Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(6):1269-1277. doi:10.7150/ijms.109770 This issue Cite
Research Paper
Genetic variants of IGF2BP2 as potential predictors for perineural invasion of prostate cancer in a Taiwanese population
1. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan.
2. Division of Urology, Department of Surgery, Tungs' Taichung Metroharbor Hospital, Taichung, Taiwan.
3. Department of Nursing, Jenteh Junior College of Medicine, Nursing and Management, Miaoli, Taiwan.
4. International Master/PhD Program in Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan.
5. Department of Urology, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan.
6. Department of Urology, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan.
7. Graduate Institute of Clinical Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan.
8. Division of Urology, Department of Surgery, Taichung Veterans General Hospital, Taichung, Taiwan.
9. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
10. School of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan.
11. Department of Mathematics and Statistics, Florida Atlantic University, Boca Raton, FL, USA.
12. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
13. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
14. Pulmonary Research Center, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan.
15. Traditional Herbal Medicine Research Center, Taipei Medical University Hospital, Taipei, Taiwan.
16. TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei, Taiwan.
Received 2025-1-2; Accepted 2025-2-8; Published 2025-2-18
Abstract
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), which binds with high affinity to numerous RNA transcripts, is known to promote tumorigenesis and metastasis, including in prostate cancer (PCa). Several case-control studies investigated associations between IGF2BP2 polymorphisms and cancer progression. However, the effects of IGF2BP2 genetic variants on clinicopathological progression and biochemical recurrence (BCR) of PCa remain unclear. In this study, we recruited 698 Taiwanese PCa patients who underwent a radical prostatectomy to investigate associations of IGF2BP2 single-nucleotide polymorphisms (SNPs) with the risk of BCR and clinicopathological progression. Using a TaqMan allelic discrimination assay, we genotyped three IGF2BP2 SNPs located in the second intron: rs11705701 (G/A), rs4402960 (G/T), and rs1470579 (A/C). Our findings revealed that these IGF2BP2 SNPs had no significant effect on initial prostate-specific antigen (iPSA) levels or postoperative BCR. However, patients with the rs1470579 A/C genotype exhibited a higher risk of developing perineural invasion (PNI) compared to those with the homozygous A/A genotype. This association was particularly pronounced in patients with an elevated iPSA level (>10 ng/mL). Clinical observations from The Cancer Genome Atlas database showed that elevated IGF2BP2 levels in PCa tissues were significantly associated with higher Gleason scores and exhibited a trend toward correlating with tumor metastasis. In conclusion, our findings highlight that the IGF2BP2 rs1470579 A>C polymorphism may increase susceptibility for PNI among PCa patients in the Taiwanese population.
Keywords: Insulin-like growth factor 2 mRNA-binding protein 2, Single-nucleotide polymorphism, Perineural invasion, Prostate cancer
Introduction
Prostate cancer (PCa) is the most commonly diagnosed cancer and the second leading cause of cancer-related deaths among men globally[1]. Clinically, approximately 15% of cases are metastatic at diagnosis, with a 5-year survival rate of 31%[2]. While tumor metastasis traditionally occurs through blood vessels and lymphatic channels, PCa usually exhibits a tendency to invade and grow along prostatic nerves, a phenomenon known as perineural invasion (PNI). This invasion extends from the prostate to the pelvic plexus[3]. The perineural space was identified as a specialized microenvironment that facilitates both the spread and growth of PCa[3, 4]. Furthermore, studies have linked PNI to higher surgical Gleason scores and an increased risk of biochemical recurrence (BCR)[5-7]. For instance, a meta-analysis involving 13,412 PCa patients revealed that those with PNI had a 1.4-fold higher risk of BCR following a radical prostatectomy (RP)[8]. Historically, clinical diagnoses relied on digital rectal examinations and measuring blood levels of prostate-specific antigen (PSA) for PCa screening. However, these methods cannot reliably differentiate aggressive tumors based solely on biopsy results and PSA levels[9]. To date, biomarkers for predicting metastatic PCa (mPCa) remain under investigation, with limited studies and insufficient evidence supporting their clinical utility.
Numerous cancer research studies have confirmed the utility of tumor-associated genetic aberration-based biomarkers in assessing risk, enabling early diagnosis, and predicting therapeutic outcomes. Genetic polymorphisms, which refer to variations in genomic sequences among individuals, occur in approximately 1% of the general population. Among these, single-nucleotide polymorphisms (SNPs) are the most frequently observed variations in repeated sequences. Recently, a growing body of research has emphasized the critical roles of SNPs and other genomic alterations in predicting, prognosticating, and determining pharmacotherapeutic outcomes in PCa[10, 11]. For example, a key regulator of androgen receptor variants, Y-box-binding protein-1, linked to resistance to androgen deprivation therapy (ADT) in PCa, possesses an intronic SNP (rs1203072) that influences gene expression and is associated with PCa metastasis[12]. Additionally, 14 SNPs across six genes—XRCC4, PMS1, GATA3, IL13, CASP8, and IGF1—were found to be significantly correlated with cancer-specific survival in patients with mPCa[13]. Genetic variants in ADAM9 were also suggested to be potential predictors of BCR in PCa patients undergoing an RP[14]. Furthermore, SNPs such as rs12422149, rs1789693, and rs1077858 in androgen transporter genes, including solute carrier organic anion transporter family member 2B1, were identified as potential pharmacogenomic markers for resistance to ADT in PCa[15].
The insulin-like growth factor 2 (IGF2) mRNA-binding protein 2 (IGF2BP2), encoded by the IGF2BP2 gene, functions as an RNA-binding protein for IGF2 messenger (m)RNA[16]. IGFBP2 expression is generally sustained postnatally and plays a crucial role in RNA localization, stability, and translation. Recent research identified IGF2BP2 as a reader of N6-methyladenosine (m6A), the most prevalent internal RNA modification in eukaryotic cells. IGF2BP2 interacts with various types of RNAs, including mRNAs, circular (circ)RNAs, microRNAs (miRNAs), and long non-coding (lnc)RNAs, regulating diverse disease processes such as diabetes and cancers[17-19]. Genome-wide association studies (GWASs) have identified a cluster of SNPs within the second intron of IGF2BP2 as being associated with type 2 diabetes (T2D). Among these, rs4402960 and rs1470579 are the most frequently reported SNPs across various ethnic groups, including Chinese Han[20], Lebanese[20, 21], and Indian[22] populations. In cancer research, the IGF2BP2 SNPs rs4402960 and rs1470579 were linked to an increased risk of developing breast cancer[23] and esophageal cancer[24]. Additionally, the SNPs rs11705701, rs4402960, and rs1470579 were associated with advanced clinical stages, larger tumors, and lymph node metastasis in oral cancer[25]. Furthermore, rs4402960 and rs6769511 were identified as strong predictors of the chemotherapeutic response in patients with metastatic gastric cancer[26]. While several studies explored the clinical significance of IGF2BP2 SNPs in various cancer types, their impacts on PCa remain unexplored. In this study, we investigated associations of SNPs in the IGF2BP2 gene with the risk of BCR and clinicopathological progression in Taiwanese PCa patients who had undergone an RP.
Materials and Methods
Study participants
This study involved the analysis of blood samples from 698 PCa patients who underwent a robotic-assisted laparoscopic RP at Taichung Veterans General Hospital (TVGH; Taichung, Taiwan), between 2012 and 2018. Written informed consent was obtained from all participants before venous blood collection, and the study protocol received approval from the TVGH Institutional Review Board (IRB no. CE19062A-2). Medical data collected at the time of diagnosis included PSA levels, pathologic Gleason grades, clinical and pathologic T (tumor) and N (node) staging, cancer invasion areas (seminal vesicle, perineural, and lymphovascular regions), D'Amico classification, and the BCR status.
Genomic DNA extraction
Whole-blood samples from PCa patients were collected in EDTA-containing tubes. Genomic DNA was then extracted from the buffy coat layer following centrifugation, using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) according to the previously described[27]. The quality of the extracted DNA was assessed with a Nanodrop-2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) before serving as a template for a polymerase chain reaction (PCR)[28].
Selection of IGF2BP2 SNPs
Three SNPs located in the second intron of IGF2BP2—rs11705701 (G/A), rs4402960 (G/T), and rs1470579 (A/C)—were selected for analysis in this study. These SNPs were chosen based on prior studies highlighting rs4402960 and rs1470579 as the most prevalent variants associated with diabetes and various cancers[17]. Additionally, rs11705701 was linked to an increased risk of T2D[29] and oral cancer progression[25].
Determination of IGF2BP2 SNPs
The TaqMan SNP Genotyping Assay on an ABI StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) was used to identify alleles of IGF2BP2 SNPs—rs11705701 (assay ID: C_31742122_10), rs4402960 (assay ID: C_2165199_10), and rs1470579 (assay ID: C_2165184_10). Results were analyzed using SDS vers. 3.0 software (Applied Biosystems, Foster City, CA, USA). Detailed procedures of DNA genotyping were outlined in our previous study[30].
Bioinformatics analysis
Clinical data and mRNA sequencing for prostate adenocarcinoma (PRAD) samples from The Cancer Genome Atlas (TCGA) were accessed via the UCSC Xena database. IGF2BP2 gene expression was analyzed in relation to clinical features, including Gleason scores and clinical M stages. Progression-free survival (PFS) was assessed by categorizing PRAD patients into high- and low-IGF2BP2 expression groups, with statistical significance determined using the log-rank test. To identify IGF2BP2-associated pathways, a gene set enrichment analysis (GSEA) was performed.
Statistical analysis
Chi-squared and Student's t-test were employed to compare demographic characteristics between the PSA ≤ 10 ng/mL and > 10 ng/mL groups. Odds ratios (ORs) and adjusted ORs (AORs) with 95% confidence intervals (CIs) were estimated using multiple logistic regression models to determine associations between genotypic frequencies and the two PSA groups, as well as the risk of different clinicopathological characteristics. All statistical analyses were performed using SAS software (vers. 9.1, 2005, for Windows; SAS Institute, Cary, NC, USA), with p values of < 0.05 considered statistically significant.
Results
Demographic characteristics of PCa patients with high and low iPSA levels
Table 1 compares demographic characteristics of PCa patients with high iPSA (> 10 ng/mL, 365 patients) and low iPSA (≤ 10 ng/mL, 333 patients). Patients with a high iPSA level were significantly more likely to present with advanced clinical T stages (T3+T4) and N stage (N1) at diagnosis compared to those with a low iPSA. Surgical pathological findings showed that high-iPSA patients more frequently exhibited higher pathologic Gleason grades (4+5), advanced pathologic T (T3+T4) and N (N1) stages, and greater evidence of seminal vesicle, perineural, and lymphovascular invasion. Additionally, based on the D'Amico risk classification, a larger proportion of high-iPSA patients fell into the high-risk category. Furthermore, these patients also demonstrated a higher recurrence rate.
Associations between IGF2BP2 SNPs and iPSA levels in PCa patients
We next investigated the potential impacts of the three selected IGF2BP2 SNPs—rs11705701 (G/A), rs4402960 (G/T), and rs1470579 (A/C)—on iPSA levels in PCa patients at diagnosis. Genotype frequencies of these SNPs were analyzed in a cohort of 698 PCa patients. The most prevalent alleles were homozygous G/G for rs11705701 and rs4402960, and homozygous A/A for rs1470579 (Table 2). After adjusting for potential confounders, no significant associations were identified of the polymorphic frequencies of rs11705701, rs4402960, and rs1470579 with iPSA levels.
Relationships between clinicopathological features and IGF2BP2 SNPs in PCa patients with high and low iPSA levels
Next, to clarify impacts of IGF2BP2 genetic polymorphisms on PCa clinicopathological characteristics, we analyzed factors such as pathologic staging, clinical staging, Gleason grade groups, tumor invasion, D'Amico classification, and BCR. Among the three IGF2BP2 loci studied, the rs1470579 AC genotype was associated with a significantly higher risk of PNI (OR = 1.455, 95% CI = 1.009-2.100; p = 0.044) compared to the wild-type (WT) AA genotype, as shown in Table 3. In contrast, no significant associations were identified for rs11705701 or rs4402960 with the clinicopathological features analyzed (data not shown). Further stratification of PCa patients into high (n = 365) and low (n = 333) iPSA groups revealed that the rs1470579 AC or AC+CC genotypes had significantly higher risks of PNI in patients with high iPSA levels (OR = 1.862, 95% CI = 1.049-3.302; p = 0.032 and OR = 1.756, 95% CI = 1.022-3.017, respectively) (Tables 4, 5). However, this association was not observed in patients with low iPSA levels (data not shown).
Associations of IGF2BP2 expression levels with clinicopathological characteristics and prognoses of PCa patients
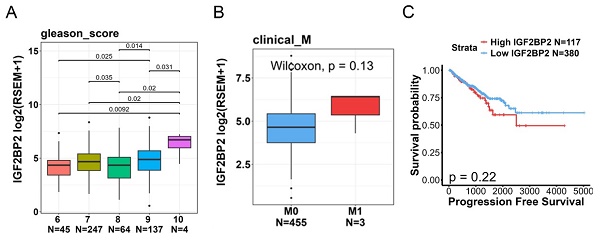
To further explore correlations between IGF2BP2 levels and disease progression or prognosis, we utilized TCGA-PRAD dataset. Our analysis revealed that IGF2BP2 expression was significantly higher in patients with high Gleason scores (Figure 1A). Moreover, we observed a trend where tumor tissues with distal metastasis exhibited higher IGF2BP2 expression compared to those without metastasis (Figure 1B). A Kaplan-Meier plot indicated that a higher IGF2BP2 expression level showed a trend to be associated with a shorter duration of PFS (Figure 1C).
Distributions of demographic characteristics among 698 prostate cancer patients
| Variable | PSA at diagnosis (ng/mL) | |||
|---|---|---|---|---|
| ≤ 10 (N=333) | > 10 (N=365) | p value | ||
| Age at diagnosis (years) | ||||
| ≤ 65 | 159 (47.7 %) | 136 (37.3 %) | p=0.005* | |
| > 65 | 174 (52.3 %) | 229 (62.7 %) | ||
| Pathologic Gleason grade group | ||||
| 1+2+3 | 303 (91.0 %) | 277 (75.9 %) | p<0.001* | |
| 4+5 | 30 (9.0 %) | 88 (24.1 %) | ||
| Clinical T stage | ||||
| 1+2 | 313 (94.0 %) | 290 (78.8 %) | p<0.001* | |
| 3+4 | 20 (6.0 %) | 75 (20.5 %) | ||
| Clinical N stage | ||||
| N0 | 330 (99.1 %) | 354 (97.0 %) | p=0.047* | |
| N1 | 3 (0.9 %) | 11 (3.0 %) | ||
| Pathologic T stage | ||||
| 2 | 230 (69.1 %) | 141 (38.6 %) | p<0.001* | |
| 3+4 | 103 (30.9 %) | 224 (61.4 %) | ||
| Pathologic N stage | ||||
| N0 | 318 (95.5 %) | 320 (87.7 %) | p<0.001* | |
| N1 | 15 (4.5 %) | 45 (12.3 %) | ||
| Seminal vesicle invasion | ||||
| No | 302 (90.7 %) | 248 (67.9 %) | p<0.001* | |
| Yes | 31 (9.3 %) | 117 (32.1 %) | ||
| Perineural invasion | ||||
| No | 114 (34.2%) | 72 (19.7 %) | p<0.001* | |
| Yes | 219 (65.8 %) | 293 (80.3 %) | ||
| Lymphovascular invasion | ||||
| No | 306 (91.9 %) | 281 (77.0 %) | p<0.001* | |
| Yes | 27 (8.1 %) | 84 (23.0 %) | ||
| D'Amico classification | ||||
| Low/Intermediate risk | 239 (71.8 %) | 107 (29.3 %) | p<0.001* | |
| High risk | 94 (28.2 %) | 258 (70.7 %) | ||
| Biochemical recurrence | ||||
| No | 266 (79.9 %) | 211 (57.8 %) | p<0.001* | |
| Yes | 67 (20.1 %) | 154 (42.2 %) | ||
* p < 0.05 indicates statistical significance. PSA, prostate-specific antigen.
Distribution frequencies of IGF2BP2 genotypes in 698 prostate cancer patients with high or low initial prostate-specific antigen (PSA)
| Variable | PSA at diagnosis (ng/mL) | |||
|---|---|---|---|---|
| ≤ 10 (N=333) (%) | > 10 (N=365) (%) | AOR (95% CI) | p value | |
| rs11705701 | ||||
| GG | 194 (58.3%) | 225 (61.6%) | 1.000 | |
| GA | 118 (35.4%) | 126 (34.5%) | 0.914 (0.637~1.313) | p=0.628 |
| AA | 21 (6.3%) | 14 (3.8%) | 0.625 (0.284~1.375) | p=0.242 |
| GA+AA | 139 (41.7%) | 140 (38.3%) | 0.871 (0.616~1.232) | p=0.435 |
| rs4402960 | ||||
| GG | 188 (56.5%) | 216 (59.2%) | 1.000 | |
| GT | 119 (35.7%) | 134 (36.7%) | 0.902 (0.630~1.293) | p=0.575 |
| TT | 26 (7.8%) | 15 (4.1%) | 0.591 (0.281~1.245) | p=0.167 |
| GT+TT | 145 (43.5%) | 149 (40.8%) | 0.850 (0.603~1.198) | p=0.354 |
| rs1470579 | ||||
| AA | 184 (55.3%) | 204 (55.9%) | 1.000 | |
| AC | 123 (36.9%) | 141 (38.6%) | 0.949 (0.663~1.357) | p=0.773 |
| CC | 26 (7.8%) | 20 (5.5%) | 0.710 (0.351~1.437) | p=0.341 |
| AC+CC | 149 (44.7%) | 161 (44.1%) | 0.909 (0.646~1.279) | p=0.583 |
The odds ratios (ORs) and their 95% confidence intervals (CIs) were estimated by logistic regression models. The adjusted ORs (AORs) with their 95% CIs were estimated by multiple logistic regression models after controlling for age at diagnosis, pathologic Gleason grade group, clinical T stage, pathologic T stage, pathologic N stage, seminal vesicle invasion, perineural invasion, lymphovascular invasion, biochemical recurrence, and D'Amico classification.
Exploration of the potential molecular mechanisms mediated by IGF2BP2 in PCa progression
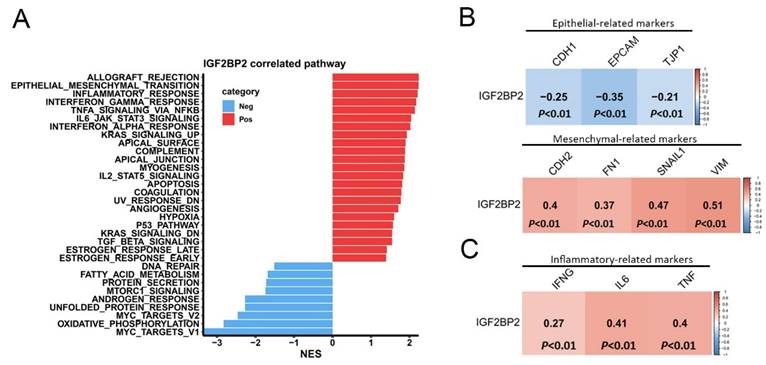
In order to investigate the mechanisms through which IGF2BP2 contributes to PCa progression, we performed a gene set enrichment analysis (GSEA) using TCGA-PRAD dataset. Results revealed that the epithelial-mesenchymal transition (EMT) pathway exhibited the second highest NES in association with IGF2BP2 expression (Figure 2A). Notably, the EMT was closely linked to PNI, as elevated levels of EMT regulators such as transforming growth factor-β (TGFB) and serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 (SERPINE1) are frequently detected in PNI-associated tissues[31]. Furthermore, the analysis identified several inflammation-related Hallmark gene sets as being enriched in patients with high IGF2BP2 expression (Figure 2A). Chronic inflammation was implicated as a key factor in PCa development and its progression to advanced metastatic stages[32]. Using the cBioPortal platform, we further analyzed correlations between EMT-related markers and IGF2BP2 expression in PCa tissues from TCGA. A strong positive correlation was observed between IGF2BP2 and mesenchymal phenotype-related genes (CDH2, FN1, SNAI1, and VIM) (Figure 2B, lower panel). Conversely, epithelial phenotype-related genes (CDH1, TJP1, and EPCAM) were negatively correlated with IGF2BP2 expression (Figure 2B, upper panel). Additionally, IGF2BP2 expression was positively associated with inflammation-related genes (IFNG, IL6, and TNF) (Figure 2C).
Clinical significance of IGF2BP2 expression in prostate cancer (PCa) patients evaluated using data from TCGA-prostate adenocarcinoma (PRAD) dataset. (A, B) IGF2BP2 expression levels were analyzed and compared according to Gleason scores (A) and clinical M stages (B) within TCGA-PRAD dataset. (C) Kaplan-Meier survival curves illustrating progression-free survival in patients with high versus low IGF2BP2 expression levels.
Odds ratios (ORs) and 95% confidence intervals (CIs) for associations between clinical characteristics and genotypic distributions of IGF2BP2 rs1470579 in a cohort of 698 prostate cancer patients
| Variable | rs1470579 | ||||||
|---|---|---|---|---|---|---|---|
| AA (N=388) | AC (N=264) | CC (N=46) | AA vs AC OR (95% CI) | p value | AA vs CC OR (95% CI) | p value | |
| Pathologic Gleason grade group | |||||||
| 1+2+3 | 321 (82.7%) | 220 (83.3%) | 39 (84.8%) | 1.000 | 0.841 | 1.000 | 0.727 |
| 4+5 | 67 (17.3%) | 44 (16.7%) | 7 (15.2%) | 0.958 (0.631~1.454) | 0.860 (0.369~2.005) | ||
| Clinical T stage | |||||||
| 1+2 | 329 (84.8%) | 232 (87.9%) | 42 (91.3%) | 1.000 | 0.265 | 1.000 | 0.236 |
| 3+4 | 59 (15.2%) | 32 (12.1%) | 4 (8.7%) | 0.769 (0.485~1.221) | 0.531 (0.184~1.537) | ||
| Clinical N stage | |||||||
| N0 | 377 (97.2%) | 262 (99.2%) | 45 (97.8%) | 1.000 | 0.062 | 1.000 | 0.796 |
| N1 | 11 (2.8%) | 2 (0.8%) | 1 (2.2%) | 0.262 (0.058~1.190) | 0.762 (0.096~6.038) | ||
| Pathologic T stage | |||||||
| 2 | 214 (55.2%) | 128 (48.5%) | 29 (63.0%) | 1.000 | 0.094 | 1.000 | 0.308 |
| 3+4 | 174 (44.8%) | 136 (51.5%) | 17 (37.0%) | 1.307 (0.955~1.788) | 0.721 (0.384~1.355) | ||
| Pathologic N stage | |||||||
| N0 | 353 (91.0%) | 241 (91.3%) | 44 (95.7%) | 1.000 | 0.892 | 1.000 | 0.283 |
| N1 | 35 (9.0%) | 23 (8.7%) | 2 (4.3%) | 0.963 (0.555~1.670) | 0.458 (0.107~1.972) | ||
| Seminal vesicle invasion | |||||||
| No | 310 (79.9%) | 200 (75.8%) | 40 (87.0%) | 1.000 | 0.209 | 1.000 | 0.252 |
| Yes | 78 (20.1%) | 64 (24.2%) | 6 (13.0%) | 1.272 (0.874~1.851) | 0.596 (0.244~1.456) | ||
| Perineural invasion | |||||||
| No | 111 (28.6%) | 57 (21.6%) | 18 (39.1%) | 1.000 | 0.044* | 1.000 | 0.140 |
| Yes | 277 (71.4%) | 207 (78.4%) | 28 (60.9%) | 1.455 (1.009~2.100) | 0.623 (0.331~1.172) | ||
| Lymphovascular invasion | |||||||
| No | 326 (84.0%) | 221 (83.7%) | 40 (87.0%) | 1.000 | 0.916 | 1.000 | 0.604 |
| Yes | 62 (16.0%) | 43 (16.3%) | 6 (13.0%) | 1.023 (0.669~1.565) | 0.789 (0.321~1.940) | ||
| D'Amico classification | |||||||
| Low risk/Intermediate risk | 194 (50.0%) | 128 (48.5%) | 24 (52.2%) | 1.000 | 0.704 | 1.000 | 0.780 |
| High risk | 194 (50.0%) | 136 (51.5%) | 22 (47.8%) | 1.063 (0.777~1.453) | 0.917 (0.497~1.690) | ||
| Biochemical recurrence | |||||||
| No | 268 (69.1%) | 177 (67.0%) | 32 (69.6%) | 1.000 | 0.585 | 1.000 | 0.945 |
| Yes | 120 (30.9%) | 87 (33.0%) | 14 (30.4%) | 1.098 (0.785~1.535) | 0.977 (0.503~1.898) | ||
ORs with their 95% CIs were estimated by logistic regression models.
* p < 0.05 indicates statistical significance.
Odds ratios (ORs) and 95% confidence intervals (CIs) of relationships between clinical characteristics and IGF2BP2 rs1470579 single-nucleotide polymorphisms under a codominant model in 365 prostate cancer patients with prostate-specific antigen of >10 ng/mL
| Variable | rs1470579 | ||||||
|---|---|---|---|---|---|---|---|
| AA (N=204) | AC (N=141) | CC (N=20) | AA vs AC OR (95% CI) | p value | AA vs CC OR (95% CI) | p value | |
| Pathologic Gleason grade group | |||||||
| 1+2+3 | 154 (75.5%) | 107 (75.9%) | 16 (80.0%) | 1.000 | 0.993 | 1.000 | 0.653 |
| 4+5 | 50 (24.5%) | 34 (24.1%) | 4 (20.0%) | 0.979 (0.593~1.615) | 0.770 (0.246~2.410) | ||
| Clinical T stage | |||||||
| 1+2 | 162 (79.4%) | 111 (78.7%) | 17 (85.0%) | 1.000 | 0.877 | 1.000 | 0.552 |
| 3+4 | 42 (20.6%) | 30 (21.3%) | 3 (15.0%) | 1.042 (0.615~1.766) | 0.681 (0.190~2.432) | ||
| Clinical N stage | |||||||
| N0 | 195 (95.6%) | 140 (99.3%) | 19 (95.0%) | 1.000 | 0.091 | 1.000 | 0.903 |
| N1 | 9 (4.4%) | 1 (0.7%) | 1 (5.0%) | 0.155 (0.019~1.236) | 1.140 (0.137~9.491) | ||
| Pathologic T stage | |||||||
| 2 | 86 (42.2%) | 48 (34.0%) | 7 (35.0%) | 1.000 | 0.128 | 1.000 | 0.535 |
| 3+4 | 118 (57.8%) | 93 (66.0%) | 13 (65.0%) | 1.412 (0.904~2.205) | 1.354 (0.518~3.535) | ||
| Pathologic N stage | |||||||
| N0 | 179 (87.7%) | 123 (87.2%) | 18 (90.0%) | 1.000 | 0.888 | 1.000 | 0.768 |
| N1 | 25 (12.3%) | 18 (12.8%) | 2 (10.0%) | 1.048 (0.548~2.003) | 0.796 (0.174~3.636) | ||
| Seminal vesicle invasion | |||||||
| No | 144 (70.6%) | 89 (63.1%) | 15 (75.0%) | 1.000 | 0.145 | 1.000 | 0.678 |
| Yes | 60 (29.4%) | 52 (36.9%) | 5 (25.0%) | 1.402 (0.889~2.212) | 0.800 (0.278~2.300) | ||
| Perineural invasion | |||||||
| No | 48 (23.5%) | 20 (14.2%) | 4 (20.0%) | 1.000 | 0.032* | 1.000 | 0.721 |
| Yes | 156 (76.5%) | 121 (85.8%) | 16 (80.0%) | 1.862 (1.049~3.302) | 1.231 (0.393~3.858) | ||
| Lymphovascular invasion | |||||||
| No | 159 (77.9%) | 107 (75.9%) | 15 (75.0%) | 1.000 | 0.655 | 1.000 | 0.763 |
| Yes | 45 (22.1%) | 34 (24.1%) | 5 (25.0%) | 1.123 (0.675~1.867) | 1.178 (0.406~3.416) | ||
| D'Amico classification | |||||||
| Low risk/Intermediate risk | 66 (32.4%) | 36 (25.5%) | 5 (25.0%) | 1.000 | 0.172 | 1.000 | 0.500 |
| High risk | 138 (67.6%) | 105 (74.5%) | 15 (75.0%) | 1.395 (0.864~2.252) | 1.435 (0.500~4.116) | ||
| Biochemical recurrence | |||||||
| No | 123 (60.3%) | 77 (54.6%) | 11 (55.0%) | 1.000 | 0.293 | 1.000 | 0.645 |
| Yes | 81 (39.7%) | 64 (45.4%) | 9 (45.0%) | 1.262 (0.818~1.948) | 1.242 (0.493~3.132) | ||
ORs with their 95% CIs were estimated by logistic regression models.
* p < 0.05 indicates statistical significance.
Pathways associated with IGF2BP2 in prostate cancer (PCa) patients. (A) Horizontal bar plot illustrating pathways linked to IGF2BP2 expression. Pathways positively associated with IGF2BP2 are shown in red, while those negatively associated are in blue. The x-axis displays normalized enrichment scores (NESs), and the y-axis lists pathways identified from the Hallmark database. (B, C) Correlation plots showing relationships of IGF2BP2 expression with biomarkers of the epithelial-mesenchymal transition (B) and with biomarkers of inflammatory responses (C). RNA sequencing data from TCGA prostate adenocarcinoma (PRAD) dataset were analyzed. A Pearson correlation analysis was conducted to assess relationships between IGF2BP2 and the biomarkers, with correlation coefficients and p values displayed in each square. The scale bar indicates the strength of the correlation.
Odds ratios (ORs) and 95% confidence intervals (CIs) for the relationship between clinical characteristics and IGF2BP2 rs1470579 single-nucleotide polymorphisms under a dominant model in 365 prostate cancer patients with prostate-specific antigen of >10 ng/mL
| Variable | Genotypic frequencies | |||
|---|---|---|---|---|
| rs1470579 | AA (N=204) | AC+CC (N=161) | OR (95% CI) | p value |
| Pathologic Gleason grade group | ||||
| 1+2+3 | 154 (75.5%) | 123 (76.4%) | 1.000 | 0.841 |
| 4+5 | 50 (24.5%) | 38 (23.6%) | 0.952 (0.587~1.544) | |
| Clinical T stage | ||||
| 1+2 | 162 (79.4%) | 128 (79.5%) | 1.000 | 0.983 |
| 3+4 | 42 (20.6%) | 33 (20.5%) | 0.994 (0.596~1.658) | |
| Clinical N stage | ||||
| N0 | 195 (95.6%) | 159 (98.8%) | 1.000 | 0.079 |
| N1 | 9 (4.4%) | 2 (1.2%) | 0.273 (0.058~1.279) | |
| Pathologic T stage | ||||
| 2 | 86 (42.2%) | 55 (34.2%) | 1.000 | 0.119 |
| 3+4 | 118 (57.8%) | 106 (65.8%) | 1.405 (0.915~2.155) | |
| Pathologic N stage | ||||
| N0 | 179 (87.7%) | 141 (87.6%) | 1.000 | 0.961 |
| N1 | 25 (12.3%) | 20 (12.4%) | 1.016 (0.542~1.903) | |
| Seminal vesicle invasion | ||||
| No | 144 (70.6%) | 104 (64.6%) | 1.000 | 0.223 |
| Yes | 60 (29.4%) | 57 (35.4%) | 1.315 (0.846~2.046) | |
| Perineural invasion | ||||
| No | 48 (23.5%) | 24 (14.9%) | 1.000 | 0.040* |
| Yes | 156 (76.5%) | 137 (85.1%) | 1.756 (1.022~3.017) | |
| Lymphovascular invasion | ||||
| No | 159 (77.9%) | 122 (75.8%) | 1.000 | 0.626 |
| Yes | 45 (22.1%) | 39 (24.2%) | 1.130 (0.692~1.843) | |
| D'Amico classification | ||||
| Low risk/Intermediate risk | 66 (32.4%) | 41 (25.5%) | 1.000 | 0.151 |
| High risk | 138 (67.6%) | 120 (74.5%) | 1.400 (0.884~2.218) | |
| Biochemical recurrence | ||||
| No | 123 (60.3%) | 88 (54.7%) | 1.000 | 0.279 |
| Yes | 81 (39.7%) | 73 (45.3%) | 1.260 (0.829~1.914) | |
ORs with their 95% CIs were estimated by logistic regression models.
* p < 0.05 indicates statistical significance.
Discussion
Given the pivotal role of IGF2BP2 as an m6A reader that stabilizes RNA and promotes oncogenic effects in PCa progression[33, 34], we examined polymorphisms in the second intron of the IGF2BP2 gene, observing distinct distributions in PCa patients with high versus low iPSA levels. Our analysis revealed that patients carrying the mutant AC genotype of rs1470579 exhibited a significantly elevated risk of developing PNI, with stronger associations observed in those with high iPSA. These findings underscore the potential impact of specific IGF2BP2 genetic variants on PNI, particularly in high-iPSA PCa patients at diagnosis. Additionally, we found that IGF2BP2 expression levels were significantly associated with pathological Gleason scores, and EMT- and inflammatory-related pathways in PCa patients.
Previous studies showed that PCa patients with PNI tend to have higher iPSA levels compared to those without PNI. Additionally, PNI was linked to elevated surgical Gleason scores[5]. We suggest that the rs1470579 SNP may regulate IGF2BP2 expression, thereby promoting PNI in PCa patients, particularly those with high iPSA. Consistent with this, we observed that IGF2BP2 expression was also associated with higher Gleason scores in PCa tissues. The rs1470579 SNP is situated within the intron of the IGF2BP2 gene. Although polymorphisms in intronic regions do not directly alter protein sequences, growing evidence indicates that such variations can lead to splicing abnormalities, potentially affecting translation and contributing to various diseases, including cancers[35]. Moreover, intronic sequences often contain numerous cis-acting regulatory elements (CREs), such as transcription factor-binding sites, enhancers, and silencers, which can modulate gene expression either positively or negatively[36]. Additionally, many lncRNAs are embedded within intronic regions and are known to regulate expressions of their corresponding host genes[36]. In this study, we proposed that the IGF2BP2 SNP rs1470579 may affect IGF2BP2 expression, contributing to the development of PNI in PCa. However, the mechanism by which the rs1470579 SNP regulates IGF2BP2 expression remains uncertain and requires further exploration in future research.
Recent studies suggested a possible role of the EMT in PNI of tumor cells, including salivary gland cystic carcinoma (SACC). The brain-derived neurotrophic factor (BDNF)/tropomyosin-related kinase B (TrkB) axis was shown to facilitate tumor progression and PNI through the EMT in SACC[37]. Similarly, in PCa, activation of the BDNF/TrkB pathway was implicated in disease progression by inducing the EMT[38]. EMT regulators are emerging as potential biomarkers for PNI. For instance, silencing the EMT-inducing factor, Slug, was demonstrated to increase E-cadherin expression, thereby inhibiting the EMT and PNI in SACC[39]. Additionally, Kakies et al. reported a PCa case featuring neuroendocrine differentiation and extensive PNI, in which reduced E-cadherin and elevated vimentin levels highlighted potential links of neuroendocrine differentiation with the, EMT and PNI in PCa[7]. In this study, we identified a positive association between IGF2BP2 expression and EMT-related gene signatures in TCGA-PRAD dataset. Analysis of human PCa samples using the cBioPortal platform revealed that IGF2BP2 expression was significantly correlated with mesenchymal phenotype-related genes and inversely correlated with epithelial phenotype-related genes. Furthermore, IGF2BP2 expression was shown to promote the EMT and metastasis in various cancers, including gastric[40] and oral[41] cancers. These findings suggest that the rs1470579 genetic variant might upregulate IGF2BP2 expression, influence the EMT process, and thereby contribute to PNI in PCa.
Several tumor studies reported a strong association between nerve invasion and an inflammatory response[42, 43]. Additionally, nerve invasion was implicated in facilitating immune escape during tumor development[44]. These observations highlight a close interplay among PNI, inflammation, and immune responses. In this study, we found that IGF2BP2 expression was significantly associated with multiple inflammation-related pathways in PCa, including INFLAMMATORY_RESPONSE, INTERFERON_GAMMA_RESPONSE, TNFA_SIGNALING_VIA_NFKB, and IL6_JAK_STAT3_SIGNALING. Analysis of TCGA dataset further revealed notable correlations between IGF2BP2 expression and levels of key inflammatory mediators, such as interferon-γ (IFNG), interleukin-6 (IL6), and tumor necrosis factor (TNF), in PCa tissues. Moreover, previous research demonstrated that IGF2BP2-knockdown suppressed inflammatory responses in gastric cancer[45]. These findings suggest that the rs1470579 genetic variant may enhance IGF2BP2 expression, thereby modulating interactions between inflammatory responses and PNI in PCa.
In summary, this is the first study to investigate distinct allelic effects of the IGF2BP2 rs1470579 SNP in a Taiwanese population, emphasizing its influence on the occurrence of PNI in PCa. Our findings suggest that IGF2BP2-related pathways, including the EMT and inflammatory responses, may act as key contributors to PCa-associated PNI. Additionally, the IGF2BP2 rs1470579 variant holds promise as a potential biomarker for PNI in PCa, particularly in patients presenting with high iPSA levels.
Acknowledgements
Funding
This research was funded by the TMU Research Center of Cancer Translational Medicine under the Featured Areas Research Center Program, supported by the Higher Education Sprout Project of the Ministry of Education (MOE) in Taiwan (grant to M.-H. Chien), as well as grant no. 114-wf-swf-01 from Taipei Medical University-Wan Fang Hospital (grant to Y.-C. Wen).
Ethics approval
Experiments involving clinical samples were approved by the Institutional Review Board at Taichung Veterans General Hospital (IRB no. CE19062A-2).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
3. Zareba P, Flavin R, Isikbay M, Rider JR, Gerke TA, Finn S. et al. Perineural Invasion and Risk of Lethal Prostate Cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:719-26
4. Ayala GE, Dai H, Ittmann M, Li R, Powell M, Frolov A. et al. Growth and survival mechanisms associated with perineural invasion in prostate cancer. Cancer Res. 2004;64:6082-90
5. Kraus RD, Barsky A, Ji L, Garcia Santos PM, Cheng N, Groshen S. et al. The Perineural Invasion Paradox: Is Perineural Invasion an Independent Prognostic Indicator of Biochemical Recurrence Risk in Patients With pT2N0R0 Prostate Cancer? A Multi-Institutional Study. Adv Radiat Oncol. 2019;4:96-102
6. Ball MW, Partin AW, Epstein JI. Extent of extraprostatic extension independently influences biochemical recurrence-free survival: evidence for further pT3 subclassification. Urology. 2015;85:161-4
7. Niu Y, Förster S, Muders M. The Role of Perineural Invasion in Prostate Cancer and Its Prognostic Significance. Cancers (Basel). 2022;14:4065
8. Zhang LJ, Wu B, Zha ZL, Qu W, Zhao H, Yuan J. et al. Perineural invasion as an independent predictor of biochemical recurrence in prostate cancer following radical prostatectomy or radiotherapy: a systematic review and meta-analysis. BMC Urol. 2018;18:5
9. Couñago F, López-Campos F, Díaz-Gavela AA, Almagro E, Fenández-Pascual E, Henríquez I. et al. Clinical Applications of Molecular Biomarkers in Prostate Cancer. Cancers (Basel). 2020;12:1550
10. Shiota M, Akamatsu S, Narita S, Terada N, Fujimoto N, Eto M. Genetic Polymorphisms and Pharmacotherapy for Prostate Cancer. Jma j. 2021;4:99-111
11. Allemailem KS, Almatroudi A, Alrumaihi F, Makki Almansour N, Aldakheel FM, Rather RA. et al. Single nucleotide polymorphisms (SNPs) in prostate cancer: its implications in diagnostics and therapeutics. Am J Transl Res. 2021;13:3868-89
12. Shiota M, Fujimoto N, Imada K, Yokomizo A, Itsumi M, Takeuchi A. et al. Potential Role for YB-1 in Castration-Resistant Prostate Cancer and Resistance to Enzalutamide Through the Androgen Receptor V7. J Natl Cancer Inst. 2016 108
13. Tsuchiya N, Matsui S, Narita S, Kamba T, Mitsuzuka K, Hatakeyama S. et al. Distinct cancer-specific survival in metastatic prostate cancer patients classified by a panel of single nucleotide polymorphisms of cancer-associated genes. Genes Cancer. 2013;4:54-60
14. Lin YW, Wen YC, Lin CY, Hsiao CH, Ho KH, Huang HC. et al. Genetic variants of ADAM9 as potential predictors for biochemical recurrence in prostate cancer patients after receiving a radical prostatectomy. Int J Med Sci. 2024;21:2934-42
15. Yang M, Xie W, Mostaghel E, Nakabayashi M, Werner L, Sun T. et al. SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J Clin Oncol. 2011;29:2565-73
16. Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262-70
17. Wang J, Chen L, Qiang P. The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers. Cancer Cell Int. 2021;21:99
18. Huang C, Xu R, Zhu X, Jiang H. m6A-modified circABCC4 promotes stemness and metastasis of prostate cancer by recruiting IGF2BP2 to increase stability of CCAR1. Cancer Gene Ther. 2023;30:1426-40
19. Lin SH, Lin CW, Lu JW, Yang WE, Lin YM, Lu HJ. et al. Cytoplasmic IGF2BP2 Protein Expression in Human Patients with Oral Squamous Cell Carcinoma: Prognostic and Clinical Implications. Int J Med Sci. 2022;19:1198-204
20. Zhang LF, Pei Q, Yang GP, Zhao YC, Mu YF, Huang Q. et al. The effect of IGF2BP2 gene polymorphisms on pioglitazone response in Chinese type 2 diabetes patients. Pharmacology. 2014;94:115-22
21. Mtiraoui N, Turki A, Nemr R, Echtay A, Izzidi I, Al-Zaben GS. et al. Contribution of common variants of ENPP1, IGF2BP2, KCNJ11, MLXIPL, PPARγ, SLC30A8 and TCF7L2 to the risk of type 2 diabetes in Lebanese and Tunisian Arabs. Diabetes Metab. 2012;38:444-9
22. Chauhan G, Spurgeon CJ, Tabassum R, Bhaskar S, Kulkarni SR, Mahajan A. et al. Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5,164 Indians. Diabetes. 2010;59:2068-74
23. Liu G, Zhu T, Cui Y, Liu J, Liu J, Zhao Q. et al. Correlation between IGF2BP2 gene polymorphism and the risk of breast cancer in Chinese Han women. Biomed Pharmacother. 2015;69:297-300
24. Qiu H, Wang Y, Kang M, Ding H, Liu C, Tang W. et al. The relationship between IGF2BP2 and PPARG polymorphisms and susceptibility to esophageal squamous-cell carcinomas in the eastern Chinese Han population. Onco Targets Ther. 2017;10:5525-32
25. Chou CH, Chang CY, Lu HJ, Hsin MC, Chen MK, Huang HC. et al. IGF2BP2 Polymorphisms Are Associated with Clinical Characteristics and Development of Oral Cancer. Int J Mol Sci. 2020;21:5662
26. Liu X, Chen Z, Zhao X, Huang M, Wang C, Peng W. et al. Effects of IGF2BP2, KCNQ1 and GCKR polymorphisms on clinical outcome in metastatic gastric cancer treated with EOF regimen. Pharmacogenomics. 2015;16:959-70
27. Weng WC, Hsieh YH, Lin CY, Liu YF, Su SC, Wang SS. et al. Functional variants of the pentraxin 3 gene are associated with the metastasis and progression of prostate cancer. J Cell Mol Med. 2024;28:e70041
28. Chen YT, Lin CW, Chou YE, Su SC, Chang LC, Lee CY. et al. Potential impact of ADAM-10 genetic variants with the clinical features of oral squamous cell carcinoma. J Cell Mol Med. 2023;27:1144-52
29. Chistiakov DA, Nikitin AG, Smetanina SA, Bel'chikova LN, Suplotova LA, Shestakova MV. et al. The rs11705701 G>A polymorphism of IGF2BP2 is associated with IGF2BP2 mRNA and protein levels in the visceral adipose tissue - a link to type 2 diabetes susceptibility. Rev Diabet Stud. 2012;9:112-22
30. Lin YW, Wang SS, Wen YC, Tung MC, Lee LM, Yang SF. et al. Genetic Variations of Melatonin Receptor Type 1A are Associated with the Clinicopathologic Development of Urothelial Cell Carcinoma. Int J Med Sci. 2017;14:1130-5
31. Pavón MA, Arroyo-Solera I, Céspedes MV, Casanova I, León X, Mangues R. uPA/uPAR and SERPINE1 in head and neck cancer: role in tumor resistance, metastasis, prognosis and therapy. Oncotarget. 2016;7:57351-66
32. Stark T, Livas L, Kyprianou N. Inflammation in prostate cancer progression and therapeutic targeting. Transl Androl Urol. 2015;4:455-63
33. Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z. et al. m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med. 2021;11:e426
34. He P, Liu X, Yu G, Wang Y, Wang S, Liu J. et al. METTL3 facilitates prostate cancer progression via inducing HOXC6 m6A modification and stabilizing its expression through IGF2BP2-dependent mechanisms. Mol Cell Biochem. 2024;479:1707-20
35. Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389-96
36. Deng N, Zhou H, Fan H, Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8:110635-49
37. Jia S, Wang W, Hu Z, Shan C, Wang L, Wu B. et al. BDNF mediated TrkB activation contributes to the EMT progression and the poor prognosis in human salivary adenoid cystic carcinoma. Oral Oncol. 2015;51:64-70
38. Li T, Yu Y, Song Y, Li X, Lan D, Zhang P. et al. Activation of BDNF/TrkB pathway promotes prostate cancer progression via induction of epithelial-mesenchymal transition and anoikis resistance. Faseb j. 2020;34:9087-101
39. Wu B, Wei J, Hu Z, Shan C, Wang L, Zhang C. et al. Slug silencing inhibited perineural invasion through regulation of EMMPRIN expression in human salivary adenoid cystic carcinoma. Tumour Biol. 2016;37:2161-9
40. Ouyang J, Li J, Li D, Jiang J, Hao T, Xia Y. et al. IGF2BP2 Promotes Epithelial to Mesenchymal Transition and Metastasis through Stabilizing HMGA1 mRNA in Gastric Cancer. Cancers (Basel). 2022;14:5381
41. Lin CW, Yang WE, Su CW, Lu HJ, Su SC, Yang SF. IGF2BP2 promotes cell invasion and epithelial-mesenchymal transition through Src-mediated upregulation of EREG in oral cancer. Int J Biol Sci. 2024;20:818-30
42. Bakst RL, Xiong H, Chen CH, Deborde S, Lyubchik A, Zhou Y. et al. Inflammatory Monocytes Promote Perineural Invasion via CCL2-Mediated Recruitment and Cathepsin B Expression. Cancer Res. 2017;77:6400-14
43. Chen SH, Zhang BY, Zhou B, Zhu CZ, Sun LQ, Feng YJ. Perineural invasion of cancer: a complex crosstalk between cells and molecules in the perineural niche. Am J Cancer Res. 2019;9:1-21
44. Cervantes-Villagrana RD, Albores-García D, Cervantes-Villagrana AR, García-Acevez SJ. Tumor-induced neurogenesis and immune evasion as targets of innovative anti-cancer therapies. Signal Transduct Target Ther. 2020;5:99
45. Liu D, Xia AD, Wu LP, Li S, Zhang K, Chen D. IGF2BP2 promotes gastric cancer progression by regulating the IGF1R-RhoA-ROCK signaling pathway. Cell Signal. 2022;94:110313
Author contact
![]() Corresponding author: Ming-Hsien Chien, PhD; E-mail: d002089002edu.tw or Shun-Fa Yang, PhD; E-mail: ysfedu.tw.
Corresponding author: Ming-Hsien Chien, PhD; E-mail: d002089002edu.tw or Shun-Fa Yang, PhD; E-mail: ysfedu.tw.

 Global reach, higher impact
Global reach, higher impact