Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(3):604-615. doi:10.7150/ijms.103834 This issue Cite
Research Paper
Glycosphingolipids-Dependent Phospholipid Metabolism Enhances Cancer Initiation and Progression through SMPD1/GLTP/B3GALT4/ST8SIA6 Signaling Axis: A Novel Therapeutic Target
Department of Gastrointestinal Surgery, The First Hospital of Quanzhou Affiliated to Fujian Medical University, Quanzhou, 362002, China.
Received 2024-9-18; Accepted 2024-12-14; Published 2025-1-6
Abstract
Colorectal cancer (CRC) is a prevalent malignancy with high morbidity and mortality rates globally. Advances in single-cell sequencing technology have enabled comprehensive analyses of tumor cells at single-cell resolution, providing valuable insights into the molecular mechanisms underlying CRC initiation and progression. In this study, we integrated single-cell sequencing data with the TCGA database to identify key molecular pathways involved in CRC pathogenesis. Our analysis revealed that dysregulation of phospholipid metabolism, particularly sphingolipid metabolism, plays a crucial role in CRC development. Specifically, we observed aberrant expression of genes involved in sphingolipid biosynthesis and degradation, as well as altered levels of various sphingolipid metabolites in CRC cells. Furthermore, we identified several potential therapeutic targets, including SMPD1, GLTP, B3GALT4, and ST8SIA6, within the sphingolipid metabolism pathway that could be exploited for the development of novel CRC treatments. Overall, our findings provide novel insights into the molecular mechanisms underlying CRC and highlight the importance of targeting phospholipid metabolism, specifically sphingolipid metabolism, as a potential therapeutic strategy for CRC.
Keywords: Colorectal cancer, Phospholipid metabolism, Molecular mechanisms, Single-cell sequencing analysis
Introduction
Cancer remains a major health challenge for humanity, with colon cancer being a common gastrointestinal tumor. Due to anatomical continuity, colon cancer data is often reported together with rectal cancer[1]. In 2020, colorectal cancer ranked third in global cancer incidence and mortality rates were also among the highest. Since the mid-1990s, rectal cancer has exhibited a long and sharp increase, with an annual increase of 1.3% in adults aged 40-49 and 0.5% in adults aged 50-54[2]. The treatment of colorectal cancer mainly involves surgery and chemotherapy, with a significant disparity in 5-year survival rates and prognosis based on different stages. The 5-year survival rate for stage IV colon cancer can be as low as 10%, posing a significant burden on human health and resulting in substantial socio-economic costs[3]. Therefore, there is crucial clinical significance in seeking new screening and treatment methods at an early stage.
The etiology of colorectal cancer is not yet fully understood, and most cases are the result of multiple risk factors acting together. In addition to gender, age, and genetic factors, the influence of lifestyle factors cannot be ignored, such as alcohol consumption, smoking, high-fat and low-fiber diets, and lack of physical activity[4, 5]. Various factors disrupt the intestinal microbiota, leading to intestinal permeability and affecting the integrity of the intestinal epithelium. Consequently, they alter intestinal mucosal barrier immunity, reduce nutrient absorption, and regulate secondary bile acid metabolites[6]. Due to the metabolic changes mentioned above, an excess of free radicals derived from secondary bile acid metabolites is produced, leading to an imbalance between endogenous antioxidants and free radicals[7]. The intestinal mucosa and epithelium undergo severe stress responses, resulting in local and even systemic inflammatory reactions and infiltration. All these factors ultimately contribute to various gastrointestinal diseases, including colorectal cancer, inflammatory bowel disease, and gastritis[8].
In summary, there are many factors contributing to the development of colon cancer, making its pathogenesis highly complex. In recent years, with the continuous optimization and development of genetic engineering techniques, high-throughput sequencing, and single-cell sequencing technologies, a large number of metabolism-related genes associated with colon cancer have been discovered, participating in crucial signaling pathways of colorectal cancer. In 2018, Wataru Sakamoto and his research team found that neutral ceramidase (nCDase), which regulates sphingolipid metabolism, is located in the Golgi apparatus of colon cancer cells and protects cells from ceramide-induced apoptosis[9]. More recently, researchers have identified specific genes, GSTA1, TONSL, and AGA, through a mixed-effect scoring test, confirming their involvement in homocysteine-related amino acid metabolism and folate interaction, thus impacting the occurrence and development of colorectal cancer[10]. Results from a study by Professor Dong Ho Lee and his team at Seoul National University in Korea showed a positive correlation between the total sum of metabolic syndrome (MetS) components and the risk of early-onset distal colon and rectal cancer, but no influence on proximal colon cancer[11]. Based on the researchers' continuous studies in recent years, the treatment of colorectal cancer not only involves early surgical intervention and traditional radiotherapy and chemotherapy but also includes targeted therapy and immunotherapy, which have shown significant efficacy. By combining comprehensive treatments and developing precise personalized treatment plans based on tumor characteristics, we aim to alleviate the physical and economic burdens on patients throughout the disease progression and treatment process. Therefore, in-depth research and exploration of relevant targets involved in the disease development process remain integral to our daily work, providing more possibilities for the diagnosis, treatment, and evaluation of the disease.
Methods
Ethical statement
This study was approved by the Ethics Committee of the First Hospital of Quanzhou Affiliated to Fujian Medical University, QuanZhou (No. GSE163974).
Single-cell sequencing data analysis
To analyze the single-cell sequencing data of colon cancer from GSE163974 using the R language, you can follow these steps: (1) Data downloading and import: Start by downloading the GSE163974 dataset from GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi)[12], and import it into the R environment. The R packages of `GEOquery` and `Seurat` were included for this purpose[13, 14]. (2) Data preprocessing: Apply quality control and preprocessing steps to the single-cell sequencing data. This involves removing low-quality cells, low-expressed genes, and outliers. Packages of `Seurat` provide useful functions like `FilterCells()`, `NormalizeData()`, and `ScaleData()` for this step. (3) Data visualization and exploratory analysis: Use scatter plots, box plots, heatmaps, and other visualization techniques to explore the single-cell data. This plot could observed cell distributions, clustering patterns, and differential gene expression. The `Seurat` package offers functions of `DimPlot()` and `FeaturePlot()` for visualization. (4) Cluster analysis: Utilize clustering algorithms to group similar cells together and assign cluster labels. Common clustering algorithms include k-means, DBSCAN, and hierarchical clustering. The `Seurat` package's `FindClusters()` function used for this analysis. (5) Differential analysis: Perform differential expression analysis to identify genes with significant differential expression among different clusters. The `Seurat` package's `FindMarkers()` function were applied. (6) Cell type annotation: Based on the results of differential analysis, annotate the cell types for each cluster using a reference genome or cell type-specific gene set. Tools of `SingleR` package and `Seurat` package's `AddModuleScore()` function used for this purpose[15]. (7) Dynamic analysis and pseudotime trajectory: If your dataset includes time-series measurements, apply dynamic analysis methods and pseudotime analysis algorithms to study the developmental patterns of colon cancer cells. The `Seurat` package provides methods of `Monocle` and `PAGA` for dynamic simulation and pseudotime analysis[14]. Herein, these methods for analyzing the GSE163974 colon cancer single-cell sequencing data using the R language.
Gene function and pathway enrichment analysis
Gene function and pathway enrichment analysis plays a crucial role in understanding the biological significance of differentially expressed genes. In this methodology, (a) Gene Ontology (GO) Enrichment Analysis: (1) Retrieving GO annotation data: Download and import the required GO annotation files or use pre-existing ones from DAVID databases (https://david.ncifcrf.gov)[16]. (2) Mapping gene identifiers: Use R packages of org.Hs.eg.db to perform mapping and annotation, to ensure that the gene identifiers in the annotation file match those in the expression dataset[17]. (3) Performing GO enrichment analysis: Utilize R packages of clusterProfiler and GOstats to identify significantly enriched GO terms among differentially expressed genes (DEGs). Perform statistical tests of Bonferroni to calculate the enrichment significance of each term with the B-H adjusted p-value less than or equal 0.05. (b) Pathway Enrichment Analysis: (1) Retrieving pathway annotation data: Download or retrieve the relevant pathway annotation files, which can be obtained from KEGG database (https://www.kegg.jp); (2) Mapping gene identifiers: Ensure the gene identifiers in the pathway annotation file match those in the expression dataset. Use R packages of clusterProfiler to perform mapping and annotation[18]. (3) Utilizing statistical approaches of Bonferroni to identify the enriched pathway with the B-H adjusted p-value less than or equal 0.05.
Colon cancer TCGA database analysis
Here, this methodology outlines the steps involved in analyzing COAD from The Cancer Genome Atlas (TCGA) database using the R language. (1) Accessing TCGA data: Obtain colon cancer data from TCGA through the Genomic Data Commons (GDC) Data Portal or using R package of TCGAbiolinks[19, 20]. Download relevant data files[21], including gene expression and clinical information[22]. (2) Data Preprocessing: (a) Data quality control: Perform quality assessment and filtering of the TCGA datasets to ensure the removal of low-quality or unreliable samples. Implement quality control checks based on specific criteria, such as sample integrity, sequencing depth, or batch effects. (b) Gene expression normalization: Apply normalization techniques of DESeq2, to account for variation in gene expression due to technical biases. This step harmonizes gene expression values for subsequent analysis. (3) Differential Expression Analysis: utilize R packages of DESeq2 to identify DEGs between COAD samples and normal controls. Adjust for multiple testing to control for false discovery rate (FDR). The statistical thresholds to determine significant DEGs based on log2|fold change| over than or equal 2 and B-H adjusted p-value less than or equal 0.05.
Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA)
GSEA and GSVA are powerful tools for identifying biological pathways or gene sets that are differentially regulated in various experimental conditions[17, 23-26]. This methodology provides a comprehensive guide for conducting GSEA and GSVA analysis to gain insights into pathway level changes in gene expression[27-30]. Gene sets were obtained from MSigDB databases (https://www.gsea-msigdb.org/gsea/index.jsp). Here, the analysis as following: (a) Preranked gene list: Rank genes based on their differential expression between different conditions or phenotypes. Use statistical methods of limma to compute fold changes and p-values. (b) Running GSEA: Utilize R packages of clusterProfiler, fgsea, and GSEABase to perform GSEA analysis. Input the ranked gene list and the gene set collection. Generate an enrichment score (ES) and a normalized enrichment score (NES) to determine the significance of gene set enrichment. (c)Visualize GSEA results using enrichment plots, showing the ranking of genes along with running enrichment scores. Identify significantly enriched gene sets based on the NES and assess their biological relevance.
For the GSVA analysis, the following steps were applied: (1) Gather the gene sets that represent biological pathways, processes, and functional gene categories. These gene sets also can be obtained from MSigDB databases[28, 29]. (2) GSVA calculation: Use R packages of GSVA to perform GSVA analysis, including the steps of (a) Input the gene expression matrix and the gene set collection; (b) Compute pathway enrichment scores for each sample, representing the activity level of each pathway[31]. (3) Differential GSVA analysis: Compare the pathway enrichment scores between different conditions or phenotypes. Use statistical methods of limma analysis to identify significantly differentially enriched pathways. (4) Visualize GSVA results using heatmaps and bar plots, showing the pathway enrichment scores for each sample. Assess the differences in pathway activity levels between conditions.
Survival analysis
Here, the hub genes and pathway enriched score survival analysis applied as following[32, 33]: (1) overall survival (OS) analysis: Assess the relationship between gene expression and patient survival. Utilize R packages including survival, survminer, and survivalROC for survival analysis and generate Kaplan-Meier curves. (2) Cox proportional hazards regression: Perform Cox regression analysis to evaluate the prognostic value of DEGs. Identify genes that significantly impact patient survival using R packages of survival and survminer[32].
Results
Single-cell sequencing data analysis
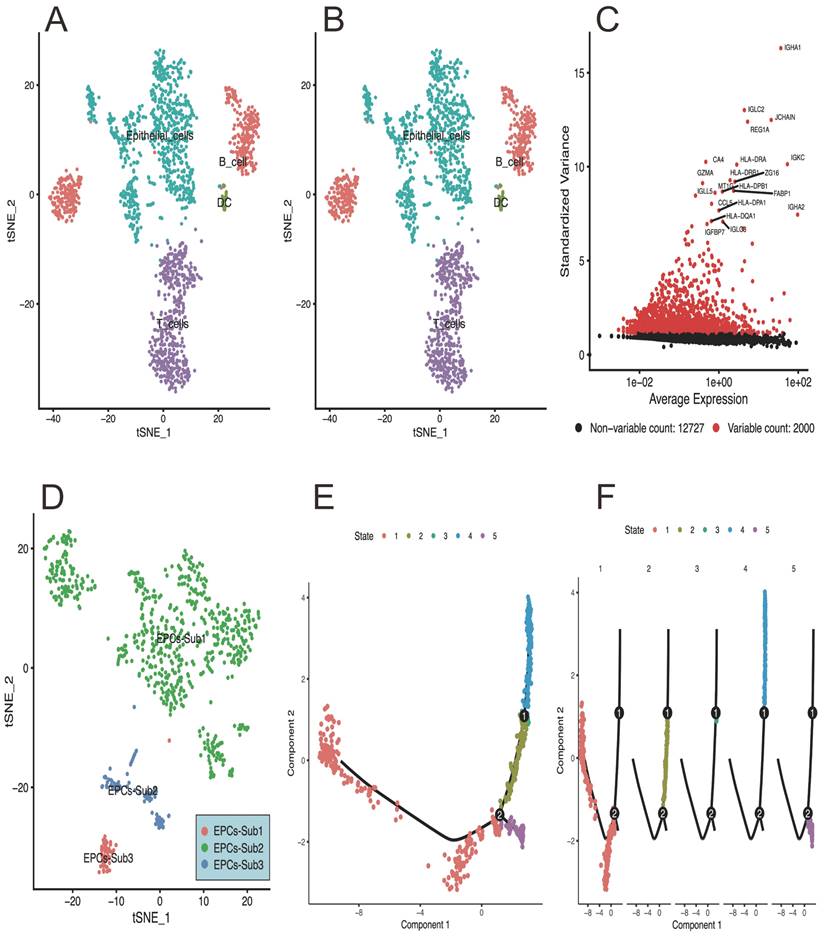
Based on the single-cell RNA-sequencing analysis, the results can be summarized as follows: Figure 1A of the PCA plot represents the clustering pattern of cells based on their gene expression profiles. It shows the distribution of cells in a reduced-dimensional space, with each point indicating an individual cell. Clusters of cells that exhibit similar gene expression profiles tend to be closer together, while cells with distinct expression patterns are located farther apart. Figure 1B visualizes the relationship between cells by projecting high-dimensional gene expression data into a two-dimensional space. Cells that are more similar in terms of gene expression are placed closer together in the plot, and providing information about cell type heterogeneity and allows for the identification of distinct subpopulations within the dataset. This plot of Figure 1C highlights the expression level of specific genes across different cell populations or clusters. It displays the enrichment or depletion pattern of selected genes in a group-wise or cell type-specific manner. This information is helpful for identifying genes that are differentially expressed and potentially associated with specific cellular functions or biological processes. Figure 1D of the tSNE map presents the distribution of epithelial cells in a reduced-dimensional space, representing distinct subpopulations with unique spatial relationships. And figure 1E-F provides insights into the pseudotime-based progression of EPCs subpopulations.
Gene Function and Pathway Enrichment Analysis
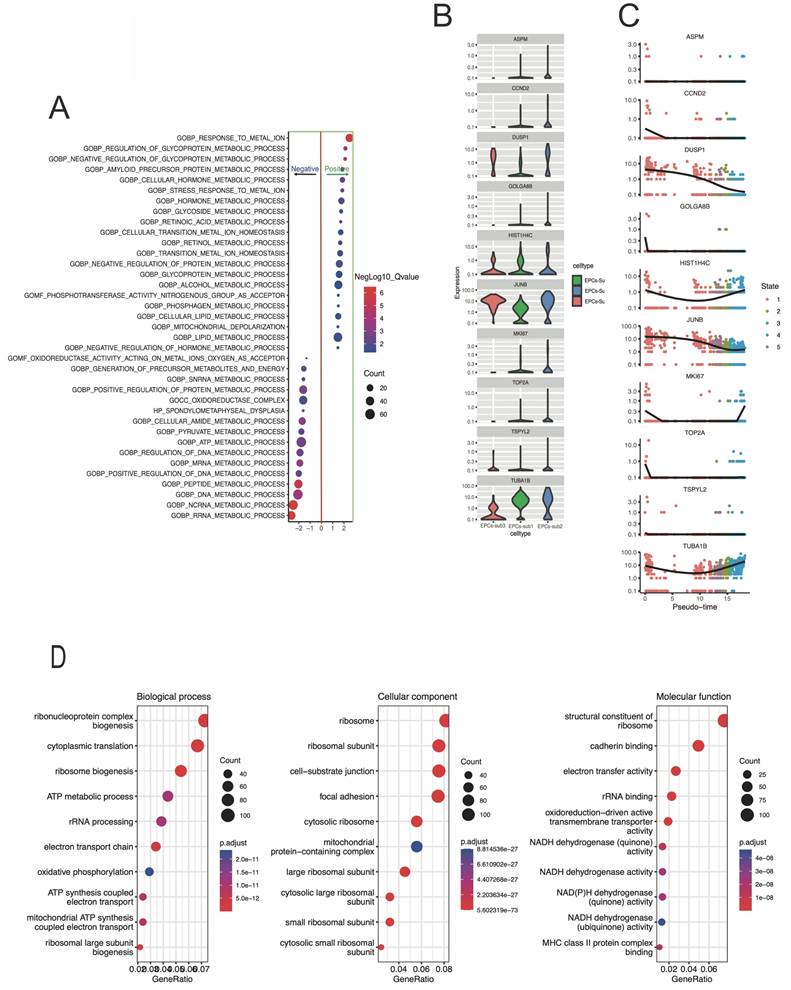
The GSEA bubble plot of Figure 2A demonstrates the enrichment level and significance of gene sets under different conditions. Here, GOBP: response to metal ion, GOBP: regulation of glycoprotein metabolic process, and GOBP: negative regul ation of glycoprotein metabolic process were positivetlly enriched. While the GOBP: DNA metabolic process, GOBP: ncRNA metabolic process, and GOBP: RRNA metabolic process were negativetlly enriched. And, clearly, metabolic related signaling pathways are significantly enriched. Figure 2B-C presents the distribution of hub gene expression levels in different subtypes. The figure 2D showing the result of a GO analysis, and the bubble plot represents a specific gene ontology term involved in biological process, molecular function, or cellular component, which provides a visual representation of enriched gene ontology terms. And the terms of ATP metabolic process, mitochondrial ATP synthesis, and oxidative phosphorylation were significantly enriched.
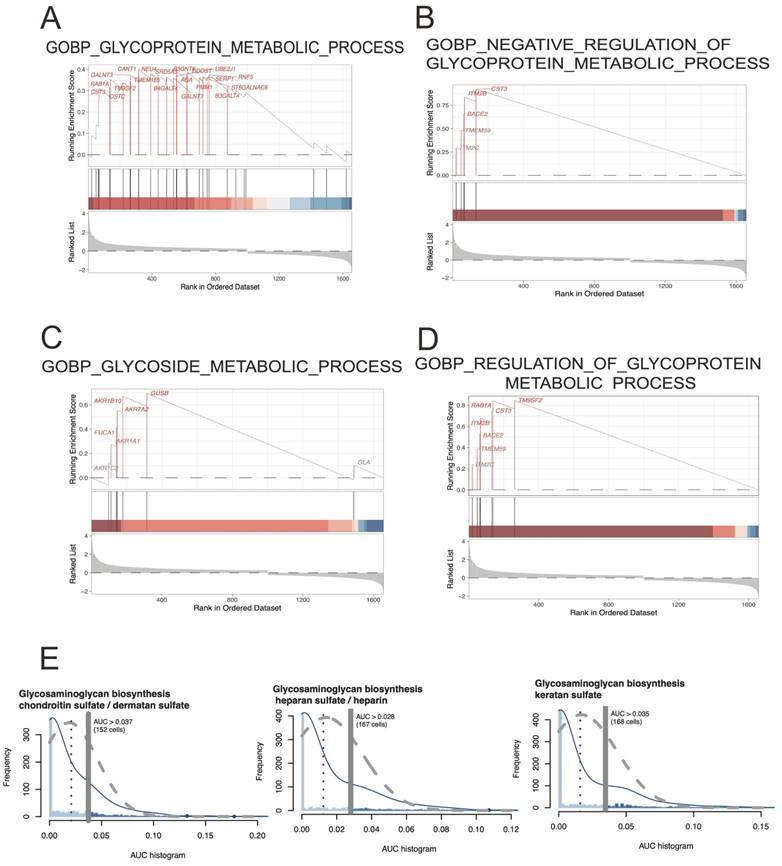
The result of a GSEA analysis typically includes a ranked list of genes, with the most strongly correlated genes at the top of the list. This list is then compared against a collection of predefined gene sets, which represent known biological pathways, molecular functions, and cellular processes. And terms including GOBP: glycoprotein metabolic process (ES=0.40; NES=1.56; p-adjusted=0.04), GOBP: negative regulation of glycoprotein metabolic process (ES=0.92; NES=2.10; p-adjusted=1.87E-05), GOBP: glycoside metabolic process(ES=0.69; NES=1.75; p-adjusted=0.019), and GOBP: regulation of glycoprotein metabolic process (ES=0.84; NES=2.13; p-adjusted=2.31E-04) were significantly detected. Here, the map of glycoprotein metabolic process may play an important role in CRC development. The figure 3E showing the Area Under the Curve (AUC)-based scoring of cell metabolic pathways involved in glycoprotein metabolic process, and showing the cell heterogeneity of glycoprotein metabolic pathway activity within EPCs cell population and detecting the regulatory mechanisms underlying cell metabolism.
Colon Cancer TCGA Database Analysis
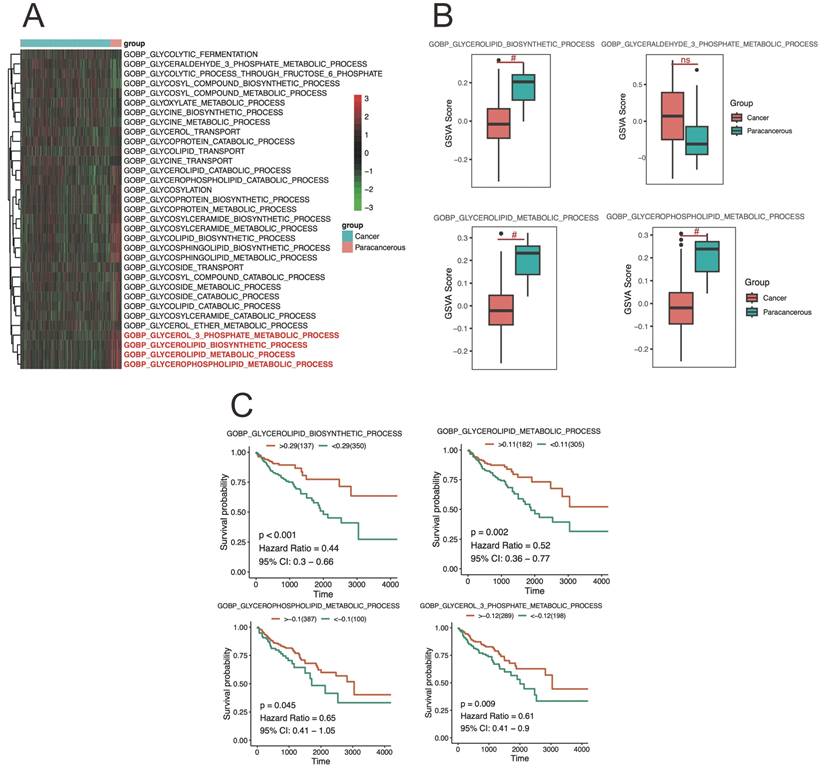
GSVA measures the enrichment of gene sets within a sample, providing an indication of pathway activity. And the differences in enrichment scores of metabolic pathways related to CRC tissue were analyzed using the GSVA method in Figure 4A-B. Here, the GSVA was employed to calculate the enrichment scores for metabolic pathways, reflecting their relative activity levels in CRC sample (Figure 4A). A differential analysis was then performed to compare the enrichment scores between different groups in figure 4B. The results revealed significant differences in the enrichment scores of glycoprotein metabolic process among the cancer and paracancerous tissues, and suggest alterations in the activity levels of glycoprotein metabolic pathways in CRC cancer tissue compared to healthy tissue.
The results of single-cell sequencing data analysis in response to CRC development. A) the PCA plot represents the clustering pattern of cells based on their gene expression profiles, and shows the distribution of cells in a reduced-dimensional space, with each point indicating an individual cell. B) showing the cells relationship by projecting high-dimensional gene expression data into a two-dimensional space. C) highlights the expression level of specific genes across different cell populations or clusters. D) presents the distribution of epithelial cells in a reduced-dimensional space, And E and F showing the pseudotime inference algorithm assigns a pseudotime value to each cell, allowing the assessment of temporal relationships and differentiation paths.
The analysis showed that glycoprotein metabolic pathways were significantly correlated with CRC patient survival. Here, high activity levels in the GOBP: glycoprotein metabolic, GOBP: negative regulation of glycoprotein metabolic process, GOBP: glycoside metabolic process, and GOBP: regulation of glycoprotein metabolic process were not favorable to overall survival, suggesting that the activation of this pathway may have a exacerbate damage for CRC progression (Figure 4C). These findings highlight the potential prognostic value of pathway activity levels in CRC cancer and provide insights into the molecular mechanisms underlying cancer progression. Further investigations are warranted to validate these results and explore the therapeutic implications of targeting specific pathways in the treatment of CRC cancer.
The gene functional enrichment results in response to pseudotime-related differently expressed genes. A) The bubble plot indicating the enrichment level and significance of the gene set under different conditions. The bubbles placed closer to the top of the plot indicate higher enrichment levels, and the size of the bubbles represents the enriched size. While x-axis representing the significance of the enrichment. B-C) the box and dot-plot presenting the expression level differences of core genes in different cell EPCs-subtypes. D) the bubble plot represents the specific GO term.
The analysis of GSEA and AUC-based scoring of cell metabolic pathways. A-D) representing the GSEA enriched results involved in glycoprotein metabolic process. E) showing the implementation of an AUC-based scoring approach allows for the quantification of cellular metabolic pathway activity using single-cell sequencing data of EPCs.
Hub regulator detection
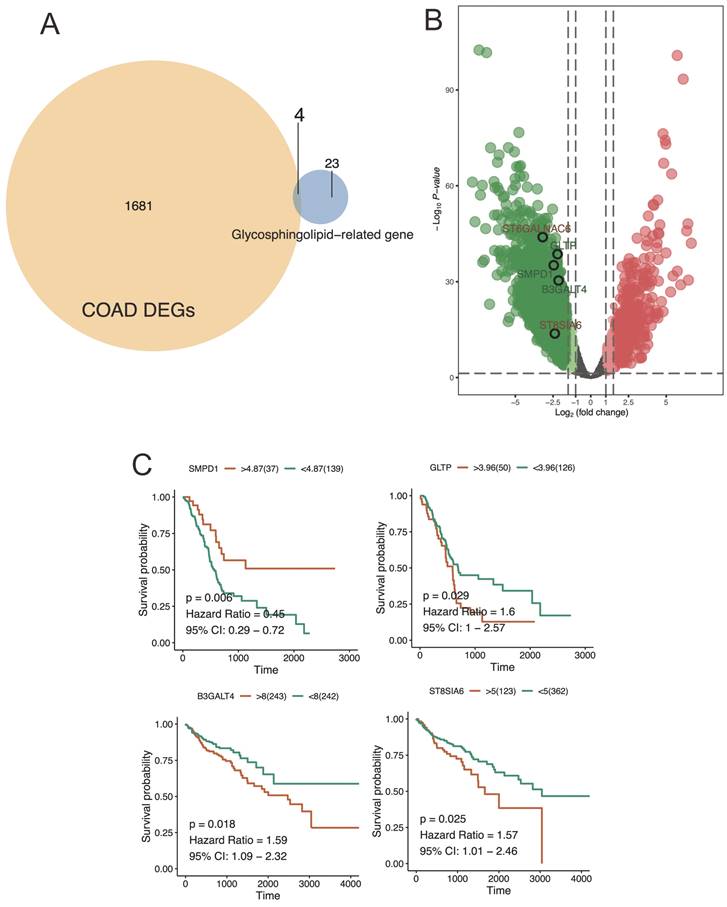
The venn diagram displayed the overlap of core genes identified from TCGA-based COAD DEGs and glycosphingolipid-related gene (Figure 5A). and the volcano plot, on the other hand, presented a representation of the statistical significance and fold change of hub gene expression between cancer and paracancerous tissue (Figure 5B). Similarly, the key regulators including SMPD1, GLTP, B3GALT4, and ST8SIA6 were significantly correlated with CRC patient survival. Here, high expression levels in SMPD1, and low expression in GLTP, B3GALT4, and ST8SIA6 were significantly correlated with worse prognosis, suggesting a significant relations to SMPD1, GLTP, B3GALT4, and ST8SIA6 expression level of the development and prognoses of the CRC patients (Figure 5C).
The differently level and survival analysis of CRC cancer tissue from TCGA. A-B) showing the results of differential enrichment scores of glycoprotein metabolic GSVA pathways in CRC cancer tissue. C presented the glycoprotein metabolic alterations associated with CRC cancer development and progression based on survival analysis.
Discussion
In this study, we conducted an analysis and screening of potential core pathways, specifically the Glycosphingolipid pathway, and identified four core genes (SMPD1, GLTP, B3GALT4, and ST8SIA6) in colorectal cancer based on data analysis from relevant databases. We also demonstrated significant differential expression of these core genes through modeling. Lipid metabolism dysregulation is one characteristic of cancer cells. Highly proliferating cancer cells not only require lipids for cell membrane synthesis (phospholipids, cholesterol, and sphingolipids) but also utilize lipids as substrates for energy metabolism (triglycerides) or as sources of signaling molecules. Bioinformatics analysis and different analytical methods applied to clinical samples have shown significant changes in the species profile of sphingomyelins (SM) and triglycerides (TG) in colorectal cancer cohorts. These changes were successfully validated in two independent cohorts and showed a significant correlation with postoperative survival rate, supporting the hypothesized clinical relevance. The dysregulation of glycerolipid metabolism mentioned in this study may be involved in the progression of CRC, with the core genes SMPD1, GLTP, B3GALT4, and ST8SIA6 playing crucial roles.
Venn diagram and volcano plot were used to illustrate the results of hub gene analysis. A) This venn plot provided a clear visual representation of the shared and unique core genes across TCGA-based COAD DEGs and glycosphingolipid-related gene. Each circle in the diagram represented a different condition or group, and the intersection of circles represented the common genes found in those conditions or groups. B) The hub genes exhibiting a significant fold change with a high statistical significance were located further away from the center of the plot, resembling the shape of a volcano. C) the survival analysis of key regulators expression level revealed significant associations between gene expression levels and patient survival outcomes.
Glycosphingolipids (GSLs) are a class of complex glycoconjugates that play crucial roles in various cellular processes, including signal transduction, cell adhesion, and modulation of immune responses[34-36]. And GPLs are composed of a hydrophobic ceramide domain linked to a hydrophilic carbohydrate moiety. They are broadly classified into ganglio-series, globo-series, isoglobo-series, and lacto/neo-lacto-series based on their carbohydrate structures[34, 37]. Dysregulation of GSL metabolism can occur through altered expression or activity of enzymes involved in GSL biosynthesis, degradation, and modification[38]. Accumulating evidence suggests that aberrant GSL metabolism contributes to tumor initiation, progression, metastasis, and drug resistance[37, 39]. Studies have demonstrated that alterations in GSL metabolism can directly impact oncogenic signaling pathways. For example, aberrant expression of glycosyltransferases involved in GSL synthesis has been observed in various cancers, leading to changes in GSL composition and functions[35, 39, 40]. Altered GSL expression can modulate key cellular events, such as cell proliferation, apoptosis, invasion, and angiogenesis, thereby promoting tumor initiation and growth. Several studies have reported the association between dysregulated GSL metabolism and tumor development. For instance, increased expression of specific GSLs, such as GM2 and GD2, has been observed in neuroblastoma and melanoma, respectively, and is associated with poor prognosis[34]. Moreover, GSLs have been implicated in cancer stem cell maintenance and epithelial-mesenchymal transition, both of which are critical processes in tumor development and metastasis[35]. Targeting dysregulated GSL metabolism shows promise as a therapeutic strategy for cancer treatment. Inhibition of GSL synthesis enzymes, such as glucosylceramide synthase, has been explored as a potential anticancer therapy[35]. Additionally, the development of antibodies targeting GSL antigens, such as GD2, has shown encouraging results in clinical trials, highlighting the therapeutic potential of targeting GSLs[41]. Here, the dysregulation of glycosphingolipid metabolism is implicated in the occurrence and development of various types of tumors. Aberrant GSL expression and altered GSL-dependent signaling pathways contribute to tumor initiation, progression, and metastasis[37]. Further investigations into the precise mechanisms underlying the relationship between dysregulated GSL metabolism and cancer are warranted to exploit GSL-related targets for novel therapeutic interventions in cancer treatment[42].
Researchers have indicated that 5-fluorouracil (5-FU)-resistant colon cancer cells predominantly acquire resistance by inhibiting cell apoptosis induced by ceramide through the sphingomyelin (SM)/ceramide pathway, with lower levels of SMPD1[43-45], compared to sensitive cell lines. SMPD1translocates to the plasma membrane to generate ceramide, promoting enhanced apoptosis signaling through FAS-FASL and TNFRSF10-TNFSF10, leading to rapid endothelial cell apoptosis. In early investigations, by comparing SMPD1 activity and tumor cell apoptosis rates, the appropriate radiation dosage for CRC radiotherapy could be selected[43-45]. And these results highlighted the upregulation of MIR196B in CRC tissues, which regulates the expression levels of GLTP during the colorectal cancer (CRC) development process[46-48]. In human colon cancer cells, GLTP overexpression interferes with cell cycle progression, induces cell death, and inhibits cell growth[49]. Recent studies have discovered that GLTP, as a differentially expressed death-related gene, is associated with poor prognosis in female cervical cancer[46-48]. Zhang et al. found that overexpression of the B3GALT4 glycosyltransferase responsible for ganglioside GM1 synthesis can induce epithelial-mesenchymal transition (EMT) in breast cancer cells[50]. Additionally, analysis shows that B3GALT4 can independently predict overall survival in osteosarcoma patients, suggesting its potential as a prognostic biomarker[15, 49, 51]. Early studies demonstrated that downregulation of B3GALT4 in neuroblastoma cells resulted in increased proliferation, invasion, and metastasis abilities in vitro[21, 31, 50, 52-59]. High expression of ST8SIA6-AS1 was detected in hepatocellular carcinoma tissues and cells, with ST8SIA6-AS1 silencing leading to weakened proliferation and migration abilities in liver cancer cells[60, 61]. ST8SIA6-AS1 displayed its oncogenic function through the absorption of the tumor suppressor miR-651-5p. Shih et al. showed that ST8SIA6-silenced colon cancer cells exhibited increased resistance to treatment with ibrutinib[42, 51, 62], as shown by cell viability assays[60, 61, 63-68]. Moreover, downregulation of ST8SIA6 was positively correlated with cancer recurrence in later stages. ST8SIA6 accelerated tumor occurrence in a genetically engineered spontaneous mouse model of colon cancer, reducing survival to approximately 67 days[60, 61, 69-72]. Thus, ST8SIA6 expression in tumors suppresses anti-tumor immune responses to enhance tumor growth[22, 33, 73]. However, this study also has limitations and drawbacks, such as the lack of large sample clinical cohort studies and experimental validation.
Acknowledgements
Funding
This work was supported by the Fujian Provincial Natural Science Foundation Projects (No. 2021J011408).
Author contributions
Liangpan Shi and Nanqi Mao: take responsibility for all aspects of the reliability and freedom from bias of the data presented and their discussed interpretations; they also drafted the article. Zhihua Zheng, Jiangrui Liu, and Jianbin Hou: take responsibility for the statistical analyses and interpretation of the data. Yibin Su: take responsibility for the full text evaluation and guidance and performed the final approval of the version to be submitted. All authors read and approved the final manuscript.
Availability of data and materials
The data and R script that support the findings of this study are available from the corresponding authors upon reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
3. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-502
4. Raphael W, Sordillo LM. Dietary polyunsaturated fatty acids and inflammation: the role of phospholipid biosynthesis. Int J Mol Sci. 2013;14:21167-88
5. Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967-76
6. Nagao-Kitamoto H, Kitamoto S, Kuffa P, Kamada N. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest Res. 2016;14:127-38
7. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191
8. Chiu HF, Venkatakrishnan K, Golovinskaia O, Wang CK. Gastroprotective Effects of Polyphenols against Various Gastro-Intestinal Disorders: A Mini-Review with Special Focus on Clinical Evidence. Molecules. 2021;26:2090
9. Sakamoto W, Coant N, Canals D, Obeid LM, Hannun YA. Functions of neutral ceramidase in the Golgi apparatus. J Lipid Res. 2018;59:2116-25
10. Haas CB, Su YR, Petersen P, Wang X, Bien SA, Lin Y. et al. Interactions between folate intake and genetic predictors of gene expression levels associated with colorectal cancer risk. Sci Rep. 2022;12:18852
11. Jin EH, Han K, Lee DH, Shin CM, Lim JH, Choi YJ. et al. Association Between Metabolic Syndrome and the Risk of Colorectal Cancer Diagnosed Before Age 50 Years According to Tumor Location. Gastroenterology. 2022;163:637-48.e2
12. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991-5
13. Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846-7
14. Slovin S, Carissimo A, Panariello F, Grimaldi A, Bouché V, Gambardella G. et al. Single-Cell RNA Sequencing Analysis: A Step-by-Step Overview. Methods Mol Biol. 2021;2284:343-65
15. Yu W, Qin X, Zhang Y, Qiu P, Wang L, Zha W. et al. Curcumin suppresses doxorubicin-induced cardiomyocyte pyroptosis via a PI3K/Akt/mTOR-dependent manner. Cardiovasc Diagn Ther. 2020;10:752-69
16. Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3
17. Wen L, Cao Y, Cheng Q, Li X, Pan L, Li L. et al. Objectively measured near work, outdoor exposure and myopia in children. Br J Ophthalmol. 2020;104:1542-7
18. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284-7
19. Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71
20. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19:A68-77
21. Jiang L, Chen T, Xiong L, Xu JH, Gong AY, Dai B. et al. Knockdown of m6A methyltransferase METTL3 in gastric cancer cells results in suppression of cell proliferation. Oncol Lett. 2020;20:2191-8
22. Lu Y, Lin Z, Wen L, Gao W, Pan L, Li X. et al. The Adaptation and Acceptance of Defocus Incorporated Multiple Segment Lens for Chinese Children. Am J Ophthalmol. 2020;211:207-16
23. Huang Z, Yu P, Tang J. Characterization of Triple-Negative Breast Cancer MDA-MB-231 Cell Spheroid Model. Onco Targets Ther. 2020;13:5395-405
24. Peng Y, Wang Y, Zhou C, Mei W, Zeng C. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front Oncol. 2022;12:819128
25. Shao Y, Zhao T, Zhang W, He J, Lu F, Cai Y. et al. Presence of the apolipoprotein E-epsilon4 allele is associated with an increased risk of sepsis progression. Sci Rep. 2020;10:15735
26. Zhao B, Li M, Su Y, Shan S, Qian W, Zhu D. et al. Role of transcription factor FOXM1 in diabetes and its complications (Review). Int J Mol Med. 2023;52:101
27. Eraso-Pichot A, Brasó-Vives M, Golbano A, Menacho C, Claro E, Galea E. et al. GSEA of mouse and human mitochondriomes reveals fatty acid oxidation in astrocytes. Glia. 2018;66:1724-35
28. Ferreira MR, Santos GA, Biagi CA, Silva Junior WA, Zambuzzi WF. GSVA score reveals molecular signatures from transcriptomes for biomaterials comparison. J Biomed Mater Res A. 2021;109:1004-14
29. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7
30. Gao P, Rao ZW, Li M, Sun XY, Gao QY, Shang TZ. et al. Tetrandrine Represses Inflammation and Attenuates Osteoarthritis by Selective Inhibition of COX-2. Curr Med Sci. 2023;43:505-13
31. Gao WL, Li XH, Dun XP, Jing XK, Yang K, Li YK. Grape Seed Proanthocyanidin Extract Ameliorates Streptozotocin-induced Cognitive and Synaptic Plasticity Deficits by Inhibiting Oxidative Stress and Preserving AKT and ERK Activities. Curr Med Sci. 2020;40:434-43
32. George B, Seals S, Aban I. Survival analysis and regression models. J Nucl Cardiol. 2014;21:686-94
33. Ding L, Lu S, Zhou Y, Lyu D, Ouyang C, Ma Z. et al. The 3' Untranslated Region Protects the Heart from Angiotensin II-Induced Cardiac Dysfunction via AGGF1 Expression. Mol Ther. 2020;28:1119-32
34. Loft LMI, Moseholm KF, Pedersen KKW, Jensen MK, Koch M, Cronjé HT. Sphingomyelins and ceramides: possible biomarkers for dementia? Curr Opin Lipidol. 2022;33:57-67
35. Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18:33-50
36. Zou R, Shi W, Tao J, Li H, Lin X, Yang S. et al. SIRT5 and post-translational protein modifications: A potential therapeutic target for myocardial ischemia-reperfusion injury with regard to mitochondrial dynamics and oxidative metabolism. Eur J Pharmacol. 2018;818:410-8
37. Slotte JP. Biological functions of sphingomyelins. Prog Lipid Res. 2013;52:424-37
38. Zou R, Shi W, Qiu J, Zhou N, Du N, Zhou H. et al. Empagliflozin attenuates cardiac microvascular ischemia/reperfusion injury through improving mitochondrial homeostasis. Cardiovasc Diabetol. 2022;21:106
39. Yu J, Hung JT, Wang SH, Cheng JY, Yu AL. Targeting glycosphingolipids for cancer immunotherapy. FEBS Lett. 2020;594:3602-18
40. Ramstedt B, Slotte JP. Membrane properties of sphingomyelins. FEBS Lett. 2002;531:33-7
41. Zou R, Tao J, Qiu J, Lu H, Wu J, Zhu H. et al. DNA-PKcs promotes sepsis-induced multiple organ failure by triggering mitochondrial dysfunction. J Adv Res. 2022;41:39-48
42. Cuny H, Bozon K, Kirk RB, Sheng DZ, Broer S, Dunwoodie SL. Maternal heterozygosity of Slc6a19 causes metabolic perturbation and congenital NAD deficiency disorder in mice. Dis Model Mech. 2023;16:dmm049647
43. Bi J, Khan A, Tang J, Armando AM, Wu S, Zhang W. et al. Targeting glioblastoma signaling and metabolism with a re-purposed brain-penetrant drug. Cell Rep. 2021;37:109957
44. Ellegaard AM, Bach P, Jäättelä M. Targeting Cancer Lysosomes with Good Old Cationic Amphiphilic Drugs. Rev Physiol Biochem Pharmacol. 2023;185:107-52
45. Mohamud Yusuf A, Hagemann N, Zhang X, Zafar M, Hussner T, Bromkamp C. et al. Acid sphingomyelinase deactivation post-ischemia promotes brain angiogenesis and remodeling by small extracellular vesicles. Basic Res Cardiol. 2022;117:43
46. Mishra SK, Gao YG, Zou X, Stephenson DJ, Malinina L, Hinchcliffe EH. et al. Emerging roles for human glycolipid transfer protein superfamily members in the regulation of autophagy, inflammation, and cell death. Prog Lipid Res. 2020;78:101031
47. Mo JS, Park YR, Chae SC. MicroRNA 196B Regulates HOXA5, HOXB6 and GLTP Expression Levels in Colorectal Cancer Cells. Pathol Oncol Res. 2019;25:953-9
48. Zou X, Gao Y, Ruvolo VR, Gardner TL, Ruvolo PP, Brown RE. Human glycolipid transfer protein gene (GLTP) expression is regulated by Sp1 and Sp3: involvement of the bioactive sphingolipid ceramide. J Biol Chem. 2011;286:1301-11
49. Chen L, Tian Q, Shi Z, Qiu Y, Lu Q, Liu C. Melatonin Alleviates Cardiac Function in Sepsis-Caused Myocarditis via Maintenance of Mitochondrial Function. Front Nutr. 2021;8:754235
50. Zhang T, Wang F, Wu JY, Qiu ZC, Wang Y, Liu F. et al. Clinical correlation of B7-H3 and B3GALT4 with the prognosis of colorectal cancer. World J Gastroenterol. 2018;24:3538-46
51. Chen L, Zhan CZ, Wang T, You H, Yao R. Curcumin Inhibits the Proliferation, Migration, Invasion, and Apoptosis of Diffuse Large B-Cell Lymphoma Cell Line by Regulating MiR-21/VHL Axis. Yonsei Med J. 2020;61:20-9
52. Ha YJ, Tak KH, Kim CW, Roh SA, Choi EK, Cho DH. et al. PSMB8 as a Candidate Marker of Responsiveness to Preoperative Radiation Therapy in Rectal Cancer Patients. Int J Radiat Oncol Biol Phys. 2017;98:1164-73
53. Sha YL, Liu Y, Yang JX, Wang YY, Gong BC, Jin Y. et al. B3GALT4 remodels the tumor microenvironment through GD2-mediated lipid raft formation and the c-met/AKT/mTOR/IRF-1 axis in neuroblastoma. J Exp Clin Cancer Res. 2022;41:314
54. Wang W, Zhai T, Luo P, Miao X, Wang J, Chen Y. Beneficial effects of silibinin on serum lipids, bile acids, and gut microbiota in methionine-choline-deficient diet-induced mice. Front Nutr. 2023;10:1257158
55. Yang Y, Wu Q, Pan W, Wen L, Luo Z, Wu H. et al. Characteristics of the Ocular Surface in Myopic Child Candidates of Orthokeratology Lens Wear. Ophthalmol Ther. 2023;12:3067-79
56. Xiong T, Xia L, Song Q. Circular RNA SPI1 expression before and after induction therapy and its correlation with clinical features, treatment response, and survival of acute myeloid leukemia patients. J Clin Lab Anal. 2023;37:e24835
57. Dong H, Li H, Wang L, Yuan Y, Zhang D, Zhou L. et al. Clinical analysis of 175 cases of vaginal intraepithelial neoplasia. Eur J Obstet Gynecol Reprod Biol. 2023;287:232-6
58. Peng Y, Yan H, Mei W, Zhang P, Zeng C. Combining Radiotherapy with Immunotherapy in Cervical Cancer: Where Do We Stand and Where Are We Going? Curr Treat Options Oncol. 2023;24:1378-91
59. Su H, Geng H, Cai L, Xu M, Xing W, Long W. et al. Immune-check blocking combination multiple cytokines shown curative potential in mice tumor model. Cancer Med. 2023;12:13573-85
60. Ko CY, Chu TH, Hsu CC, Chen HP, Huang SC, Chang CL. et al. Bioinformatics Analyses Identify the Therapeutic Potential of ST8SIA6 for Colon Cancer. J Pers Med. 2022;12:401
61. Zhang Y, Yang Y, Zhang Y, Liu Z. lncRNA ST8SIA6-AS1 facilitates proliferation and invasion in liver cancer by regulating miR-142-3p. Exp Ther Med. 2021;22:1348
62. Chen Z, Jin M, He H, Dong J, Li J, Nie J. et al. Mesenchymal stem cells and macrophages and their interactions in tendon-bone healing. J Orthop Translat. 2023;39:63-73
63. Zhang Z, Ni P, Tang M, Song Y, Liu C, Zhao B. Dapagliflozin alleviates renal podocyte pyroptosis via regulation of the HO-1/NLRP3 axis. Mol Med Rep. 2023;28:200
64. Liu Y, Shen D, Wang HY, Qi MY, Zeng QY. Development and validation to predict visual acuity and keratometry two years after corneal crosslinking with progressive keratoconus by machine learning. Front Med (Lausanne). 2023;10:1146529
65. Cao Y, Lan W, Wen L, Li X, Pan L, Wang X. et al. An effectiveness study of a wearable device (Clouclip) intervention in unhealthy visual behaviors among school-age children: A pilot study. Medicine (Baltimore). 2020;99:e17992
66. Liu J, Lu Y, Huang D, Yang J, Fan C, Chen C. et al. The Efficacy of Defocus Incorporated Multiple Segments Lenses in Slowing Myopia Progression: Results from Diverse Clinical Circumstances. Ophthalmology. 2023;130:542-50
67. Dang X, Fan C, Cui F, He Y, Sun G, Ruan J. et al. Interactions between ultrasonographic cervical length and placenta accreta spectrum on severe postpartum hemorrhage in women with placenta previa. Int J Gynaecol Obstet. 2023;161:1069-74
68. Zhou J, Chen S, Liu J, Du J, Li J. Knockdown of hnRNPAB reduces the stem cell properties and enhances the chemosensitivity of human colorectal cancer stem cells. Oncol Rep. 2023;49:129
69. Li J, Liang X, Wang X, Yang P, Jian X, Fu L. et al. A missense GDF5 variant causes brachydactyly type A1 and multiple-synostoses syndrome 2. JOR Spine. 2024;7:e1302
70. Zhang L, Jiang B, Zhu N, Tao M, Jun Y, Chen X. et al. Mitotic checkpoint kinase Mps1/TTK predicts prognosis of colon cancer patients and regulates tumor proliferation and differentiation via PKCalpha/ERK1/2 and PI3K/Akt pathway. Med Oncol. 2019;37:5
71. Li H, Shi W, Shen T, Hui S, Hou M, Wei Z. et al. Network pharmacology-based strategy for predicting therapy targets of Ecliptae Herba on breast cancer. Medicine (Baltimore). 2023;102:e35384
72. Lin L, Wu Q, Lu F, Lei J, Zhou Y, Liu Y. et al. Nrf2 signaling pathway: current status and potential therapeutic targetable role in human cancers. Front Oncol. 2023;13:1184079
73. Zhang Y, Zhang Y, Liu S, Li B, Song Z, Han Q. et al. Acupuncture for cancer pain: a scoping review of systematic reviews and meta-analyses. Front Oncol. 2023;13:1169458
Author contact
![]() Corresponding author: Yibin Su, email: syb3266com. Department of Gastrointestinal Surgery, The First Hospital of Quanzhou Affiliated to Fujian Medical University, Quanzhou, 362002, China.
Corresponding author: Yibin Su, email: syb3266com. Department of Gastrointestinal Surgery, The First Hospital of Quanzhou Affiliated to Fujian Medical University, Quanzhou, 362002, China.

 Global reach, higher impact
Global reach, higher impact