Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(6):1363-1374. doi:10.7150/ijms.106867 This issue Cite
Research Paper
LncRNA FTX accelerates the progression of hepatocellular carcinoma by FTX/miR-374a-3p/HMGB1 pathway
1. Department of Infectious Diseases, Hunan Key Laboratory of Viral Hepatitis, Xiangya Hospital, Central South University, Changsha, China.
2. National Clinical Research Centre for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China.
Received 2024-11-3; Accepted 2025-1-22; Published 2025-2-18
Abstract
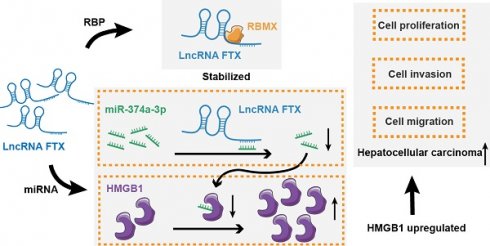
The situation regarding Hepatocellular carcinoma (HCC) is severe, with high incidence and mortality rates worldwide. Abnormal expression of long noncoding RNAs (LncRNAs) has been implicated in the progression of malignant tumors. Although there are reports on LncRNA FTX (Lnc-FTX) in HCC, the findings are still contradictory, leaving its role unclear. This research aims to examine the relationship between Lnc-FTX expression and clinical prognosis in HCC, assess its impact on HCC cell biological functions, and elucidate the underlying mechanisms involved. Our findings demonstrate that patients in the high-expression group exhibit more severe TNM staging and poorer overall survival (OS) rates based on data from HCC patients in cohort 1 (n=27). Lnc-FTX expression is upregulated according to both the GSE 77314 and TCGA databases. This suggests that Lnc-FTX may serve as a potential prognostic biomarker for HCC. Overexpressed level of Lnc-FTX promotes proliferation, migration, and invasion while reducing the apoptotic rate in Hep3B and MHCC-LM3 cells. Conversely, the knockdown of Lnc-FTX yields opposite effects. RNA pulldown assay and mass spectrometry reveal that Lnc-FTX combines with RNA binding motif protein X-linked (RBMX), which in turn enhances the RNA stability of Lnc-FTX. Additionally, Lnc-FTX can sponge miR-374a-3p, thereby targeting High mobility group box 1 protein (HMGB1). In summary, high expression of Lnc-FTX possesses clinical value in predicting poor prognosis in HCC. As an oncogene, it promotes the malignant functions of HCC through the RBMX/Lnc-FTX interaction and the Lnc-FTX/miR-374a-3p/HMGB1 signaling pathway.
Keywords: Lnc-FTX, hepatocellular carcinoma, RBMX, HMGB1, miR-374a-3p
Introduction
Primary hepatic cancer (PHC) is the sixth most commonly diagnosed cancer, and ranks as the third leading cause of cancer-related deaths globally in 2022, accounting for 866,136 new cases and 758,725 deaths [1,2]. HCC, the predominant type of PHC, constitutes about 75%-85% [3]. The difficulty in early diagnosis of HCC, along with its high recurrence and metastasis rates, is the main reason for high mortality [4,5]. For early-stage HCC patients (stage 0 or stage A), curative treatment options include liver transplantation, surgical resection, and radiofrequency ablation [6]. For advanced HCC, systemic therapy is recommended. First-line agents consist of small molecule inhibitors such as Sorafenib and Lenvatinib [7,8], while second-line treatment options include immune checkpoint inhibitors, such as Nivolumab (PD-1) [9]. Additionally, epigenetic therapies are under further investigation, including DNA methyltransferase inhibitors (zebularine) and miRNA-targeted therapies (miR-124) [6]. Currently, emerging research suggests that LncRNAs play a role in HCC and may serve as novel therapeutic targets. SiHSAL3 nanoparticles loaded with oncogenic lncRNA HSAL3 inhibitors could be a potential therapeutic drug for HCC [10].
LncRNAs are a group of non-coding RNA transcripts (> 200 nt) [11], which are now recognized as epigenetic regulators that play a role in tumor malignant processes, such as proliferation [12], invasion and metastasis [13], as well as metabolism [14]. Our team's preliminary research highlights the significant role of lncRNA in HCC. SEMA 6A-AS1 has been identified as a potential prognostic biomarker for HBV-related HCC [15]. LncRNA TP73-AS1 functions as an oncogene in HCC by promoting M2 macrophage infiltration via the miR-539/MMP-8/TGF-β1 signaling pathway [16] and regulates HCC cell proliferation through the miR-200a/HMGB1 pathway [17].
Lnc-FTX is derived from the FTX gene, a highly conserved gene, which is located on the human X chromosome q13.2.[18] Originally discovered as an important regulator of X-chromosome inactivation, it can regulate X-inactive-specific Transcript (XIST) activation and expression. Subsequently, numerous studies have demonstrated up-regulation of Lnc-FTX in diverse cancers, including gastric cancer [19], colorectal cancer [20], and other digestive cancers. The research on Lnc-FTX in HCC remains controversial. In the study by Liu F, et al., Lnc-FTX acts as a tumor suppressor gene in HCC by inhibiting epithelial-mesenchymal transition (EMT) and invasion of HCC cells through the miR-374a-5p/Wnt/β-catenin axis [21] and regulating the M1/M2 polarization of Kupffer cells [22], and inhibits cell proliferation by binding with MCM2 [23]. However, other studies have shown that Lnc-FTX expression is upregulated in HCC, where it contributes to aerobic glycolysis via the PPARγ pathway [24] and promotes HCC progression through the miR-545/RIG-I/PI3K/Akt axis [25]. The abnormal expression of Lnc-FTX exhibits tissue heterogeneity, and the molecular mechanism of tumor regulation needs to be further investigated.
HMGB1 has been extensively reported to exhibit pro-carcinogenic effects [26]. It can promote tumor invasion, metastasis [27], and proliferation [28] by interacting with receptors (such as RAGE and TLRs), thereby activating downstream signaling pathways including NF-κB/IL-6 [29], caspase-1 [30], PI3K/AKT [31] and cyclin D1 [32]. Additionally, nuclear HMGB1 inhibits apoptosis by regulating heat shock protein β-1 [33]. This study hypothesizes that HMGB1 is a potential target for FTX in regulating HCC.
In this research, the expression of Lnc-FTX in tumor and adjacent non-tumor liver tissues from HCC patients were examined, and its impact on the malignant phenotype of HCC cells was investigated. The potential molecular mechanisms were explored by focusing on RNA-binding proteins (RBPs) and microRNAs that bind to Lnc-FTX. Our study identifies Lnc-FTX as a potential prognostic biomarker and a therapeutic target, thereby enhancing our understanding of the developmental mechanisms underlying HCC.
Material and Methods
Additional methodologies are listed in Supplementary Material and Methods Doc S1.
Tissue samples and public databases
Between 2016 and 2021, 27 paired HCC samples and their respective adjacent non-tumor tissues were obtained from tumor surgeries conducted at the Third Xiangya Hospital (Table S1) (HCC cohort 1). All samples were collected strictly in accordance with standard procedures, and stored at -80°C. FTX transcript level data for 371 HCC cases from the The Cancer Genome Atlas (TCGA) database were obtained online from The University of Alabama at Birmingham Cancer data analysis Portal (UALCAN) and Lnc-FTX expression data for 50 HCC patients were obtained from Gene Expression Omnibus (GEO) database.
Cell culture
MHCC-LM3, Hep3B, MHCC97H, MHCC97L (HCC cell lines) and HEK-293T (embryonic kidney cell line) cells were cultivated in Dulbecco's Modified Eagle Medium (Gibco, USA). HepG2 cells were cultivated in MEM (Gibco, USA). All the cell lines were obtained from the American Type Culture Collection. L02 (the hepatocyte cell line) was purchased from the Culture Collection of the Chinese Academy of Sciences and was maintained in RPMI 1640 medium (Gibco, USA). All cells were cultured in complete medium (basal medium + 10% Fetal Bovine Serum (Gibco, USA) + 1% penicillin-streptomycin (Gibco, USA)). The cell incubator was set to 5% CO2 and 37 °C.
RNA pulldown assays
Lnc-FTX RNA was obtained by transcription of the FTX gene template containing the T7 promoter using the TranscriptAid T7 High Yield Transcription Kit (K0441, Thermo, USA). Lnc-FTX RNA pulldown was conducted using the RNA-protein PullDown kit (20164, Thermo, USA). Subsequently, biotin-labelled Lnc-FTX was co-incubated with streptavidin magnetic beads for 30 min (room temperature) and further co-incubated with whole cell lysate for 1 hour at 4°C. Following washes with wash buffer for 3 times, proteins were pulled down by the probe-encapsulated magnetic beads. Finally, the eluted proteins samples were analyzed by nanoLC-MS/MS (BiOTREE, China). We performed Tagged RNA affinity purification (TRAP) assay using the TRAP kit (BersinBio, GuangZhou, China) to detect microRNAs that bind to Lnc-FTX. All the relative primers were detailed in Table S2.
Dual-luciferase reporter assay
Luciferase reporter plasmids should be prepared in advance. Cells were seeded into the 24-well plates (5×104/well). Then the miR-374a-3p or NC mimics were transfected into the cells. Collected cell lysates were used to detect the activities of Renilla and firefly luciferase (RG027, Beyotime, China). The absorbance value was detected using a PerkinElmer spectrophotometer.
RNA stability assay
Cells were treated with actinomycin D (sigma, USA) after transfection with shNC or shRNA-RBMX. Cells were collected at 0, 4 and 8h after actinomycin D treatment, followed by RT-PCR analysis to measure the expression level of Lnc-FTX. The half-life (t1/2) of Lnc-FTX was determined as the time when the expression of Lnc-FTX decreased to 50% after treatment with actinomycin D. The decay rate of Lnc-FTX was determined by non-linear regression curve fitting.
Statistical analysis
For categorical variables between the two groups, P values were statistically assessed using the χ2 test, while Student's t-test was applied for quantitative data. ANOVA was utilized for comparisons involving more than two groups. Statistical values were analyzed and presented using GraphPad Prism 9.0 (CA, USA). Data with p value < 0.05 was considered as statistically significant.
Results
The overexpression of Lnc-FTX is linked to poor prognosis in HCC
To explore the expression of Lnc-FTX in HCC, RT-PCR experiments were performed on tumor tissues and adjacent non-tumor tissues collected from 27 HCC patients. The results showed no significant difference between tumor and adjacent non-tumor tissue. (Figure 1A). Based on data from GEO GSE77314 (Figure 1B) and TCGA (Figure 1C), we observed that Lnc-FTX is significantly upregulated in HCC tissues. Remarkably, the high Lnc-FTX group exhibited significantly poorer OS compared to the low Lnc-FTX group (P<0.05) (Figure 1D). Additionally, clinicopathological analysis showed that patients in high Lnc-FTX group exhibited higher TNM stage (Figure 1E), with detailed data presented in Table 1. Furthermore, expression levels of Lnc-FTX were elevated in HCC cell lines, including MHCC-97L, HepG2, MHCC-97H, Hep3B (Figure 1F). In summary, overexpression of Lnc-FTX was associated with higher TNM stage and poor clinical prognosis.
Association of Lnc-FTX with clinicopathologic characteristics of patients with hepatocellular carcinoma.
| Variable | Lnc-FTX expression level | p | ||
|---|---|---|---|---|
| High (n=11) | Low (n=16) | |||
| Age group | ≤55 | 5 | 9 | 0.704 |
| >55 | 6 | 7 | ||
| Sex | Male | 9 | 15 | 0.549 |
| Female | 2 | 1 | ||
| Edmondson grade | II | 7 | 12 | 0.452 |
| II-Ⅲ | 3 | 4 | ||
| Ⅲ | 1 | 0 | ||
| TNM classification | I | 4 | 9 | 0.027* |
| II | 4 | 0 | ||
| Ⅲ | 1 | 6 | ||
| Ⅳ | 2 | 1 | ||
| Tumor size (cm) | ≤5 | 6 | 7 | 0.704 |
| >5 | 5 | 9 | ||
| Tumors | Multiple | 5 | 7 | >0.999 |
| Solitary | 6 | 9 | ||
| BCLC stage | 0+A | 1 | 1 | >0.999 |
| B+C | 11 | 15 | ||
| Vascular invasion | Yes | 1 | 1 | >0.999 |
| No | 10 | 15 | ||
| Capsular invasion | Yes | 4 | 5 | >0.999 |
| No | 7 | 11 | ||
| Cirrhosis, No. (%) | Yes | 9 | 7 | 0.109 |
| No | 2 | 9 | ||
* TNM = tumor node metastasis; BCLC = Barcelona Clinic liver cancer. The low expression group included 16 patients whose Lnc-FTX expression in tumor tissues was < or equal to the normal tissues. The high expression group included 11 patients whose Lnc-FTX expression in tumor tissues was > the normal tissues. The Pearson χ2 test and Chi-Squared Test were used for the analysis of the correlation between Lnc-FTX expression levels and clinical characteristics. *P < 0.05
Lnc-FTX promotes cell proliferation and inhibits apoptosis in HCC
High Lnc-FTX-expressing LM3 and Hep3B cell clines were selected to study the function of FTX through overexpression or knockout. We utilized MHCC-LM3 and Hep3B cell lines to construct stable cell lines. Among the various ShRNA constructs, ShRNA2-Lnc-FTX was selected for subsequent experiments due to its highest inhibition efficiency compared to ShRNA1-Lnc-FTX through ShRNA2-Lnc-FTX (Figure S1). We then established cell lines with either downregulated or overexpressed Lnc-FTX in both MHCC-LM3 and Hep3B, and confirmed the expression levels using RT-PCR (Figure S1). To evaluate the influence of Lnc-FTX on cell proliferation, Cell counting kit-8 (CCK-8) and colony formation assays were performed. We observed that cell proliferation (Figure 2A-D) and colony formation (Figure 2E-G) were considerably decreased upon Lnc-FTX knockdown, whereas both were markedly increased with Lnc-FTX overexpression. Further, we conducted flow cytometry analyses on cell cycle distribution and apoptosis. The Proliferation Index (PI) refers to the proportion of cells in the S phase and G2/M phase relative to the total number of cells. In Figure 2, the total percentage of cells in the S phase and G2/M phase in FTX knockdown cell line (shRNA2-Lnc-FTX) is lower than that of the control group (sh NC) (Figure 2H-J). The DNA content of cells in the S phase and G2/M phase exceeds the diploid amount, indicating that the cells have entered the next round of division. The reduced percentage of cells in these phases suggests that the proliferation index and proliferation activity decrease after FTX knockdown. In FTX overexpression cell line, the total percentage of cells in the S phase and G2/M phase is higher than that of the control group (Figure 2K-M), indicating that FTX overexpression increases the proliferation index and enhances proliferation activity. These results demonstrate that FTX enhances the proliferation ability of HCC cells, while FTX knockdown suppresses proliferation. Additionally, Lnc-FTX knockdown led to increased apoptosis in both MHCC-LM3 and Hep3B cells (Figure 2N), while Lnc-FTX overexpression resulted in reduced apoptosis (Figure 2O). Overall, Lnc-FTX promotes cell proliferation in HCC by accelerating the cell cycle and inhibiting cell apoptosis.
Lnc-FTX promotes invasion and migration of HCC cells
The wound healing assays established that Lnc-FTX knockdown significantly impaired the healing rate of cells, while upregulation of Lnc-FTX significantly accelerated it (Figure 3A, B). The Transwell assays demonstrated that, compared to their respective control cell lines, the shRNA2-FTX cell line had fewer cells passing through the Transwell chamber, whereas the cell lines with upregulated Lnc-FTX had more cells migrating through the chamber (Figure 3C, D). The upper chamber of the Transwell was coated with a layer of Matrigel for the same experimental procedure to investigate the effect of FTX on the invasion ability of HCC cells, which is consistent with the previous results (Figure 3C, D). All these findings conclude that Lnc-FTX enhances HCC cell invasion and migration.
The overexpression of Lnc-FTX is related to the poor clinical prognosis of HCC patients. (A) Lnc-FTX levels in 27 pairs HCC and adjacent tissues were measured by RT-PCR; (B) Lnc-FTX is significantly upregulated in 50 HCC tissues compared with the matched adjacent normal liver tissues obtained from GEO database GSE77314; (C) Lnc-FTX expression level is higer in HCC (n = 371) than in the normal liver tissues (n = 50) obtained from TCGA (UCLCAN); (D) Kaplan-Meier survival analysis suggests that HCC patients with higher Lnc-FTX levels had worse overall survival (OS) than those with lower lnc-FTX expression; (E) Chi-Squared Test suggests that HCC patients with higher Lnc-FTX levels had higher TNM classification than those with lower Lnc-FTX expression; (F) LncRNA FTX expression was detected by RT-PCR in normal human hepatocytes L02 and HCC cell lines, including MHCC-97L, HepG2, MHCC-97H, Hep3B, MHCC-LM3.
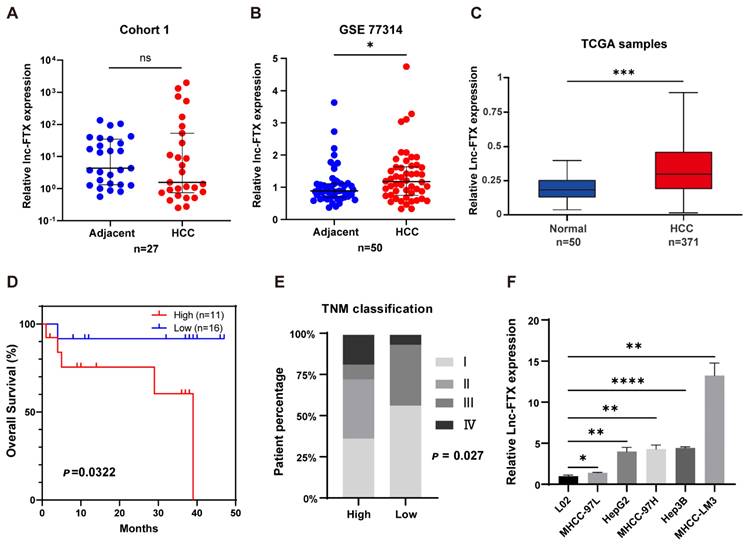
The effect of Lnc-FTX on HCC cell proliferation and apoptosis. (A-D) CCK8 assay was performed to measure the cell viability;(E-G) MHCC-LM3 cells and Hep3B cells, transfected with shRNA2-Lnc-FTX/shNC and vector/vector-Lnc-FTX, were cultured for 2 weeks to allow colony formation; (H-J) Cell cycle distribution of shRNA2-Lnc-FTX or sh NC transfected MHCC-LM3 Hep3B was evaluated by flow cytometry. The percentage of cells in each phase were shown; (K-M) Cell cycle distribution of pc-vector or pc-FTX transfected MHCC-LM3 and Hep3B was evaluated by flow cytometry. The percentage of cells in each phase were shown; (N-O) The apoptotic level of HCC cells was determined by flow cytometry.
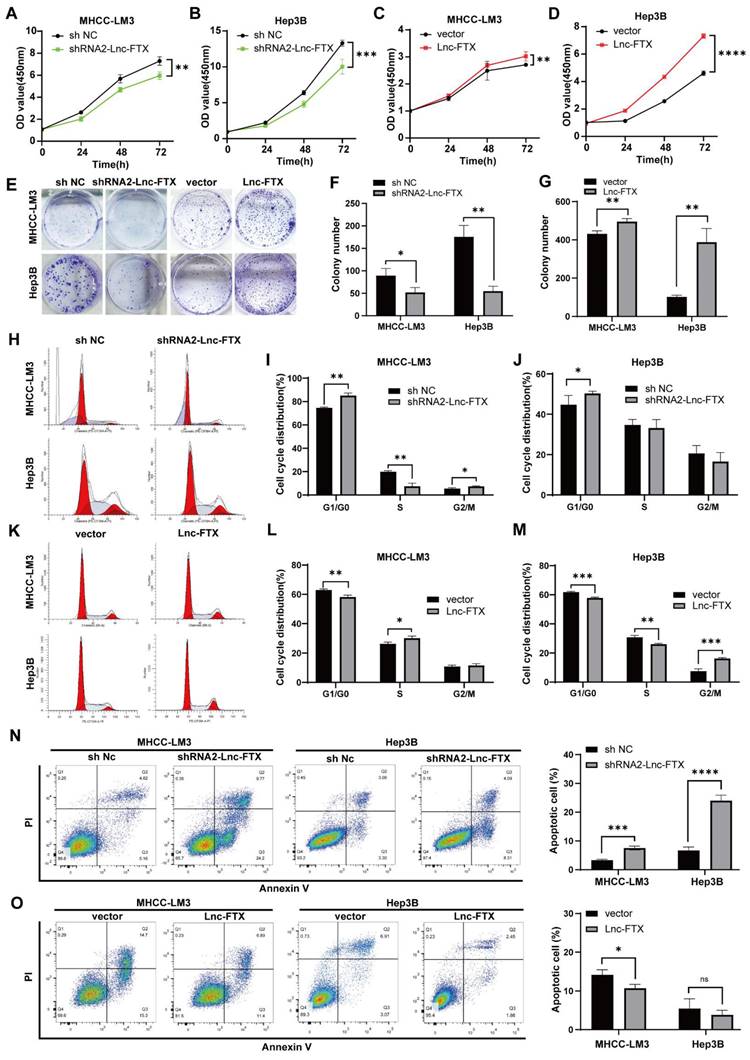
Lnc-FTX expression promotes migration and invasion in HCC. (A, B) The expression of Lnc-FTX was silenced treated with shRNAs and exogenously overexpressed in MHCC-LM3 and Hep3B cells. Wound-healing assays showed the increased movement capacity of HCC cells in-duced by Lnc-FTX; (C, D) Transwell migration and invasion assays demonstrated the effect of Lnc-FTX on the migration and invasion of HCC cells. Cells were stained using 0.1% crystal violet. The representative images and the corresponding statistical results were shown.
RBMX interacts with Lnc-FTX to remain its RNA stability. (A, B) RNA pulldown was performed using biotin-labeled Lnc-FTX probe, followed by mass spectrometry analysis; (C) Western Blot assay was performed to verify the interaction between Lnc-FTX and RBMX in HepG2 cells. (D) The mRNA level of RBMX in HCC (n = 369) and adjacent normal (n = 50) tissues was analyzed using TCGA database; (E) Kaplan-Meier analysis (log-rank test) of overall survival according to RBMX expression level (P < 0.01) (GEPIA); (F) Lnc-FTX expression level of MHCC-LM3 transfected with sh-NC or shRNA-RBMX was detected by RT-PCR analysis; (G, H) Measurement of Lnc-FTX RNA stability. MHCC-LM3 and Hep3B cells transfected with sh-NC or shRNA-RBMX were treated with actinomycin D to block de novo transcription and the levels of Lnc-FTX were assessed by RT-PCR analysis.
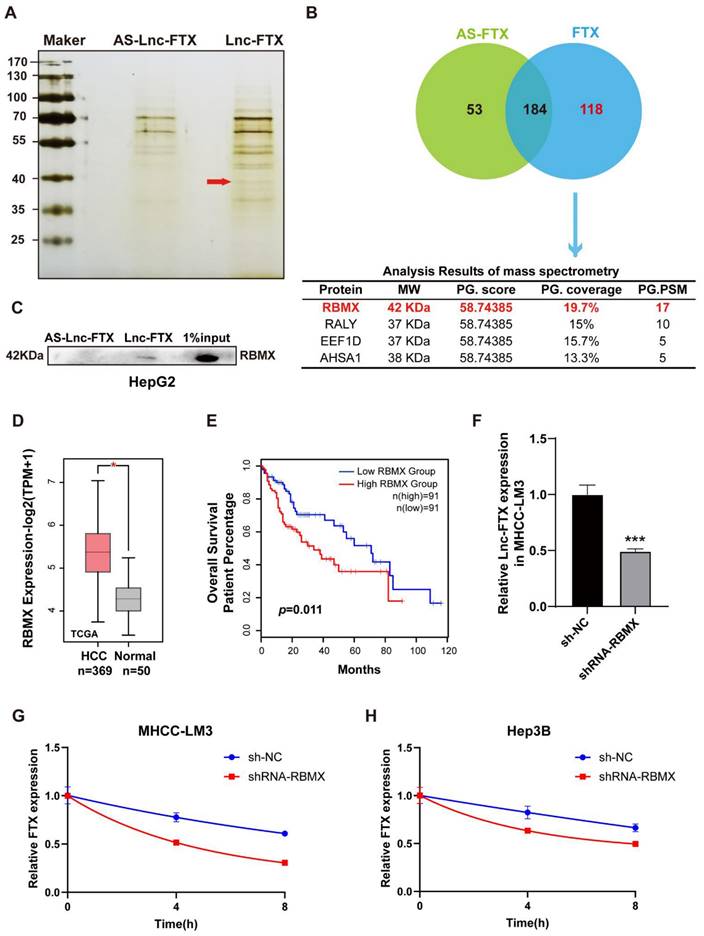
RNA-binding protein RBMX enhances the stability of Lnc-FTX
To explore the molecular mechanisms by which FTX regulates HCC, RNA pulldown assays (Figure 4A) were conducted to obtain candidate RBPs that directly interact with Lnc-FTX. The results revealed that 118 proteins with a molecular weight of approximately 40 kDa may specifically bind to Lnc-FTX. Among these proteins, RBMX emerged as the top candidate with the highest score (Figure 4B), thought to be crucial in functions such as alternative splicing [34] and maintaining genome stability [35, 36]. Western blot analysis confirmed that RBMX existed in the pulldown protein of Lnc-FTX but not in the pulldown protein of the antisense of Lnc-FTX (Figure 4C), indicating that Lnc-FTX can directly bind to RBMX. Meanwhile, the expression level of RBMX in HCC tissues was markedly upregulated with the data from the TCGA database (Figure 4D). Moreover, the upregulated of RBMX is associated with the lower OS rates in HCC patients (Figure 4E), suggesting a potential tumor-promoting role of RBMX in HCC. RBMX knockout cells were constructed by using shRNA to explore its effect on Lnc-FTX. (Figure S2). Our findings revealed that knockdown of RBMX decreased Lnc-FTX expression (Figure 4F). Additionally, RNA stability assays demonstrated an accelerated decay rate of Lnc-FTX after RBMX knockdown (Figure 4G, H). These results collectively confirm RBMX can bind and stabilize Lnc-FTX to accelerates the progression of HCC.
Lnc-FTX serves as a miRNA sponge miR-374-3p in HCC cells
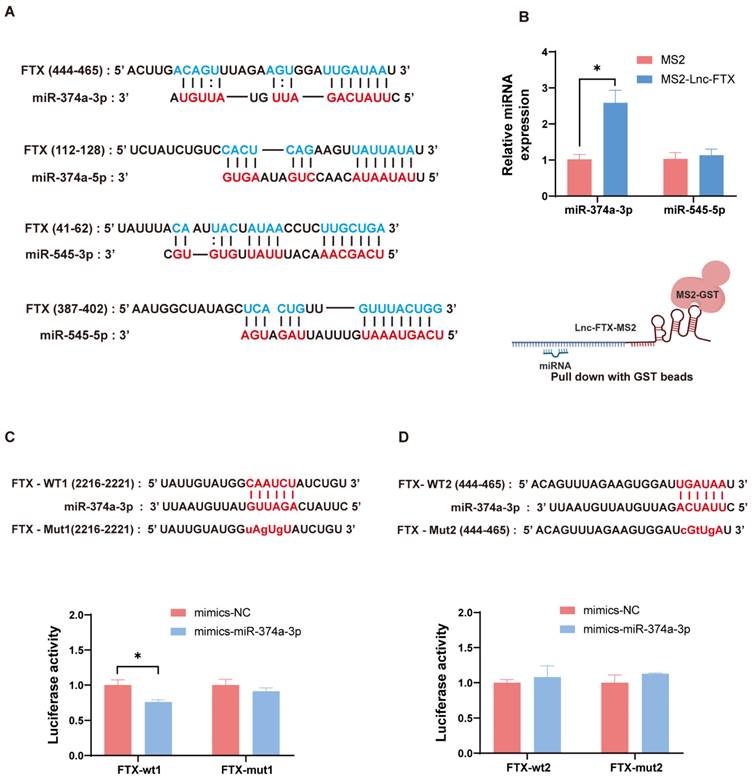
To further investigate the downstream molecular mechanisms by which FTX promotes HCC progression, we approached the study from the competitive endogenous RNA (ceRNA) hypothesis for subsequent research. We utilized the prediction website LncBase V.2 (https://dianalab.e-ce.uth.gr) and found the binding sites between Lnc-FTX and miR-374a-3p, miR-374a-5p, miR-545-3p, and miR-545-5p. (Figure 5A). Next, we conducted a TRAP assay, and observed that miR-374a-3p was enriched in the pulldown samples of Lnc-FTX, while miR-545-5p showed no notable difference between the MS2-Lnc-FTX group and the control group (Figure 5B). Other miRNAs were undetectable. These findings indicate that Lnc-FTX can bind to miR-374a-3p. In addition, we constructed dual-luciferase gene reporter plasmids (PmiRGLO-Lnc-FTX-WT1/WT2) and their corresponding mutant plasmids (PmiRGLO-Lnc-FTX-Mut1/Mut2) for the Dual-luciferase reporter assay. We co-transfected the mentioned reporter plasmids in MHCC-LM3 cells with mimics NC and mimics miR-374a-3p, respectively. In FTX-WT1 transfected cells, the luciferase signal of the mimics miR-374a-3p group exhibited a significant decrease compared to mimics NC group. However, this effect was disappeared in FTX-mut1 transfected cells (Figure 5C). Conversely, there was no notable disparity in luciferase signal in FTX-WT2 transfected cells (Figure 5D). In conclusion, we demonstrate that Lnc-FTX binds to miR-374a-3p at FTX-WT1 site and functions as a sponge.
Lnc-FTX upregulates HMGB1 through miR-374a-3p
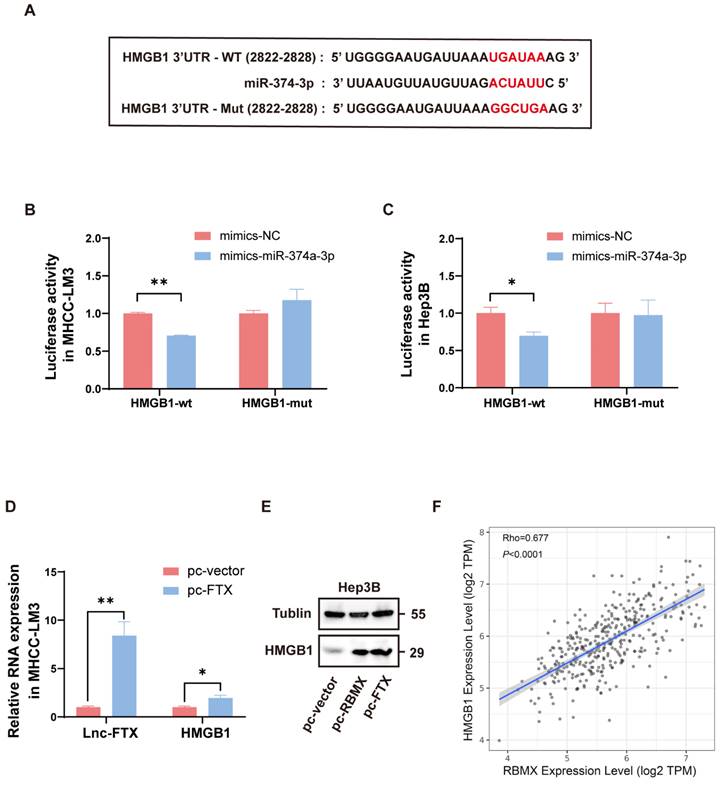
We predicted that there were binding sites between miR-374a-3p and HMGB1 3'UTR by analyzing online database TargetScan (http://www.targetscan.org/) (Figure 6A). Dual-luciferase reporter assay indicated that luciferase activity was notably decreased when co-transfected with HMGB1-WT and miR-374a-3p mimics, suggesting that miR-374a-3p can directly bind to the 3'UTR of HMGB1 (Figure 6B, C). Combined with the arguments above, Lnc-FTX can sponge miR-374a-3p, which can interact with its target gene HMGB1. Thus, we further investigated the effect of Lnc-FTX overexpression on the expression levels of HMGB1. We observed that HMGB1 expression was increased when Lnc-FTX was overexpressed (Figure 6D, E). These results suggest that Lnc-FTX can upregulate HMGB1 expression. Interestingly, we analyzed the TCGA database and found that there was a strong correlation between HMGB1 and RBMX expression in HCC (Figure 6F) and validated that RBMX could upregulate HMGB1 expression (Figure 6E). All these supports the existence of the Lnc-FTX/miR-374a-3p/HMGB1 regulatory axis.
Discussion
LncRNA has been demonstrated to regulate a range of biological processes, including chromosome inactivation, nuclear transport, carcinogenesis, and disease mechanisms [37,38,39]. Given the insidious and recurrent nature of cancer, it poses a significant threat to human health, highlighting the importance of identifying effective therapeutic targets [40,41]. Recently, lncRNA has gained considerable attention for its roles in cancer, with many, such as HOTAIR, HULC, HOTTIP, and NEAT1, identified as oncogenes [42]. Overall, this study confirms that the overexpression of Lnc-FTX in HCC is correlated with dismal prognosis, providing valuable guidance for clinical risk assessment. Additionally, the promotion of proliferation, invasion, and migration by Lnc-FTX in HCC further supports its role as an oncogene.
Lnc-FTX functions as an oncogene across various cancers, facilitating the malignant behaviors of cancer cells. A study on lung adenocarcinoma demonstrated that Lnc-FTX is upregulated in tumor tissues and accelerates malignant characteristics of Lung adenocarcinoma by activating the miR-335-5p/NUCB2 pathway [43]. Research by Kui Zhao et al. revealed that Lnc-FTX is positively regulates miR-192-5p to target EIF5A2, thereby enhancing cancer cell proliferation ability in colorectal cancer cells [44]. Consistent with these observations, our research identified that Lnc-FTX expression level was markedly elevated in HCC tissues with the data from GSE77314 and TCGA. Clinical data from a cohort of 27 HCC patients (cohort 1) showed that high Lnc-FTX expression is strongly associated with advanced TNM classification and reduced OS, indicating a correlation between high Lnc-FTX levels and poor prognosis. Nonetheless, in cohort 1, the expression of lnc-FTX did not show a significant difference between cancerous and adjacent non-cancerous tissues. It may be attributed to: (1) Tumor heterogeneity: Variability in tumor cells across individuals. (2) Sample size impact: A larger clinical sample size will help more accurately reflect the expression levels of Lnc-FTX. (3) TNM staging relevance: Lnc-FTX levels correlate with TNM staging, with lower Lnc-FTX expression observed in patients with lower TNM stages (Stage I). Thus, the higher number of patients in lower TNM stages in our cohort likely explains the increased prevalence of lower Lnc-FTX expression in tumor tissues.
Lnc-FTX functions as a sponge of miR-374a-3p in HCC. (A) Schematic of predicted binding sites of miR-374a-3p, miR-374a-5p, miR-545-3p and miR-545-5p on Lnc-FTX. (B) RT-PCR analysis of miR-374a-3p and miR-545-5p expression in the MS2-TRAP RNA pull-down sample. (C, D) The predicted miR-374a-3p binding site in the FTX gene. The corresponding sequence in the mutated (mut) version is also shown. Luciferase activity of MHCC-LM3 cells transfected with NC/miR-374a-3p mimics and FTX-wt1, FTX-mut1, FTX-wt2, FTX-mut2 plasmids. Data are presented as the ratio of Firefly luciferase activity to Renilla luciferase activity.
Lnc-FTX upregulates HMGB1 through miR-374a-3p. (A) The predicted miR-374a-3p binding site in the 3′UTR of the human HMGB1 gene. The corresponding sequence in the mutated version is also shown; (B, C) Luciferase activity of MHCC-LM3 and Hep3B cells transfected with NC/miR-374a-3p mimics and HMGB1-wt/mut plasmids. Data are presented as the ratio of Firefly luciferase activity to Renilla luciferase activity; (D) RT-PCR analysis of HMGB1 mRNA expression of MHCC-LM3 cells transfected with Lnc-FTX plasmids(pc-FTX) or control plasmids(pc-vector); (E) Western Blot analysis of HMGB1 expression of Hep3B cells transfected with pc-FTX/pc-RBMX or pc-vector. (F) The Pearson correlation analysis of the expression of RBMX and HMGB1 in TCGA (LIHC).
LncRNAs exert their influence through a variety of mechanisms: decoys, scaffolds, guidance, stabilization, ceRNA or "RNA sponge" [45]. It is summarized in two ways: (1) direct binding to regulatory regions of genes, such as promoters, enhancers and transposons, to recruit the relevant protein complexes, thereby affecting chromatin remodeling and transcription [45]. (2) Binding to intermediate regulators, including RBPs or microRNAs [46], which is the mainstream molecular mechanism of LncRNA regulation in tumor progression [47].
RNA-protein interactions participate in many essential cellular functions, such as splicing, polyadenylation, stability, and translation [48]. RBMX, commonly referred to as hnRNP G, is a widely expressed nuclear RBP. It plays a role in normal developmental processes, such as brain development, and is associated with the regulation of multiple diseases, including cancer (activation in liver cancer), systemic lupus erythematosus, and viral infections [49]. Its primary biological functions include involving in chromatin modification, splicing and alternative splicing regulation [50], transcriptional control, and maintenance of genomic stability [51]. RBMX is vital to the progression of HCC, although research into the underlying mechanisms is still limited. Recent research indicates that RBMX can bind with SOCS5 to co-stimulate the SREBP1 promoter, and lipid accumulation induced by SREBP1 may promote the metastasis of steatotic HCC [52]. Another study on HCC shows that RBMX specifically binds to and stabilizes Lnc-BLACAT1, thereby promoting malignant phenotypes of HCC and resistance to chemotherapeutic drugs [53]. Our findings are in accord with these studies indicating that RBMX binds to Lnc-FTX and enhances Lnc-FTX stability and suggesting that RBMX is of clinical significance to be one of potential therapeutic targets for HCC.
Among the multiple models of lncRNA-RNA interactions, the ceRNA hypothesis has garnered significant attention. This hypothesis posits that LncRNAs, miRNAs, and mRNAs engage in a complex network of interactions. LncRNAs serves as ceRNAs or natural microRNA sponges, which regulate mRNAs by competitively inhibiting miRNAs in the presence of microRNA response elements [54]. To further identify the downstream targets of Lnc-FTX, LncBase Predicted V2 was used to predict the associated candidate microRNAs, with miR-374a-3p confirmed as one of the potential downstream targets. HMGB1, a well-known oncogene, plays a critical role in regulating breast cancer, gastrointestinal stromal tumor and melanoma [55]. A significant amount of evidence demonstrates that HMGB1 is engaged in tumor cell malignant biological processes through signaling pathways such as NF-κB, JAK/STAT and PI3K/AKT [56]. This study predicted and confirmed a miR-374a-3p binding site in HMGB1. Up-regulation of Lnc-FTX led to elevated HMGB1 expression. Coupled with the previous finding, Lnc-FTX regulates HMGB1 through miR-374a-3p. The Lnc-FTX/miRNA-374a-3p/HMGB1 axis regulation for HCC also brings new perspectives for clinical treatment.
This study innovatively explored the interaction between Lnc-FTX and HMGB1 and discovered a strong association between the expression of RBMX and HMGB1. Overexpression of RBMX in HCC cells led to a significant upregulation of HMGB1, indicating that RBMX can enhance HMGB1 expression. In summary, the establishment of FTX/miR-374a-3p/HMGB1 axis via the RBMX stabilizing the FTX provides new directions for future research into the molecular mechanisms of HCC.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
The authors would like to thank Professor Yang Wang for the suggestions provided in the experimental design.
Funding
This research was funded by National Natural Science Foundation of China (81970550); Natural Science Foundation of Hunan province (2021JJ31067, 2021JJ41048).
Data availability
The data used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics statement
Approval of the research protocol by an Institutional Reviewer Board: The ethical consent was granted from the Medical Ethics Committee of Xiangya hospital. All patients provided written informed consent for tissue analysis before the procedure.
Author contributions
Min Zhang: Investigation; methodology; formal analysis; visualization; writing-original draft; writing-review and editing. Songman Yu: Methodology; data curation; formal analysis; writing-review and editing. Shang Gao: Methodology; data curation. Haiyan Bu: visualization; validation. Lihua Duan: Data curation; writing-review and editing. Yan Huang: Investigation; project administration; supervision; writing-review and editing; funding acquisition.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Ervik M, Lam F. et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. (accessed on 6th March 2024).
2. Qiu H, Cao S, Xu R. Cancer incidence, mortality, and burden in China: a time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun (Lond). 2021;41(10):1037-1048
3. Sung H, Ferlay J, Siegel R L. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209-49
4. Jemal A, Ward E M, Johnson C J. et al. Annual Report to the Nation on the Status of Cancer, 1975-2014, Featuring Survival. J Natl Cancer Inst. 2017;109(9):djx030
5. Wu M Z, Yuan Y C, Huang B Y. et al. Identification of a TGF-β/SMAD/lnc-UTGF positive feedback loop and its role in he-patoma metastasis. Signal Transduct Target Ther. 2021;6(1):395
6. Couri T, Pillai A. Goals and targets for personalized therapy for HCC. Hepatol Int. 2019;13(2):125-137
7. Singal AG, Yarchoan M, Yopp A, Sapisochin G, Pinato DJ, Pillai A. Neoadjuvant and adjuvant systemic therapy in HCC: Current status and the future. Hepatol Commun. 2024;8(6):e0430
8. Singal AG, Kudo M, Bruix J. Breakthroughs in Hepatocellular Carcinoma Therapies. Clin Gastroenterol Hepatol. 2023;21(8):2135-2149
9. El-Khoueiry AB, Sangro B, Yau T. et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492-2502
10. Yuan XQ, Zhou N, Wang JP. et al. Anchoring super-enhancer-driven oncogenic lncRNAs for anti-tumor therapy in hepatocellular carcinoma. Mol Ther. 2023;31(6):1756-1774
11. Farzaneh M, Ghasemian M, Ghaedrahmati F. et al. Functional roles of lncRNA-TUG1 in hepatocellular carcinoma. Life Sci. 2022;308:120974
12. Kitagawa M, Kitagawa K, Kotake Y. et al. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. 2013;70(24):4785-94
13. Wang H, Huo X, Yang X R. et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136
14. Bridges M C, Daulagala A C, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021 220(2)
15. Yu S, Li N, Wang J. et al. Correlation of Long Noncoding RNA SEMA6A-AS1 Expression with Clinical Outcome in HBV-Related Hepatocellular Carcinoma. Clin Ther. 2020;42(3):439-447
16. Chen J, Huang ZB, Liao CJ. et al. LncRNA TP73-AS1/miR-539/MMP-8 axis modulates M2 macrophage polarization in hepatocellular carcinoma via TGF-β1 signaling. Cell Signal. 2020;75:109738
17. Li S, Huang Y, Huang Y. et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36(1):51
18. Zhang W, Bi Y, Li J. et al. Long noncoding RNA FTX is upregulated in gliomas and promotes proliferation and invasion of glioma cells by negatively regulating miR-342-3p. Lab Invest. 2017;97(4):447-57
19. Zhang F, Wang X S, Tang B. et al. Long non-coding RNA FTX promotes gastric cancer progression by targeting miR-215. Eur Rev Med Pharmacol Sci. 2020;24(6):3037-48
20. Pan L, Du M, Liu H. et al. LncRNA FTX promotes the malignant progression of colorectal cancer by regulating the miR-214-5p-JAG1 axis. Ann Transl Med. 2021;9(17):1369
21. Liu F, Yuan JH, Huang JF. et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35(41):5422-5434
22. Wu H, Zhong Z, Wang A. et al. LncRNA FTX represses the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via regulating the M1/M2 polarization of Kupffer cells. Cancer Cell Int. 2020;20:266
23. Sun Y, Cheng Z, Liu S. MCM2 in human cancer: functions, mechanisms, and clinical significance. Mol Med. 2022;28(1):128
24. Li X, Zhao Q, Qi J. et al. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARγ pathway in hepatocellular carcinoma. Int J Oncol. 2018;53(2):551-566
25. Liu Z, Dou C, Yao B. et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7(18):25350-25365
26. Wang S, Zhang Y. HMGB1 in inflammation and cancer. J Hematol Oncol. 2020;13(1):116
27. Yan W, Chang Y, Liang X. et al. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma in-vasiveness and metastases. Hepatology. 2012;55(6):1863-1875. 2012;55(6):1863-1875
28. Amornsupak K, Thongchot S, Thinyakul C. et al. HMGB1 mediates invasion and PD-L1 expression through RAGE-PI3K/AKT signaling pathway in MDA-MB-231 breast cancer cells. BMC Cancer. 2022;22(1):578
29. Jiang M, Liu L, Huang W. et al. HMGB1-activated tumor-associated macrophages promote migration and invasion via NF-κB/IL-6 signaling in oral squamous cell carcinoma. Int Immunopharmacol. 2024;126:111200
30. Yan W, Chang Y, Liang X. et al. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology. 2012;55(6):1863-1875
31. El-Sahar AE, Bekhit N, Eissa NM. et al. Targeting HMGB1/PI3K/Akt and NF-κB/Nrf-2 signaling pathways by vildagliptin mitigates testosterone-induced benign prostate hyperplasia in rats. Life Sci. 2023;322:121645
32. Gong H, Zuliani P, Komuravelli A. et al. Analysis and verification of the HMGB1 signaling pathway. BMC Bioinformatics. 2010 11 Suppl 7(Suppl 7): S10
33. Kang R, Livesey Km, Zeh Hj 3rd. et al. Metabolic regulation by HMGB1-mediated autophagy and mitophagy. Autophagy. 2011;7(10):1256-1258
34. Yan Q, Zeng P, Zhou X. et al. RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing. Oncogene. 2021;40(15):2635-50
35. Liu J, Zheng T, Chen D. et al. RBMX involves in telomere stability maintenance by regulating TERRA expression. PLoS Genet. 2023;19(9):e1010937
36. Munschauer M, Nguyen C T, Sirokman K. et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561(7721):132-6
37. Su M, Xiao Y, Ma J. et al. Long non-coding RNAs in esophageal cancer: molecular mechanisms, functions, and potential applications. J Hematol Oncol. 2018;11(1):118
38. Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108(10):1927-1933
39. Shi X, Sun M, Liu H. et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159-166
40. Bright Cj, Reulen Rc, Winter Dl. et al. Risk of subsequent primary neoplasms in survivors of adolescent and young adult cancer (Teenage and Young Adult Cancer Survivor Study): a population-based, cohort study. Lancet Oncol. 2019;20(4):531-545
41. Patel Sa, Demichele A. Adding Adjuvant Systemic Treatment after Neoadjuvant Therapy in Breast Cancer: Review of the Data. Curr Oncol Rep. 2017;19(8):56
42. Shah M, Sarkar D. HCC-Related lncRNAs: Roles and Mechanisms. Int J Mol Sci. 2024;25(1):597
43. Huo X, Wang H, Huo B. et al. FTX contributes to cell proliferation and migration in lung adenocarcinoma via targeting miR-335-5p/NUCB2 axis. Cancer Cell Int. 2020;20:89
44. Zhao K, Ye Z, Li Y. et al. LncRNA FTX Contributes to the Progression of Colorectal Cancer Through Regulating miR-192-5p/EIF5A2 Axis. Onco Targets Ther. 2020;13:2677-2688
45. Wang Kc, Chang Hy. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904-914
46. Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17(10):1169-1174
47. Qian X, Zhao J, Yeung P Y. et al. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem Sci. 2019;44(1):33-52
48. Lukong K E, Chang K W, Khandjian E W, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24(8):416-25
49. Climente-González H, Porta-Pardo E, Godzik A. et al. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017;20(9):2215-2226
50. Moursy A, Allain F H, Cléry A. Characterization of the RNA recognition mode of hnRNP G extends its role in SMN2 splicing regulation. Nucleic Acids Res. 2014;42(10):6659-72
51. Matsunaga S, Takata H, Morimoto A. et al. RBMX: a regulator for maintenance and centromeric protection of sister chromatid cohesion. Cell Rep. 2012;1(4):299-308
52. Wang Y, Zhao Z, Guo T. et al. SOCS5-RBMX stimulates SREBP1-mediated lipogenesis to promote metastasis in steatotic HCC with HBV-related cirrhosis. NPJ Precis Oncol. 2024;8(1):58
53. Song Y, He S, Ma X. et al. RBMX contributes to hepatocellular carcinoma progression and sorafenib resistance by specifically binding and stabilizing BLACAT1. Am J Cancer Res. 2020;10(11):3644-3665
54. Tay Y, Rinn J, Pandolfi P P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344-52
55. Ellerman J E, Brown C K, De Vera M. et al. Masquerader: high mobility group box-1 and cancer. Clin Cancer Res. 2007;13(10):2836-48
56. Pu Z, Duda D G, Zhu Y. et al. VCP interaction with HMGB1 promotes hepatocellular carcinoma progression by activating the PI3K/AKT/mTOR pathway. J Transl Med. 2022;20(1):212
Author contact
![]() Corresponding author: Yan Huang; Email: drhyanedu.cn.
Corresponding author: Yan Huang; Email: drhyanedu.cn.

 Global reach, higher impact
Global reach, higher impact