulating miR-545-3p/SLC7A11 axis. Autoimmunity. 2022;55:203-213
116. Wang Y, Zhang F, Wu D. et al. A novel circ_0099999/miR-330-5p/FSCN1 ceRNA crosstalk in pancreatic cancer. Autoimmunity. 2021;54:471-482
117. Zhou X, Liu K, Cui J. et al. Circ-MBOAT2 knockdown represses tumor progression and glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer. J Exp Clin Cancer Res. 2021;40:124
118. Rong Z, Xu J, Yang J. et al. CircRREB1 Mediates Metabolic Reprogramming and Stemness Maintenance to Facilitate Pancreatic Ductal Adenocarcinoma Progression. Cancer research. 2024;84:4246-4263
119. Yao X, Mao Y, Wu D. et al. Exosomal circ_0030167 derived from BM-MSCs inhibits the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/β-catenin axis. Cancer Lett. 2021;512:38-50
120. Zhao Q, Zhu Z, Xiao W. et al. Hypoxia-induced circRNF13 promotes the progression and glycolysis of pancreatic cancer. Exp Mol Med. 2022;54:1940-1954
121. Chen Y, Li Z, Zhang M. et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2019;38:466
122. Liu B, Gong Y, Jiang Q. et al. Hsa_circ_0014784-induced YAP1 promoted the progression of pancreatic cancer by sponging miR-214-3p. Cell Cycle. 2023;22:1583-1596
123. Liu X, Zhong L, Jiang W. et al. Repression of circRNA_000684 inhibits malignant phenotypes of pancreatic ductal adenocarcinoma cells via miR-145-mediated KLF5. Pancreatology. 2021;21:406-417
124. Zhang X, Tan P, Zhuang Y. et al. hsa_circRNA_001587 upregulates SLC4A4 expression to inhibit migration, invasion, and angiogenesis of pancreatic cancer cells via binding to microRNA-223. Am J Physiol Gastrointest Liver Physiol. 2020;319:G703-G717
125. Zhang J, Li J, Xiong Y. et al. Circ_0000284 upregulates RHPN2 to facilitate pancreatic cancer proliferation, metastasis, and angiogenesis through sponging miR-1179. J Biochem Mol Toxicol. 2023;37:e23274
126. Zhao R, Ni J, Lu S. et al. CircUBAP2-mediated competing endogenous RNA network modulates tumorigenesis in pancreatic adenocarcinoma. Aging (Albany NY). 2019;11:8484-8501
127. Ou ZL, Luo Z, Wei W. et al. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: role of circ_0000977/miR-153 axis. RNA Biol. 2019;16:1592-1603
128. He Y, Han P, Chen C. et al. circPTPN22 attenuates immune microenvironment of pancreatic cancer via STAT3 acetylation. Cancer Gene Ther. 2023;30:559-566
129. Fu X, Sun G, Tu S. et al. Hsa_circ_0046523 Mediates an Immunosuppressive Tumor Microenvironment by Regulating MiR-148a-3p/PD-L1 Axis in Pancreatic Cancer. Front Oncol. 2022;12:877376
130. Guan H, Tian K, Luo W. et al. m6A-modified circRNA MYO1C participates in the tumor immune surveillance of pancreatic ductal adenocarcinoma through m6A/PD-L1 manner. Cell Death Dis. 2023;14:120
131. Song Y, Wang J, Xu J. et al. Circ_0018909 knockdown inhibits the development of pancreatic cancer via the miR-545-3p/FASN axis and reduces macrophage polarization to M2. J Biochem Mol Toxicol. 2023;37:e23293
132. Gao G, Wang L, Li C. Circ_0006790 carried by bone marrow mesenchymal stem cell-derived exosomes regulates S100A11 DNA methylation through binding to CBX7 in pancreatic ductal adenocarcinoma. Am J Cancer Res. 2022;12:1934-1959
133. Lin L, Xiao L, Jin C. et al. Circ_0058058 Drives the Malignant Phenotypes and Immune Evasion of Pancreatic Cancer by the MicroRNA-557-Dependent Regulation of PDL1. Pancreas. 2022;51:1444-1454
134. Wang S, Liu D, Wei H. et al. The hsa_circRNA_102049 mediates the sorafenib sensitivity of hepatocellular carcinoma cells by regulating Reelin gene expression. Bioengineered. 2022;13:2272-2284
135. Qin H, Ni H, Liu Y. et al. RNA-binding proteins in tumor progression. J Hematol Oncol. 2020;13:90
136. Liu Y, Ao X, Yu W. et al. Biogenesis, functions, and clinical implications of circular RNAs in non-small cell lung cancer. Mol Ther Nucleic Acids. 2021;27:50-72
137. Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-264
138. Li X, Zhang JL, Lei YN. et al. Linking circular intronic RNA degradation and function in transcription by RNase H1. Sci China Life Sci. 2021;64:1795-1809
139. Kong R, Wei W, Man Q. et al. Hypoxia-induced circ-CDYL-EEF1A2 transcriptional complex drives lung metastasis of cancer stem cells from hepatocellular carcinoma. Cancer Lett. 2023;578:216442
140. Ma J, Du WW, Zeng K. An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol Ther. 2021;29:2754-2768
141. Shang Q, Yang Z, Jia R. et al. 2019 The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6
142. Pamudurti NR, Bartok O, Jens M. et al. Translation of CircRNAs. Mol Cell. 2017;66:9-21.e7
143. Qian L, Yu S, Chen Z. et al. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer. 2018;1870:247-260
144. Zhang M, Huang N, Yang X. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805-1814
145. Wang X, Jian W, Luo Q. et al. CircSEMA4B inhibits the progression of breast cancer by encoding a novel protein SEMA4B-211aa and regulating AKT phosphorylation. Cell Death Dis. 2022;13:794
146. Xiong L, Liu HS, Zhou C. et al. A novel protein encoded by circINSIG1 reprograms cholesterol metabolism by promoting the ubiquitin-dependent degradation of INSIG1 in colorectal cancer. Mol Cancer. 2023;22:72
147. Yang Y, Fan X, Mao M. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-641
148. Zhong J, Wu X, Gao Y. et al. Circular RNA encoded MET variant promotes glioblastoma tumorigenesis. Nat Commun. 2023;14:4467
149. Chu X, Tian W, Ning J. et al. Cancer stem cells: advances in knowledge and implications for cancer therapy. Signal Transduct Target Ther. 2024;9:170
150. Zeng S, Pöttler M, Lan B. et al. Chemoresistance in Pancreatic Cancer. Int J Mol Sci. 2019;20:4504
151. Tao J, Yang G, Zhou W. et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J Hematol Oncol. 2021;14:14
152. Liu ZL, Chen HH, Zheng LL. et al. Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal Transduct Target Ther. 2023;8:198
153. Shi X, Wang M, Zhang Y. et al. Hypoxia activated HGF expression in pancreatic stellate cells confers resistance of pancreatic cancer cells to EGFR inhibition. EBioMedicine. 2022;86:104352
154. Zheng S, Tian Q, Yuan Y. et al. Extracellular vesicle-packaged circBIRC6 from cancer-associated fibroblasts induce platinum resistance via SUMOylation modulation in pancreatic cancer. Journal of experimental & clinical cancer research: CR. 2023;42:324
155. Loh JJ, Ma S. Hallmarks of cancer stemness. Cell stem cell. 2024;31:617-639
156. Qu S, Yang X, Li X. et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141-148
157. De Fraipont F, Gazzeri S, Cho WC. et al. Circular RNAs and RNA Splice Variants as Biomarkers for Prognosis and Therapeutic Response in the Liquid Biopsies of Lung Cancer Patients. Front Genet. 2019;10:390
158. Yang X, Ye T, Liu H. et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol Cancer. 2021;20:4
159. Wang T, Shigdar S, Shamaileh HA. et al. Challenges and opportunities for siRNA-based cancer treatment. Cancer Lett. 2017;387:77-83
160. Frazier KS. Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist's perspective. Toxicol Pathol. 2015;43:78-89
161. Zhang Y, Xue W, Li X. et al. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15:611-624
Author contact
![]() Corresponding author: Institute of Hepatopancreatobiliary Surgery, Chingqing General Hospital, Chongqing University, Chongqing, 401147, China. E-mail addresses: 1041523161com (Hang Chen), Becihamcom (Pijiang Sun), whuaizhicom (Huaizhi Wang).
Corresponding author: Institute of Hepatopancreatobiliary Surgery, Chingqing General Hospital, Chongqing University, Chongqing, 401147, China. E-mail addresses: 1041523161com (Hang Chen), Becihamcom (Pijiang Sun), whuaizhicom (Huaizhi Wang).

 Global reach, higher impact
Global reach, higher impact