Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(6):1301-1312. doi:10.7150/ijms.107038 This issue Cite
Research Paper
NADPH oxidase activator 1 (NOXA1) suppresses ferroptosis and radiosensitization in colorectal cancer
1. The Second School of Clinical Medicine, Zhejiang Chinese Medical University, Hangzhou 310053, China.
2. Department of Radiation Oncology, Zhejiang Cancer Hospital, Hangzhou 310000, Zhejiang, China.
3. Hangzhou Institute of Medicine (HIM), Chinese Academy of Sciences, Hangzhou 310000, China.
4. Zhejiang Key Laboratory of Radiation Oncology, Hangzhou 310000, China.
Received 2024-11-15; Accepted 2025-2-5; Published 2025-2-18
Abstract
Radiotherapy is one of the main treatments for colorectal cancer (CRC), but due to the intrinsic resistance of cells or resistance caused by long-term radiotherapy, the effectiveness of this treatment is limited for some CRC patients. Consequently, identifying novel sensitization strategies is essential. This study identifies Noxa1 as a marker linked to radiotherapy resistance in CRC, suggesting its potential as a prognostic biomarker for patients with CRC. The study found that Noxa1 was significantly overexpressed in radiotherapy-resistant colorectal cancer patients, correlating with a poor prognosis. Additionally, we discovered that the high expression of Noxa1 was negatively correlated with ferroptosis and primarily played a role through the glutathione metabolic pathway, as indicated by GSVA analysis. Experimental data indicated that the expression levels of NOXA1, SLC7A11, and GPX4 were significantly elevated in CRC cell lines resistant to radiotherapy. The expression of SLC7A11 and GPX4 decreased after the knockdown of Noxa1, leading to an increase in cellular ROS levels, which induced ferroptosis and sensitized the cells to radiotherapy. Therefore, Noxa1 might influence the radiotherapy sensitivity of CRC via regulating ferroptosis. Targeting Noxa1 could enhance radiotherapy sensitization and improve the prognosis of CRC patients.
Keywords: Noxa1, colorectal cancer, ferroptosis, radiotherapy sensitization, ROS
Background
Colorectal cancer ranks among the most prevalent malignancies worldwide, holding the third highest incidence and mortality rate globally[1]. In the management of CRC, radiotherapy is a crucial tool for addressing unresectable tumors, postoperative residual tumors, and recurrent tumors. Its benefits encompass primary tumor size reduction, tumor grade reduction, enhanced local tumor control, and the preservation of vital organs[2-4]. Nonetheless, the efficacy of radiotherapy can be limited in patients with inherently radiotherapy-resistant colorectal cancer or those who develop treatment-induced resistance[5, 6]. Radiotherapy for CRC can therefore be enhanced by identifying potential targets that can enhance the cells' radiosensitivity. Understanding the associated molecular mechanisms holds promise for significantly improving patient prognosis.
The NOXA1 enzyme is a key component of the activation of NOX1, a member of the NADPH oxidase family known to regulate ROS production[7, 8]. NOXA1 has been shown to play an important role in hypertension, vascular inflammation, stenosis, and atherosclerosis through its role in regulating ROS[9-11]. Notably, Epithelial-Mesenchymal Transition, hypoxia, angiogenesis, and activated inflammatory responses contribute to the poor prognosis of colorectal cancer, and NOXA1 has been found to be involved in vascular inflammation; thus, the expression of Noxa1 may predict poor prognosis in CRC patients[12].
Radiotherapy is based on ionizing radiation that directly or indirectly induces ROS production, resulting in DNA double-strand breaks that lead to tumor cell death[13]. Tumor cells adapt to oxidative stress by limiting ROS production or activating their antioxidant systems, a phenomenon linked to radiotherapy resistance[14-18]. ROS is a key regulator of ionizing radiation, and NOXA1 has the function of regulating ROS production, so our aim is to explore whether Noxa1 influences the radioresistance of CRC by regulating ROS levels.
Ferroptosis, a modality of programmed cell death, characterized by iron accumulation and ROS generation, which catalyze lipid peroxidation of unsaturated fatty acids and decrease levels of glutathione peroxidase 4 (GPX4), thereby inducing cell death[19-23]. Increasing evidence indicates that tumor cells can upregulate SLC7A11 and GPX4 to protect cells from radiotherapy-induced ferroptosis, suggesting ferroptosis is crucial to tumor radiosensitivity[24-26]. As a result, targeting ferroptosis in radiotherapy may be a potential therapeutic strategy for overcoming radiotherapy resistance in tumors.
The study aimed to investigate the functional role of Noxa1 in CRC radioresistance and its underlying mechanisms. Specifically, we explored the mechanism of interaction between Noxa1 and ferroptosis in relation to radioresistance in CRC and explored the feasibility of using Noxa1 as a biomarker for radioresistance.
Materials and methods
Identification of RTDEGs
Transcriptome data and associated clinical information were obtained from the Cancer Genome Atlas (TCGA) database. Raw gene expression counts were collected from the GSE35452 dataset in GEO. Differential gene expression analysis between CRC patients who responded to radiotherapy and those who did not was performed using the R package "limma" (version 4.2.1) with a standard comparison model. RTDEGs were identified with a threshold of absolute log2-fold change (FC) ≥ 0.3 and adjusted P-value < 0.05.
Functional enrichment analysis of RTDEGs
Functional enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was carried out via the Metascape database (http://metascape.org/) to comprehensively understand the biological processes and pathways linked to RTDEGs. The top 20 critical signaling pathways were selected for further exploration (P<0.05).
Survival analysis of RTDEGs
The median expression level of the Noxa1 gene was utilized to establish an optimized cut-off value, stratifying colorectal cancer patients into high- and low-risk groups. We employed Kaplan-Meier survival curves and time-dependent ROC curves to demonstrate significant prognostic differences between the two groups and assess the Noxa1 gene's efficacy as an overall survival (OS) biomarker.
Predictive nomogram construction and evaluation
Univariate and multivariate COX analyses were conducted to assess Noxa1's independent predictive ability in comparison to T stage, M stage, N stage, age, and gender (P < 0.05). For each factor, hazard ratios and their 95% confidence intervals were computed. A predictive nomogram was then established, combining the expression level of the Noxa1 gene with clinical characteristics as a quantitative tool for predicting survival risk in colorectal cancer patients. Meanwhile, the accuracy of the nomogram's predictions was assessed using a calibration curve, where closer alignment with the 45° line indicates better predictive performance. The nomogram and calibration curve were generated using the "rms" R package.
Correlation between ferroptosis and Noxa1 expression
The ferroptosis score for each of the 64 ferroptosis regulator gene sets was calculated using the GSVA algorithm. This process utilized two input files: a gene set of 64 ferroptosis regulators and a standardized gene expression matrix of patient samples. We then analyzed the correlation between Noxa1 and ferroptosis, as well as the relationships between Noxa1 and key pathways such as iron ion metabolism, lipid metabolism, etc. Gene sets from the Molecular Signatures Database were analyzed using the GSVA package in R.
Hematoxylin and Eosin (H&E) and immunohistochemistry (IHC) staining in CRC tissues
Colorectal cancer (CRC) samples were formalin-fixed, paraffin-embedded, and sectioned into 4-μm slices. For H&E staining, tissue sections underwent deparaffinization and rehydration before being stained with hematoxylin for 4 minutes and eosin for 90 seconds. The sections underwent dehydration using ascending ethanol concentrations, were cleared with xylene, mounted in neutral resin, and then examined microscopically. Immunohistochemistry (IHC) involved staining tissue sections using an anti-NOXA1 antibody, followed by hematoxylin counterstaining. IHC images were obtained at 400× magnification using a microscope, and semi-quantitative analysis of the staining was performed on three random fields with Image J software.
Cell lines and culture conditions
Colorectal cancer (CRC) cell lines HCT8, HCT15, HCT116, and DLD-1 were sourced from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China. The radio-resistant CRC cell lines HCT8R, HCT15R, HCT116R, and DLD-1R, were derived from their respective parental cell lines through 25 cycles of exposure to 2 Gy X-rays at a dose rate of 1 Gy/min. All cell lines were maintained in high-glucose DMEM (Gibco, USA) with 10% fetal bovine serum (Gibco, NY, USA) and 1% penicillin/streptomycin (Gibco, USA) under 5% CO2 and 95% air humidity. Regular testing for mycoplasma contamination was conducted every two months to ensure cell line integrity.
Three-dimensional (3D) cell culture
A three-dimensional lrECM on-top culture was performed following established protocols. A 24-well plate was coated with 150 µl of Matrigel (Corning, NY, USA; 6347014). HCT15R and HCT8R cells were seeded onto the Matrigel layer (2×104 cells per well), followed by the addition of medium with 10% Matrigel on top. Cells were incubated at 37°C for one week, with the medium replaced every 2-3 days. Colony cell numbers and the diameter of 3D cells were observed and analyzed using Image J software.
Cell irradiation
CRC cells were subjected to varying radiation doses with an X-ray irradiator (SARRP III, Precision X-Ray, Inc, X Strahl, USA), operating at 13mA and employing 0.15mm copper for treatment and 1.0mm aluminum for imaging.
Small interfering RNA (siRNA) transfection
CRC cells were transfected with 20μM Noxa1-specific siRNA using Lipofectamine 2000 (Invitrogen, 11668019), according to the manufacturer's instructions. Transfection efficiency was assessed by Western blot analysis 48-72 hours post-transfection. The siRNA sequences were as follows: si-Noxa1 1# (sense: 5′-ACC AUG AUG CCA GGU CCC UAA TT-3′, antisense: 5′-UUA GGG ACC UGG CAU CAU GGU TT-3′); si-Noxa1 2# (sense: 5′-CCA GCU UGG GCA ACU CAG UUA TT-3′, antisense: 5′-UAA CUG AGU UGC CCA AGC UGG TT-3′); si-Noxa1 3# (sense: 5′-CCU GCG GUU CAA GCU GCA ATT-3′, antisense: 5′-UUG CAG CUU GAA CCG CAG GTT-3′).
CCK8 assay
Cell Counting Kit-8 (CCK-8) assays were performed according to the protocol provided by the manufacturer (Beyotime Biotechnology, Shanghai, China). In summary, 2000 cells in 100μl of culture medium were placed in each well of a 96-well plate and incubated for 1 to 7 days.
Quantification of intracellular ROS levels
To evaluate the production of intracellular ROS, CRC resistant cells with si-Noxa1 transfection underwent 6Gy X-ray irradiation. Six hours after irradiation, cells were collected, resuspended at 0.5×105 cells/mL, and exposed to 10μM DCFH-DA fluorescence probe. The cell samples were incubated at 37°C for 30 minutes in the dark. The levels of mitochondrial ROS were visualized using a fluorescence microscope.
After 6 hours of irradiation, cells were incubated with the DCFH-DA fluorescence probe, diluted 1:1000 in serum-free medium, at 37°C for 30 minutes. The ROS levels were quantified using flow cytometry (Beckman, USA) and analyzed using FlowJo software. The results are expressed as mean percentages ± standard deviation (SD) of ROS-positive cells compared to the total cell population in the plots.
Statistical analysis
Continuous variables are presented as the mean ± SD. Group differences were assessed using the Wilcoxon test in R software. Survival time variations were evaluated using the log-rank test, with a significance threshold set at P < 0.05. Kaplan-Meier plots were generated to visualize differences in survival time. Results are expressed as means ± SD from three independent experiments, and statistical analyses were conducted using GraphPad Prism 8.0c Software. Group differences were determined using the t-test, with statistical significance defined as a p-value < 0.05.
Results
Identification and functional annotation of RTDEGs associated with radiotherapy resistance
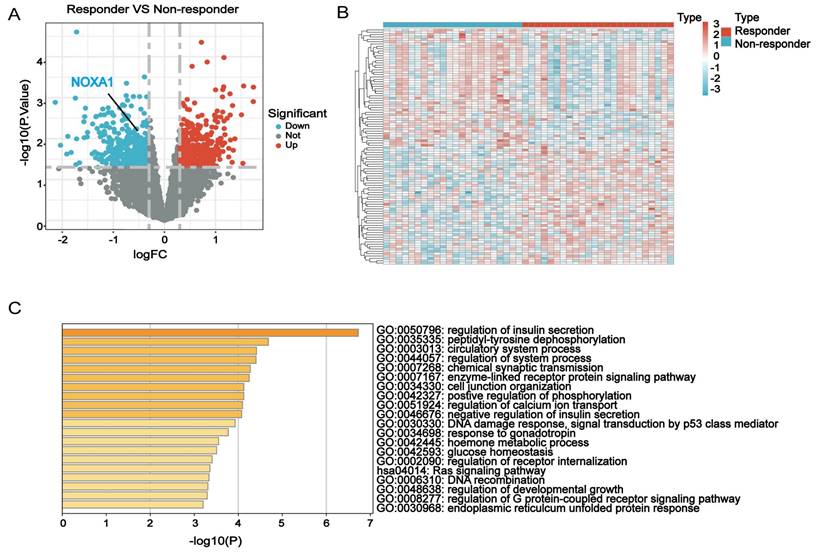
This study analyzed RTDEGs in CRC cells with diverse radiation responses using the GSE35452 dataset from the GEO database. RTDEGs were determined with a significance level of P<0.05 and an absolute log2 fold change (FC) of at least 0.3. A total of 846 RTDEGs were identified, comprising 423 up-regulated and 423 down-regulated genes. The differences in gene expression were visually represented through volcano and heatmap plots (Figure 1A-B). Metascape pathway enrichment analysis was performed to understand RTDEGs functions. The findings indicated that RTDEGs were predominantly enriched in biological process terms, including DNA damage response, p53-mediated signal transduction, positive regulation of phosphorylation, and DNA recombination (Figure 1C). The results indicate that genes linked to radiation resistance are pivotal in CRC malignant progression, primarily affecting DNA damage response and repair mechanisms.
Identification of hub genes
To identify core genes influencing radiotherapy resistance among RTDEGs, we selected RTDEGs from the TCGA-READ and TCGA-COAD cohorts for random survival forest analysis. Genes with relative importance greater than 0.45 were selected. Ultimately, 14 genes met our screening threshold: Krt84, Lhx8, Pdcl2, Larp6, Magec3, Tmem88, Noxa1, Chat Dnm1p35, Ngf, Tnnt1, Ccnd3, Tm4sf4, and Enfa5 (Fig. 2A-B). We then conducted a copy number variation (CNV) analysis on these genes. The top 5 genes with the highest CNV frequency were Enfa5, Ccnd3, Noxa1, Ngf, and Lhx8 (except for Ccnd3, which has a gain-of-function mutation; the other four have loss-of-function mutations). No relevant mutations were detected in Magec3 and Dnm1p35 (Figure 2C-D). Kaplan-Meier survival analysis revealed that low expression of Noxa1, Tmem88, Larp6, and Enfa5 were significantly linked to improved OS compared to higher expression levels (P<0.001 for each) (Figure 2E-H). While the expression of Ccnd3 was not statistically significantly associated with the prognosis of colorectal cancer. Therefore, it was excluded from further analysis (Figure S1). Although Enfa5 expression was linked to the prognosis of colorectal cancer, its variable relative importance was lower than that of other genes. As a result, Enfa5 was not the main focus of our investigation.
Identification and functional annotation of RTDEGs in CRC. A-B: Volcano and heatmap plots showing differentially expressed genes related to radiotherapy resistance in colorectal cancer. C: Bar chart indicating GO and KEGG analyses of the differential network.
Analysis of RTDEGs in CRC. A-B: Random survival forest analysis of RTDEGs. C-D: CNV variations in RTDEGs from the TCGA cohort, with green and red dots representing gain and loss of function, respectively. E-H: Kaplan-Meier survival analysis showing that low expression of Noxa1, Tmem88, Larp6, and Enfa5 is associated with better overall survival (P < 0.001). I: Gene expression analysis of DEGs in radiotherapy non-responder and responder CRC organoids. J: Gene expression analysis of DEGs in normal and cancerous colorectal tissues (*P < 0.05).
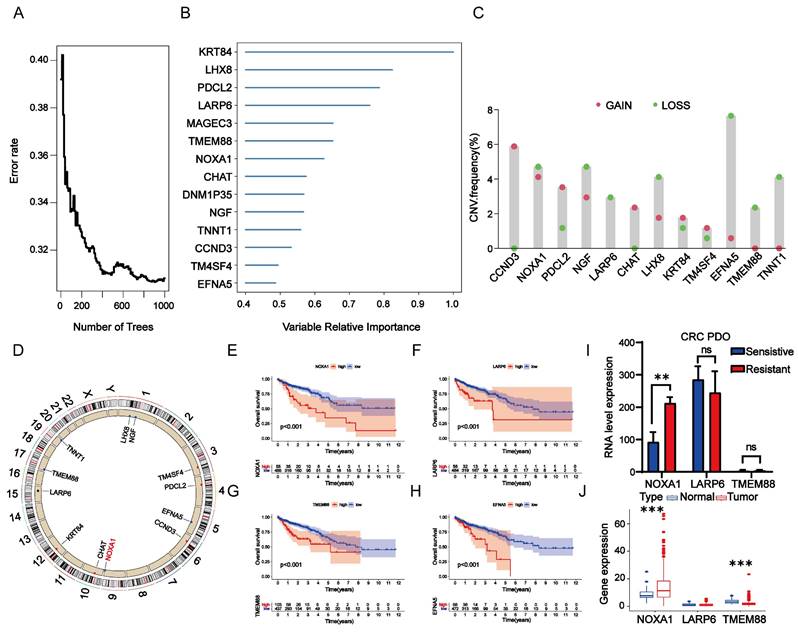
Hence, we then investigated the differential expression of Noxa1, Larp6, and Tmem88 in radio-sensitive and radio-resistant organoids. Our findings revealed that Noxa1 and Larp6 were notably overexpressed in radio-resistant organoids, while Tmem88 showed no significant variation. Notably, only Noxa1's differential expression reached statistical significance (Figure 2I). In a comparative analysis, Noxa1 was primarily expressed in colorectal cancer tissues, Tmem88 was more prevalent in normal tissues, and Larp6 showed no significant expression differences between the two (Figure 2J). These findings emphasize the significant role of the Noxa1 gene in contributing to colorectal cancer's radioresistance.
Assessment of Noxa1 as a standalone prognostic indicator
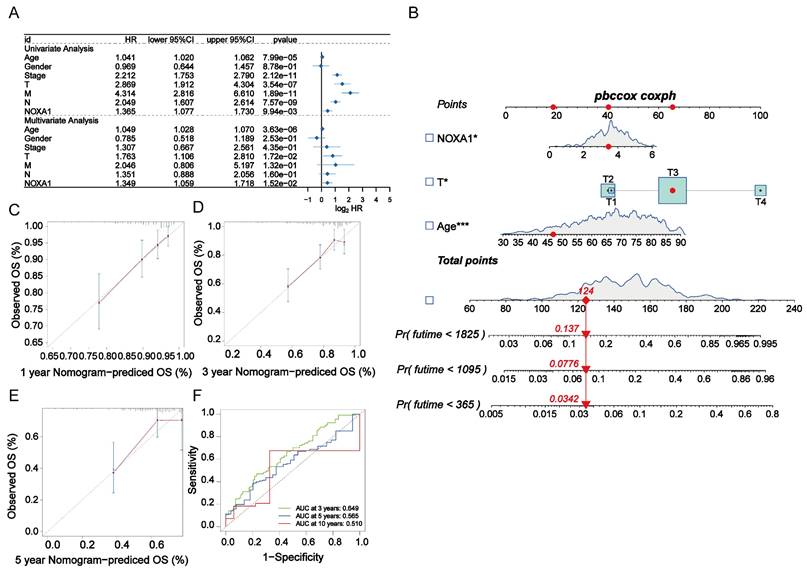
Considering the link between elevated Noxa1 expression and unfavorable outcomes in colorectal cancer patients, along with its loss-of-function mutations, we performed a comprehensive analysis to confirm its role as a standalone prognostic marker. We conducted univariate and multivariate Cox regression analyses to compare Noxa1 with clinical features such as age, gender, and stages (T, M, N). Our findings revealed that age (P=3.63×10-6, HR=1.049), T stage (P=1.72×10-2, HR=1.763), and Noxa1 (P=1.52×10-2, HR=1.349) independently predicted colorectal cancer prognosis (Figure 3A). We developed a prognostic nomogram model incorporating these key factors to improve prognostic accuracy (Figure 3B). A calibration plot was used to evaluate the model's precision, revealing strong concordance between predicted and observed overall survival outcomes, thus demonstrating reliable predictive consistency (Figure 3C-E). Additionally, ROC and AUC analyses of the TCGA cohort highlighted Noxa1's potential as a standalone prognostic feature, with AUC values of 0.649 at 3 years, 0.565 at 5 years, and 0.510 at 10 years (Figure 3F), reinforcing Noxa1's significance in predicting long-term outcomes in colorectal cancer patients.
Overexpression of Noxa1 enhances radioresistance in CRC
We explored the relationship between Noxa1 expression and the radioresistance in CRC. Western blot assay indicated that NOXA1 levels increased in various radioresistant CRC cell lines, including HCT8R, HCT15R, DLD1R and HCT116R cells. Notably, HCT15R and HCT8R exhibited higher NOXA1 expression than DLD1R and HCT116R, prompting us to use HCT15R and HCT8R for further experiments (Figures 4A-B).
Additionally, we assessed NOXA1 expression in CRC patient samples, distinguishing between those sensitive and resistant to radiotherapy. Our analysis revealed that NOXA1 expression was markedly elevated in radio-resistant CRC tissues compared to radio-sensitive ones, indicating its potential as a diagnostic marker for CRC radiotherapy sensitivity (Figures 4C-D).
We then exposed HCT8 and HCT15 parental cells to 6Gy radiation. The experimental results showed that radiation did not induce an upregulation of NOXA1 expression in the short term (Figure S2A). On the other hand, we observed that NOXA1 expression was significantly higher in HCT15 cells compared to HCT8 cells, and HCT15 cells exhibited stronger radiation resistance (Figure S2A-B). Based on the expression patterns of Noxa1 in both radioresistant and radiosensitive cell lines, we speculate that Noxa1 expression may be closely related to both primary and acquired radiation resistance in colorectal cancer.
To further clarify Noxa1's function, we silence Noxa1 in HCT15R and HCT8R cells (Figure 5A-B). siNoxa1#3 exhibited the highest knockdown efficiency and was selected for further experiments. Knockdown of Noxa1 led to a marked decrease in the survival fraction of HCT15R and HCT8R cells after radiation exposure (Figures 5C-F).
Construction of the radiotherapy-related prognostic signature in CRC. A: Forest plot of univariate and multivariate Cox regression analyses for Noxa1 and clinical characteristics in the TCGA cohort. B: Nomogram based on Noxa1 expression and clinical prognostic factors. C-E: Calibration curves predicting 1-, 3-, and 5-year survival rates. F: Time-dependent ROC curve for the radiosensitivity-related signature at 3, 5, and 10 years.
Noxa1 as a radioresistant oncogene in CRC. A-B: Western blot analysis of CRC cell lines (original blots/gels in Supplementary Fig. 4). C-D: H&E and IHC images of radiosensitive vs. radioresistant CRC tissues.
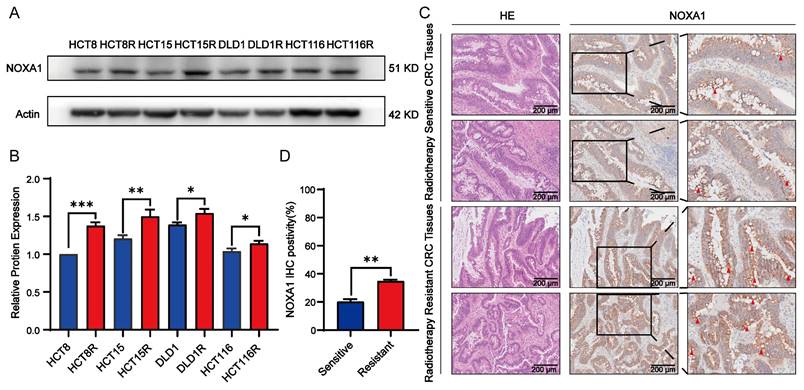
Impact of Noxa1 knockdown on radiosensitization in colorectal cancer. A-B: Western blot analysis of HCT15R and HCT8R cells transfected with siNoxa1 (original blots/gels in Supplementary Fig. 2). C-F: Dose-response survival fractions and colony formation assays for HCT15R and HCT8R cells with/without siNoxa1 transfection.
Noxa1 promotes tumor growth in CRC
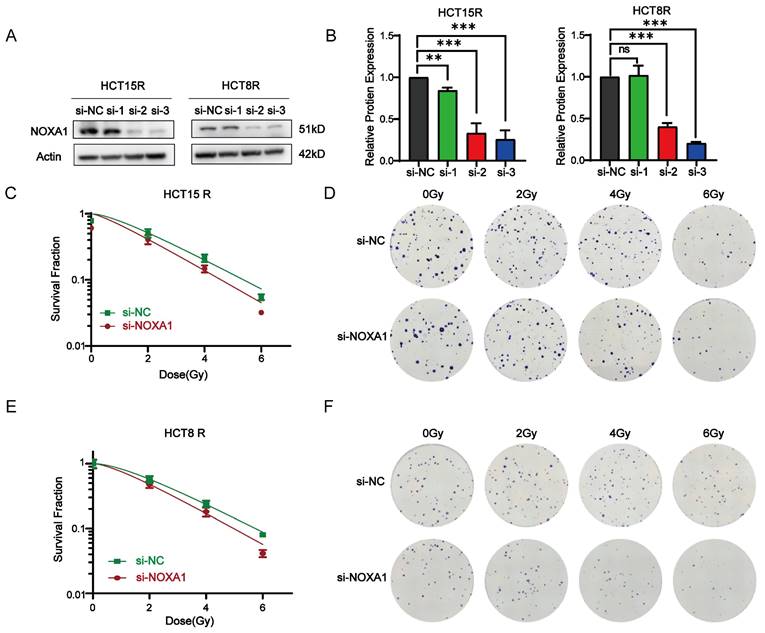
To investigate Noxa1's role in CRC cell viability, we first knocked down Noxa1 in HCT15R and HCT8R cells and conducted CCK8 assays. Noxa1 knockdown markedly reduced cell viability (Figures 6A-B). Furthermore, results from three-dimensional (3D) cell culture showed that both the number and diameter of 3D colonies in HCT15R and HCT8R cells transfected with siNoxa1 were notably decreased, indicating an inhibition of tumor growth capabilities (Figures 6C-G).
Noxa1 enhanced radiosensitivity of CRC through ferroptosis pathway
Ferroptosis, an iron-dependent programmed cell death caused by lipid peroxidation, plays a crucial role in cell death and tumor suppression during radiotherapy. This process encompasses iron metabolism, lipid peroxidation, and glutathione metabolism. Our study identified a negative correlation between Noxa1 expression and the ferroptosis score, highlighting Noxa1's primary impact on glutathione metabolism pathways (Figure 7A-D).
Western blot assay results showed elevated expression of SLC7A11 and GPX4 in HCT15R and HCT8R cells, alongside increased NOXA1 expression (Figure 7E). Additionally, knockdown of Noxa1 in HCT15R and HCT8R cells resulted in decreased expression of SLC7A11 and GPX4, suggesting that overexpression of Noxa1 inhibits ferroptosis in radio-resistant CRC cells (Figure 7F). Furthermore, we measured ROS levels in HCT15R and HCT8R cells and found significantly higher ROS levels in cells transfected with siNoxa1 (Figures 7G-N), while the ROS levels were significantly higher in the parental cells compared to the radioresistant cells (Figure S3A-D). We then treated Noxa1-knockdown cells with Ferrostatin-1 (10µmol). The results showed that cell survival was significantly improved in the Noxa1-knockdown cells upon treatment with Ferrostatin-1(Figures 7O-P).
Noxa1 promotes the occurrence and development of colorectal cancer. A-B: CCK8 assay of CRC cell viability after Noxa1 knockdown. C-G: The images of three-dimensional (3D) cell culture showed that both the number and diameter of 3D colonies in HCT15R and HCT8R cells with or without siNoxa1 transfected.
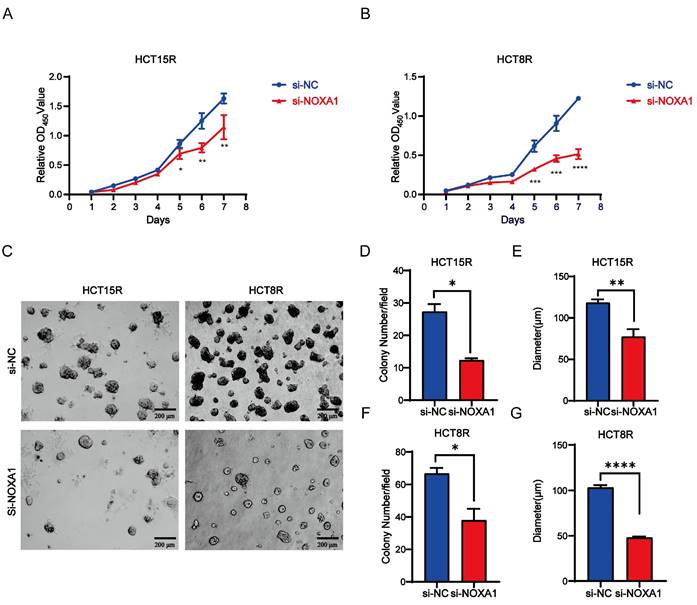
Noxa1's association with ferroptosis in CRC. A-D: Correlation of Noxa1 expression with ferroptosis, iron homeostasis, lipid, and glutathione metabolism. E: Western blot of HCT15, HCT15R, HCT8, and HCT8R cells (original blots/gels in Supplementary Fig. 4). F: Western blot of HCT15R and HCT8R cells with/without siNoxa1. G-N: Flow cytometry and immunofluorescence assays of ROS in HCT15R and HCT8R cells transfected with siNoxa1 post-irradiation. O-P: CCK8 analysis of Noxa1 knockdown cells treated with Fer-1.
Discussion
For stage II~III low-to-medium rectal cancer (tumor < 12 cm from the anus) neoadjuvant radiotherapy is the standard treatment[27]. However, due to individual differences, the pathological complete response rate following neoadjuvant chemoradiotherapy for rectal cancer is only 15-27%[28], and the proportion of patients exhibiting radioresistance ranges from 20% to 40%[29]. This subset of radioresistant colorectal cancer patients experiences poor radiotherapy efficacy due to endogenous cellular resistance or acquired resistance resulting from prolonged radiotherapy[30]. Consequently, we conducted research on radiotherapy-related genes in colorectal cancer. This study identified the radioresistance-associated gene Noxa1 from the GEO database, revealing that elevated Noxa1 expression is linked to poor prognosis in colorectal cancer patients. Noxa1 knockdown suppressed the proliferation of radiotherapy-resistant colorectal cancer cells, indicating its potential role as an oncogene in colorectal cancer development and progression.
In our analyses of colorectal cancer cells, tissues, and organoids, we observed that NOXA1 expression was higher in radioresistant tissues compared to radiosensitive ones. Notably, HCT15R and HCT8R cells exhibited significant radiosensitization following Noxa1 knockdown after irradiation treatment. The experimental findings robustly indicate that Noxa1 could serve as a predictive biomarker for radioresistance in CRC.
Radiotherapy damages cell DNA via direct high-energy ionizing radiation and indirectly through ROS generated by radiation hydrolysis. This process leads to cell cycle arrest, ultimately resulting in cell death via apoptosis or necrosis, thereby facilitating tumor cell eradication[31, 32]. Consequently, a tumor cell's ability to manage ROS levels significantly influences its radiosensitivity[33]. Enhancing the clearance of intracellular ROS can increase the radiosensitivity of hypoxic tumor cells by disrupting survival signals linked to redox imbalance, thereby protecting cells from ROS damage and facilitating cancer progression[34]. In radiotherapy, elevated NOX levels significantly contribute to ROS production, with tumor cells showing greater NOX expression compared to normal tissues[35]. Therefore, targeting NOXs to effectively downregulate intracellular ROS levels may impact the survival of tumor cells post-ionizing irradiation without affecting normal cells[36-40]. NOXA1, as an activating subunit of NOX1, has been demonstrated to influence the production of intracellular ROS. Our measurements indicated that ROS levels were reduced in HCT15R and HCT8R cells compared to HCT15 and HCT8 cells. Research indicates that the upregulation of NOX in cancer cells causes sustained intracellular ROS elevation, facilitating adaptation to elevated ROS and contributing to radiotherapy resistance[41]. Thus, we hypothesize that Noxa1 overexpression may contribute to radioresistance by chronically elevating ROS production within tumor cells, mediated by antioxidant stress genes induced by the adaptation to high ROS levels.
We employed GSVA to investigate the relationship between Noxa1 expression and tumor-related signaling pathways, aiming to clarify how Noxa1 influences radioresistance in colorectal cancer. We discovered that Noxa1 expression was negatively correlated with ferroptosis, primarily affecting this process through glutathione metabolism. Ferroptosis is negatively regulated by SLC7A11 and GPX4. In HCT15R and HCT8R cells, we observed increased expression of NOXA1, SLC7A11, and GPX4, which inhibited ferroptosis. Following Noxa1 knockdown, SLC7A11 and GPX4 levels decreased, triggering ferroptosis. Additionally, we found that ROS levels in HCT15R and HCT8R cells significantly increased after Noxa1 knockdown and subsequent irradiation treatment. High SLC7A11 expression promotes GSH synthesis by mediating cystine transport, upregulating GPX4 activity, enhancing the antioxidant capacity of tumor cells, scavenging ROS, and inhibiting ferroptosiss[42-44]. Studies have indicated that radiotherapy reduces the expression of SLC7A11 and GPX4 proteins, thereby triggering ferroptosis[45]. These findings suggest that Noxa1 might inhibit ferroptosis via upregulating the SLC7A11/GSH/GPX4 axis to eliminate ROS produced by its own overexpression, ultimately leading to radioresistance in colorectal cancer. However, the intrinsic biological regulatory mechanisms linking NOXA1 to the SLC7A11/GSH/GPX4 axis require further investigation.
Supplementary Material
Supplementary materials and methods, figures.
Acknowledgements
This work was supported by Key Research Development Program of Zhejiang (2022C03015), National Health Commission Research Foundation (WKJ-ZJ-2305), National Natural Science Foundation of China (NSFC) Youth Project (82303688), Natural Science Foundation of Zhejiang Province of China (LQ23H220002 and LQ23H220003), China Postdoctoral Science Foundation (2022M723204) and Zhejiang Students' Technology and Innovation Program (2024R410B058).
Author contributions
Qingyu Jiang: data operation, writing original paper; Qianping Chen, Quanquan Sun, Dong Liu, Ji Zhu, Wei Mao: Material preparation, precise analysis, project design; All authors have read and approved the final manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. The datasets generated during and/or analyses during the current study are available on TCGA and GEO websites.
Ethics approval and consent to participate
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of Zhejiang Cancer Hospital (IRB-2022-677). Written informed consent was obtained from individual or guardian participants.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48
2. van der Valk MJM, Hilling DE, Bastiaannet E, Meershoek-Klein Kranenbarg E, Beets GL, Figueiredo NL. et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): an international multicentre registry study. Lancet. 2018;391:2537-45
3. Deng S, Vlatkovic T, Li M, Zhan T, Veldwijk MR, Herskind C. Targeting the DNA Damage Response and DNA Repair Pathways to Enhance Radiosensitivity in Colorectal Cancer. Cancers (Basel). 2022;14:4874
4. Chen Y, Zhou S, Wan K, Yu L, Zhao C, Deng H. et al. RIOK1 mediates p53 degradation and radioresistance in colorectal cancer through phosphorylation of G3BP2. Oncogene. 2022;41:3433-44
5. Yao PA, Wu Y, Zhao K, Li Y, Cao J, Xing C. The feedback loop of ANKHD1/lncRNA MALAT1/YAP1 strengthens the radioresistance of CRC by activating YAP1/AKT signaling. Cell Death Dis. 2022;13:103
6. Yu X, Zhou L, Liu W, Liu L, Gao F, Li W. et al. Skp2 stabilizes Mcl-1 and confers radioresistance in colorectal cancer. Cell Death Dis. 2022;13:249
7. Vermot A, Petit-Härtlein I, Smith SME, Fieschi F. NADPH Oxidases (NOX): An Overview from Discovery, Molecular Mechanisms to Physiology and Pathology. Antioxidants (Basel, Switzerland). 2021;10:890
8. Kwon J, Wang A, Burke DJ, Boudreau HE, Lekstrom KJ, Korzeniowska A. et al. Peroxiredoxin 6 (Prdx6) supports NADPH oxidase1 (Nox1)-based superoxide generation and cell migration. Free Radic Biol Med. 2016;96:99-115
9. Vendrov AE, Stevenson MD, Lozhkin A, Hayami T, Holland NA, Yang X. et al. Renal NOXA1/NOX1 Signaling Regulates Epithelial Sodium Channel and Sodium Retention in Angiotensin II-induced Hypertension. Antioxid Redox Signal. 2022;36:550-66
10. Orr AW, Woolard MD. Cardiovascular disease is obNOXious: New insights into NoxA1 in smooth muscle phenotype. Redox biology. 2019;22:101081
11. Vendrov AE, Sumida A, Canugovi C, Lozhkin A, Hayami T, Madamanchi NR. et al. NOXA1-dependent NADPH oxidase regulates redox signaling and phenotype of vascular smooth muscle cell during atherogenesis. Redox biology. 2019;21:101063
12. Feng J, Fu F, Nie Y. Comprehensive genomics analysis of aging related gene signature to predict the prognosis and drug resistance of colon adenocarcinoma. Front Pharmacol. 2023;14:1121634
13. Dong X, Li X, Gan Y, Ding J, Wei B, Zhou L. et al. TRAF4-mediated ubiquitination-dependent activation of JNK/Bcl-xL drives radioresistance. Cell Death Dis. 2023;14:102
14. Sun X, Dong M, Li J, Sun Y, Gao Y, Wang Y. et al. NRF2 promotes radiation resistance by cooperating with TOPBP1 to activate the ATR-CHK1 signaling pathway. Theranostics. 2024;14:681-98
15. Chaiswing L, St Clair WH, St Clair DK. Redox Paradox: A Novel Approach to Therapeutics-Resistant Cancer. Antioxid Redox Signal. 2018;29:1237-72
16. Kim W, Youn H, Kang C, Youn B. Inflammation-induced radioresistance is mediated by ROS-dependent inactivation of protein phosphatase 1 in non-small cell lung cancer cells. Apoptosis. 2015;20:1242-52
17. Lu L, Dong J, Wang L, Xia Q, Zhang D, Kim H. et al. Activation of STAT3 and Bcl-2 and reduction of reactive oxygen species (ROS) promote radioresistance in breast cancer and overcome of radioresistance with niclosamide. Oncogene. 2018;37:5292-304
18. Liu R, Li W, Tao B, Wang X, Yang Z, Zhang Y. et al. Tyrosine phosphorylation activates 6-phosphogluconate dehydrogenase and promotes tumor growth and radiation resistance. Nat Commun. 2019;10:991
19. Lu B, Chen XB, Ying MD, He QJ, Cao J, Yang B. The Role of Ferroptosis in Cancer Development and Treatment Response. Front Pharmacol. 2017;8:992
20. Li J, Hu C, Du Y, Tang X, Shao C, Xu T. et al. Identification of Iron Metabolism-Related Gene Signatures for Predicting the Prognosis of Patients With Sarcomas. Front Oncol. 2020;10:599816
21. Wang Y, Yu L, Ding J, Chen Y. Iron Metabolism in Cancer. Int J Mol Sci. 2018;20:95
22. Torti SV, Manz DH, Paul BT, Blanchette-Farra N, Torti FM. Iron and Cancer. Annu Rev Nutr. 2018;38:97-125
23. Yang W, Wang Y, Zhang C, Huang Y, Yu J, Shi L. et al. Maresin1 Protect Against Ferroptosis-Induced Liver Injury Through ROS Inhibition and Nrf2/HO-1/GPX4 Activation. Front Pharmacol. 2022;13:865689
24. Lei G, Mao C, Yan Y, Zhuang L, Gan B. Ferroptosis, radiotherapy, and combination therapeutic strategies. Protein & cell. 2021;12:836-57
25. Lei G, Zhang Y, Koppula P, Liu X, Zhang J, Lin SH. et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30:146-62
26. Zhang W, Sun Y, Bai L, Zhi L, Yang Y, Zhao Q. et al. RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A11. J Clin Invest. 2021;131:e152067
27. Wo JY, Anker CJ, Ashman JB, Bhadkamkar NA, Bradfield L, Chang DT. et al. Radiation Therapy for Rectal Cancer: Executive Summary of an ASTRO Clinical Practice Guideline. Pract Radiat Oncol. 2021;11:13-25
28. Maas M, Nelemans PJ, Valentini V, Das P, Rödel C, Kuo LJ. et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. The Lancet Oncology. 2010;11:835-44
29. Teng H, Wang Y, Sui X, Fan J, Li S, Lei X. et al. Gut microbiota-mediated nucleotide synthesis attenuates the response to neoadjuvant chemoradiotherapy in rectal cancer. Cancer Cell. 2023;41:124-38.e6
30. Wu Y, Song Y, Wang R, Wang T. Molecular mechanisms of tumor resistance to radiotherapy. Mol Cancer. 2023;22:96
31. Mittal A, Nenwani M, Sarangi I, Achreja A, Lawrence TS, Nagrath D. Radiotherapy-induced metabolic hallmarks in the tumor microenvironment. Trends in cancer. 2022;8:855-69
32. Schaue D, McBride WH. Opportunities and challenges of radiotherapy for treating cancer. Nat Rev Clin Oncol. 2015;12:527-40
33. Jiang H, Wang H, De Ridder M. Targeting antioxidant enzymes as a radiosensitizing strategy. Cancer Lett. 2018;438:154-64
34. Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579-91
35. Tang Y, Zhang Z, Chen Y, Qin S, Zhou L, Gao W. et al. Metabolic Adaptation-Mediated Cancer Survival and Progression in Oxidative Stress. Antioxidants (Basel, Switzerland). 2022;11:1324
36. Reis J, Gorgulla C, Massari M, Marchese S, Valente S, Noce B. et al. Targeting ROS production through inhibition of NADPH oxidases. Nature chemical biology. 2023;19:1540-50
37. Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiological reviews. 2007;87:245-313
38. Espinosa-Sotelo R, Fusté NP, Peñuelas-Haro I, Alay A, Pons G, Almodóvar X. et al. Dissecting the role of the NADPH oxidase NOX4 in TGF-beta signaling in hepatocellular carcinoma. Redox biology. 2023;65:102818
39. Konaté MM, Antony S, Doroshow JH. Inhibiting the Activity of NADPH Oxidase in Cancer. Antioxid Redox Signal. 2020;33:435-54
40. Desouki MM, Kulawiec M, Bansal S, Das GM, Singh KK. Cross talk between mitochondria and superoxide generating NADPH oxidase in breast and ovarian tumors. Cancer Biol Ther. 2005;4:1367-73
41. Zhu L, Zhao Y, Liu T, Chen M, Qian WP, Jiang B. et al. Inhibition of NADPH Oxidase-ROS Signal using Hyaluronic Acid Nanoparticles for Overcoming Radioresistance in Cancer Therapy. ACS nano. 2022;16:18708-28
42. Wang Y, Zhang Z, Sun W, Zhang J, Xu Q, Zhou X. et al. Ferroptosis in colorectal cancer: Potential mechanisms and effective therapeutic targets. Biomed Pharmacother. 2022;153:113524
43. Rochette L, Dogon G, Rigal E, Zeller M, Cottin Y, Vergely C. Lipid Peroxidation and Iron Metabolism: Two Corner Stones in the Homeostasis Control of Ferroptosis. Int J Mol Sci. 2022;24:449
44. Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17:2054-81
45. Chen Q, Zheng W, Guan J, Liu H, Dan Y, Zhu L. et al. SOCS2-enhanced ubiquitination of SLC7A11 promotes ferroptosis and radiosensitization in hepatocellular carcinoma. Cell Death Differ. 2023;30:137-51
Author contact
![]() Corresponding authors: Prof. Mao Wei, E-mail: maoweiac.cn; Prof. Ji Zhu, E-mail: zhujiorg.cn.
Corresponding authors: Prof. Mao Wei, E-mail: maoweiac.cn; Prof. Ji Zhu, E-mail: zhujiorg.cn.

 Global reach, higher impact
Global reach, higher impact