Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(2):328-340. doi:10.7150/ijms.102331 This issue Cite
Research Paper
Comprehensive multi-omics analysis identifies NUSAP1 as a potential prognostic and immunotherapeutic marker for lung adenocarcinoma
1. Department of Laboratory Medicine, Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China.
2. Zhengzhou Key Laboratory for In vitro Diagnosis of Hypertensive Disorders of Pregnancy, Zhengzhou, Henan, China.
Received 2024-8-14; Accepted 2024-10-22; Published 2025-1-1
Abstract
While NUSAP1's association with various tumors is established, its predictive value for prognosis and immunotherapy in lung adenocarcinoma (LUAD) remains unconfirmed. We analyzed Nucleolar Spindle-Associated Protein 1 (NUSAP1) gene expression in TCGA and GTEx datasets and validated it in clinicopathological tissues using qRT-PCR and immunohistochemistry. Additionally, we investigated NUSAP1's relationship with patient prognosis across TCGA and five GEO cohorts. The IMvigor210 cohort was utilized to explore NUSAP1's association with immunotherapy efficacy. Furthermore, single-cell RNA-sequencing data was used to examine the correlation between NUSAP1 and immune cell infiltration. Finally, we analyzed the relationship between NUSAP1 and m6A methylation. NUSAP1 expression was significantly elevated in tumor tissues, correlating with poorer prognosis in LUAD patients. It exhibited a significant correlation with immune cell infiltration in the tumor microenvironment, predominantly expressed in Tprolif cells. LUAD patients with heightened NUSAP1 expression may derive greater benefit from anti-PD-L1 treatment. Additionally, NUSAP1 was tightly linked with m6A methylation. Enrichment analysis revealed its association with key biological functions, including lipid metabolism and cell cycle regulation. Our comprehensive analysis underscores NUSAP1's potential as a prognostic and immunotherapeutic biomarker for LUAD, warranting further investigation.
Keywords: NUSAP1, LUAD, immunotherapy, m6A, scRNA-seq
Introduction
Nucleolar spindle-associated protein 1 (NUSAP1) serves as a key cell cycle regulatory protein, primarily engaged in mitotic progression, spindle formation, and stabilization [1]. Dysregulation of NUSAP1 results in spindle structural abnormalities, culminating in chromosome misaggregation and aberrant cell division, thereby contributing to tumorigenesis [2]. Extensive evidence suggests NUSAP1's involvement in the onset and progression of various cancers [3]. NUSAP1 plays a pivotal role in driving the progression of pancreatic ductal adenocarcinoma by facilitating epithelial-mesenchymal transition and AMPK phosphorylation [4]. Moreover, it serves as a shared genetic signature linking chronic HBV infection to HBV-associated HCC, while also contributing to cisplatin resistance in hepatocellular carcinoma [5, 6]. NUSAP1 profoundly influences the proliferation and invasive potential of basal cell carcinoma [7], glioblastoma [8], ovarian cancer [9], as well as breast [10] and nephroblastoma cells [11]. Regrettably, there is currently no documented research exploring the correlation between NUSAP1 and the response to immunotherapy in LUAD, highlighting the need for further investigation in this area.
In this study, we delved into the expression profiles and potential prognostic significance of NUSAP1 across multiple publicly available datasets. Furthermore, we meticulously validated these findings in clinical samples using qRT-PCR and immunohistochemistry (IHC). Additionally, we assessed the predictive value of NUSAP1 expression for immunotherapy response in LUAD patients. Lastly, our study explored the correlation between NUSAP1 expression and immune infiltration as well as m6A-related genes. These discoveries offer valuable insights into the potential relevance of NUSAP1 in LUAD.
Methods
Data collection
For the TCGA-LUAD and GTEx datasets, RNA sequencing data and related clinicopathologic details were acquired from the UCSC Xena database (https://xenabrowser.net/). Transcriptome data underwent normalization using the log2 (FPKM+1) transformation, and batch effects were adjusted using Combat from the R package "SVA". Normalized expression data from the GSE10072, GSE31210, GSE37745, GSE50081, GSE68465, and GSE42127 cohorts were obtained from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo) database. Data pertaining to the cohort receiving anti-PD-L1 immunotherapy were retrieved by downloading and installing the “IMvigor210CoreBiologies” package from http://research-pub.gene.com/.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Following established protocols [12], NUSAP1 mRNA expression was evaluated via qRT-PCR in tumor and paracancerous tissues from 30 LUAD patients who did not receive any radiotherapy or chemotherapy. The following primer-synthesized NUSAP1 sequences were utilized: forward 5ʹ-AGCCCATCAATAAGGGAGGG-3ʹ, reverse 5ʹ-ACCTGACACCCGTTTTAGCTG-3ʹ. Ultimately, the relative expression of NUSAP1 mRNA was determined using the 2-ΔΔCT method.
Human tissue samples
Paraffin-embedded human lung adenocarcinoma tissues were obtained from The First Affiliated Hospital of Zhengzhou University. The specimens were collected under an Institutional Review Board (IRB) approved protocol (2019-KY-258) with obtained informed consent.
Immunohistochemistry (IHC) assay
Independent IHC tests were conducted on paraffin-embedded tissues using anti-NUSAP1 (Invitrogen, US). Images were captured using a Leica DM 2500 microscope following the application of secondary antibodies to the slides. Immunostaining intensity was evaluated and scored by two independent observers. A final score, distinguishing between negative (≤4) and positive (>4) NUSAP1 protein expression, was derived by summing the extent of expression and intensity scores.
Immune cell infiltration analysis
Various databases including the ESTIMATE [13], CIBERSORT [14], xCELL [15], TIMER databases [16], and single-sample genome enrichment analysis (ssGSEA) algorithm were employed to assess the infiltration of different immune cell types in the tumor microenvironment (TME). Furthermore, we scrutinized the expression profile of NUSAP1 across various cell types within the TME using the TISCH database (http://tisch.comp-genomics.org/) [17], including GSE139555, GSE148071, GSE162498, and GSE99254 cohorts.
Prediction of potential tumor-sensitive drugs targeting NUSAP1
The CellMiner database [18] was utilized to explore potential tumor-sensitive drugs targeting the NUSAP1 gene. Screening criteria included an adjusted P value of less than one in a thousand and a Pearson correlation coefficient exceeding four-tenths. Differences in half-maximal inhibitor dose (IC50) of drugs across different NUSAP1 gene expression levels were analyzed to demonstrate therapeutic sensitivity.
The pathway richness analysis
Gene Set Variation Analysis (GSVA) and Gene Set Enrichment Analysis (GSEA) were employed using the R packages "GSVA" and "ClusterProfiler", respectively, to evaluate differences in biological processes between NUSAP1-high and NUSAP1-low expression subgroups.
Statistical analysis
All statistical analyses were conducted using R-4.2.1. Hazard ratios (HRs) and associated 95% confidence intervals were calculated using univariate survival analysis. Kaplan-Meier analysis categorized patients based on high or low NUSAP1 expression levels to assess patient survival. Statistical significance was attributed to changes with a p-value less than 0.05, and all tests were two-sided.
Results
High elevated NUSAP1 expression in LUAD
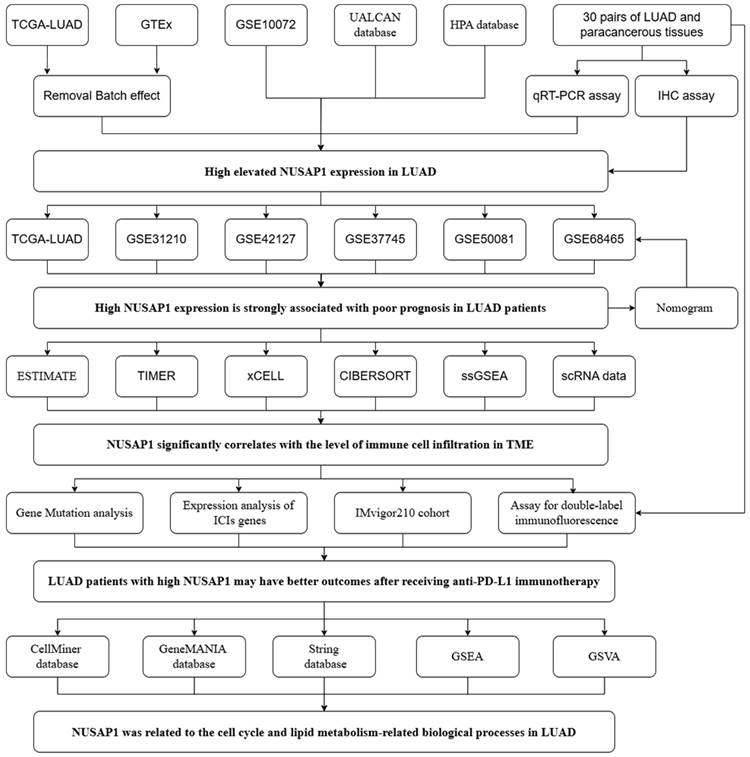
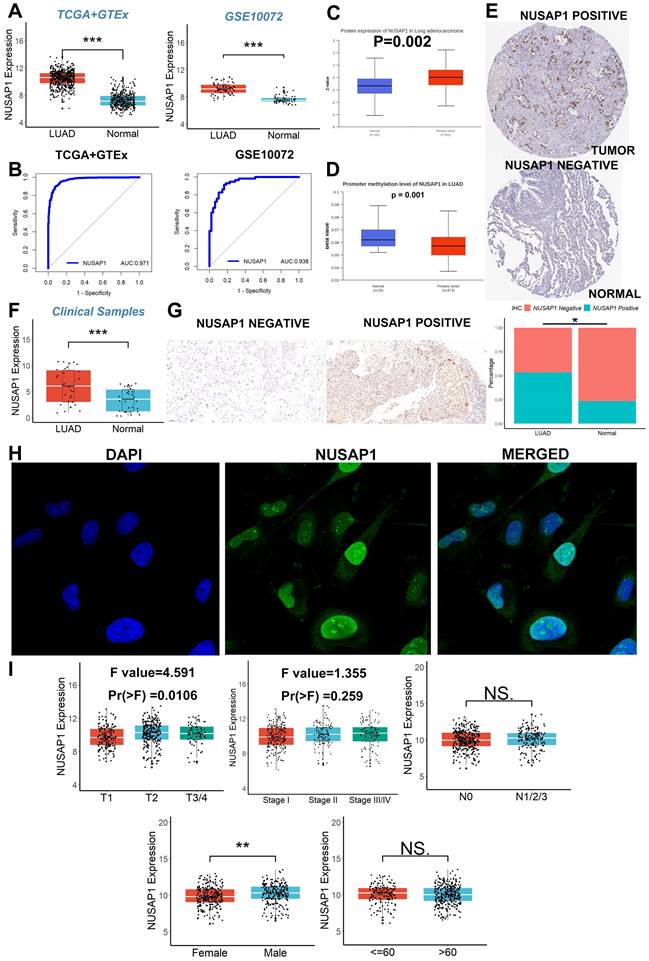
The flow chart of this study is shown in Figure 1. We initiated our investigation by examining the expression patterns of NUSAP1 within LUAD across multiple cohorts, including TCGA, GTEx, and GSE10072. Our analysis revealed a significant upregulation of NUSAP1 in tumor tissues (Figure 2A), with a remarkable capacity to distinguish between tumor and normal samples, boasting an area under the curve (AUC) value exceeding 0.930 (Figure 2B). Additionally, leveraging the UALCAN database (http://ualcan.path.uab.edu/index.html), we confirmed a substantial elevation of NUSAP1 expression (Figure 2C) but with lower promoter methylation levels (Figure 2D) in tumor tissues compared to normal tissues. Subsequent exploration utilizing the Human Protein Atlas database (https://www.proteinatlas.org/) underscored a pronounced discrepancy in NUSAP1 expression between tumor and normal samples (Figure 2E). Remarkably, our experimental validations through qRT-PCR (Figure 2F) and IHC assays (Figure 2G) on clinical specimens corroborated the heightened expression of NUSAP1 in tumor tissues. Immunofluorescence staining analysis from the Human Protein Atlas database highlighted the predominant localization of NUSAP1 within the nucleus (Figure 2H). Expanding our inquiry, we delved into the relationship between NUSAP1 expression and clinicopathological features, revealing a significant association with advanced T stage and male gender, while no substantial correlations were observed with N stage, TNM stage, or age among LUAD patients (Figure 2I).
The flow chart.
The potential prognostic power of NUSAP1 in LUAD
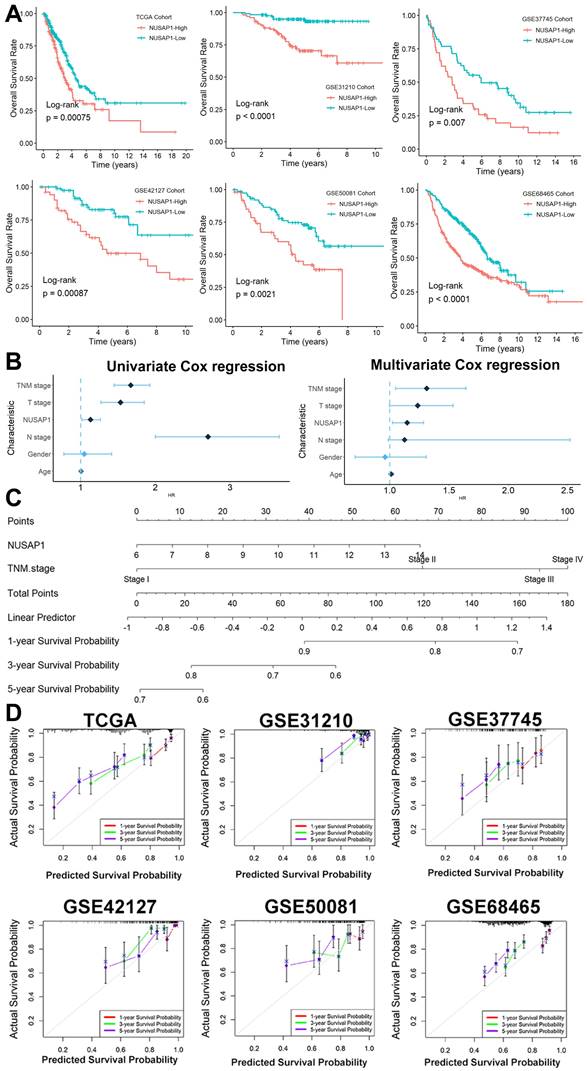
The Kaplan-Meier survival analysis unveiled a robust correlation between elevated NUSAP1 expression and unfavorable survival outcomes across six distinct LUAD cohorts, with a directly proportional relationship between NUSAP1 expression levels and patients' shortened survival duration (Figure 3A). Furthermore, through both univariate and multivariate Cox regression analyses (Figure 3B), we identified NUSAP1 expression along with TNM stage as independent predictors of poor prognosis in LUAD patients. To provide clinicians with a practical tool for prognostic assessment, we developed an overall survival nomogram model capable of predicting 1-, 3-, and 5-year survival probabilities for patients (Figure 3C). Impressively, as illustrated in Figure 3D, this nomogram model exhibited excellent fitting performance, indicating its reliability in prognostic estimations.
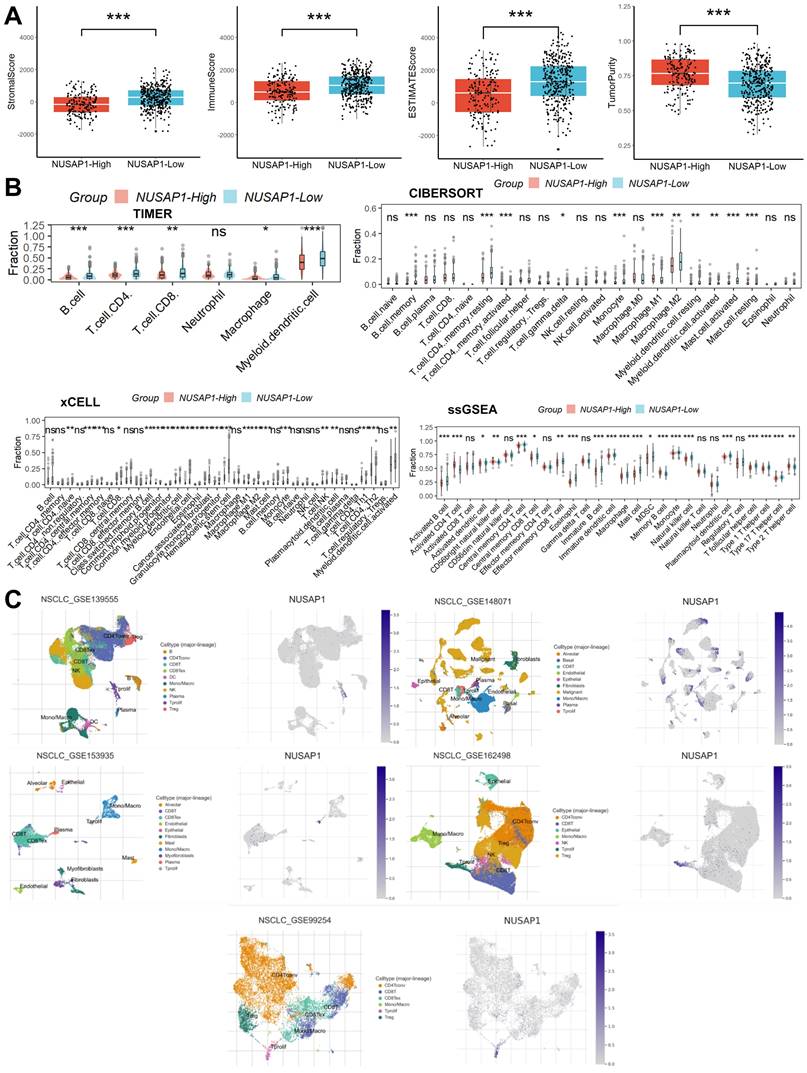
Relationship between NUSAP1 expression and TME
Initially, we evaluated the composition of immune and stromal cells as well as tumor purity in samples from LUAD patients using the ESTIMATE algorithm. This analysis revealed that patients with high NUSAP1 expression exhibited a lower proportion of immune and stromal cells but higher tumor cell purity compared to those with low NUSAP1 expression (Figure 4A). Subsequently, to delve deeper into the distribution of immune cells within the TME, we employed the CIBERSORT, xCELL, TIMER, and ssGSEA algorithms. Remarkably, we observed a reduced proportion of most immune cell types in patients with high NUSAP1 expression compared to those with low NUSAP1 expression (Figure 4B). Lastly, insights gleaned from analyses of five single-cell RNA sequencing datasets unveiled the distribution of NUSAP1 expression among Tprolif cells (Figure 4C).
Relationship between NUSAP1 expression and immunotherapy efficacy
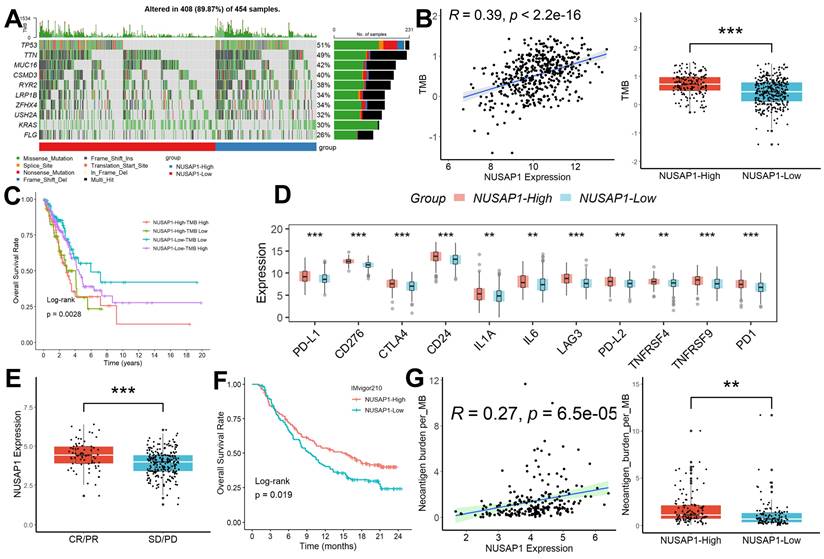
We conducted a comprehensive analysis of mutation data retrieved from LUAD patients using the R package “TCGAmutations,” comparing the mutational landscape between subgroups characterized by different levels of NUSAP1 expression. Notably, LUAD samples exhibiting high NUSAP1 expression (96.3%) displayed a higher frequency of somatic mutations compared to those with low NUSAP1 expression (77.8%) (Figure 5A). Furthermore, NUSAP1 expression exhibited a significant positive correlation with tumor mutation burden (TMB), with LUAD samples featuring high NUSAP1 expression showing markedly elevated TMB values compared to those with low NUSAP1 expression (Figure 5B). Intriguingly, patients with high NUSAP1 expression and elevated TMB values exhibited the poorest survival outcomes (Figure 5C). Additionally, we explored variations in the expression levels of multiple immune checkpoint inhibitors (ICIs) genes across different NUSAP1 expression subgroups. Our analysis revealed heightened expression of ICIs in LUAD samples with high NUSAP1 expression compared to those with low NUSAP1 expression (Figure 5D). Furthermore, we investigated the association between NUSAP1 expression and response to anti-PD-L1 immunotherapy in the IMvigor210 cohort. Remarkably, NUSAP1 expression levels were significantly higher in LUAD samples exhibiting complete response (CR) and partial response (PR) to treatment compared to those with stable disease (SD) and progressive disease (PD) (Figure 5E). Intriguingly, patients with high NUSAP1 expression demonstrated improved survival outcomes relative to those with low NUSAP1 expression (Figure 5F). Ultimately, we observed a significant positive correlation between NUSAP1 expression and neoantigen burden, coupled with higher neoantigen burden values in LUAD samples with high NUSAP1 expression compared to those with low NUSAP1 expression (Figure 5G).
Prediction of potential tumor-sensitive drugs targeting NUSAP1
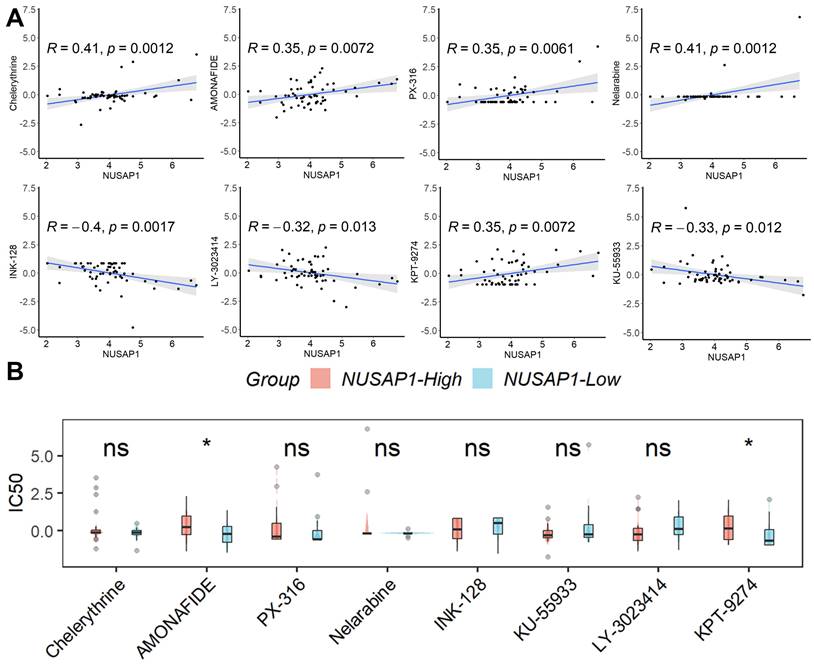
Based on the findings from CellMiner analysis, we identified eight tumor-sensitive drugs (Figure 6A). Notably, Chelerythrine, AMONAFIDE, PX-316, and Nelarabine exhibited significant positive correlations with NUSAP1 expression, whereas INK-128, KU-55933, LY-3023414, and KPT-9274 showed significant negative correlations with NUSAP1 expression. Of these eight drugs, only the IC50 values of AMONAFIDE and KPT-9274 displayed differentiation between samples with high or low NUSAP1 expression (Figure 6B). These results suggest that amonafide and KPT-9274 may offer heightened efficacy in antitumor therapy for patients with low NUSAP1 expression.
Relationship between NUSAP1 expression and m6A-related genes
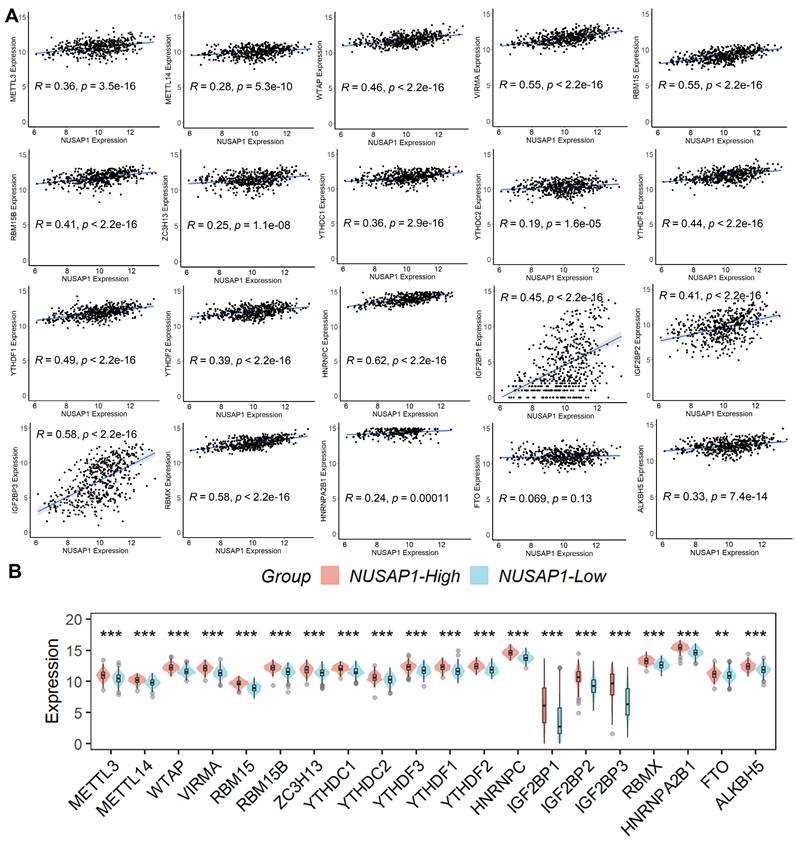
Recognizing the pivotal role of m6A modification in LUAD progression [19], we explored the correlation between the expression of various m6A-related genes and NUSAP1 expression. Our analysis revealed a significant positive correlation between NUSAP1 expression and the expression of these m6A-related genes (except FTO) (Figure 7A). Additionally, we observed a notable elevation in the expression of these m6A-related genes in samples with high NUSAP1 expression compared to those with low NUSAP1 expression (Figure 7B). These findings suggest a potential involvement of NUSAP1 in LUAD progression through the mediation of m6A modification pathways.
High elevated NUSAP1 expression in LUAD. (A) The significant upregulation of NUSAP1 in tumor tissues. (B) The remarkable capacity of NUSAP1 to distinguish between tumor and normal samples. (C) NUSAP1 expression in the UALCAN database. (D) The promoter methylation levels of NUSAP1. (E) NUSAP1 expression between tumor and normal samples in the Human Protein Atlas database. (F) The result of qRT-PCR. (G) The result of IHC assays. (H) The result of immunofluorescence staining analysis. (I) The relationship between NUSAP1 expression and clinicopathological features. ns, not significant; *p < 0.05; **p <0.01; ***p < 0.001.
The potential prognostic power of NUSAP1 in LUAD. (A) Kaplan-Meier survival analysis across six distinct LUAD cohorts (In TCGA Date, NUSAP1-High=156, NUSAP1-Low=336; In GSE31210 Cohort, NUSAP1-High=102, NUSAP1-Low=124; In GSE37745 Cohort, NUSAP1-High=50, NUSAP1-Low=56; In GSE50081 Cohort, NUSAP1-High=50, NUSAP1-Low=77; In GSE68465 Cohort, NUSAP1-High=225, NUSAP1-Low=214; In GSE42127 Cohort, NUSAP1-High=52, NUSAP1-Low=79). (B) The univariate and multivariate Cox regression analyses. (C) We developed an overall survival nomogram model capable of predicting 1-, 3-, and 5-year survival probabilities for patients. (D) This nomogram model exhibited excellent fitting performance, indicating its reliability in prognostic estimations.
Relationship between NUSAP1 expression and TME. (A) Patients with high NUSAP1 expression exhibited a lower proportion of immune and stromal cells but higher tumor cell purity compared to those with low NUSAP1 expression (In TCGA Date, NUSAP1-High=156, NUSAP1-Low=336). (B) The reduced proportion of most immune cell types in patients with high NUSAP1 expression compared to those with low NUSAP1 expression. (C) The distribution of NUSAP1 expression among Tprolif cells. ns, not significant; *p < 0.05; **p <0.01; ***p < 0.001.
Relationship between NUSAP1 expression and immunotherapy efficacy. (A) LUAD samples exhibiting high NUSAP1 expression displayed a higher frequency of somatic mutations compared to those with low NUSAP1 expression. (B) NUSAP1 expression exhibited a significant positive correlation with TMB, with LUAD samples featuring high NUSAP1 expression showing markedly elevated TMB values compared to those with low NUSAP1 expression (NUSAP1-High=153, NUSAP1-Low=301). (C) Patients with high NUSAP1 expression and elevated TMB values exhibited the poorest survival outcomes (NUSAP1-High-TMB-High =101, NUSAP1-High-TMB-Low=52; NUSAP1-Low-TMB-High =126, NUSAP1-Low-TMB- Low=175). (D) The heightened expression of ICIs in LUAD samples with high NUSAP1 expression compared to those with low NUSAP1 expression. (E) NUSAP1 expression levels were significantly higher in LUAD samples exhibiting CR/PR to treatment compared to those with SD/PD (CR/PR=68, SD/PD=227). (F) Patients with high NUSAP1 expression demonstrated improved survival outcomes relative to those with low NUSAP1 expression (NUSAP1-High=147, NUSAP1-Low=148). (G) The significant positive correlation between NUSAP1 expression and neoantigen burden, coupled with higher neoantigen burden values in LUAD samples with high NUSAP1 expression compared to those with low NUSAP1 expression. **p <0.01; ***p < 0.001.
Function enrichment analyses
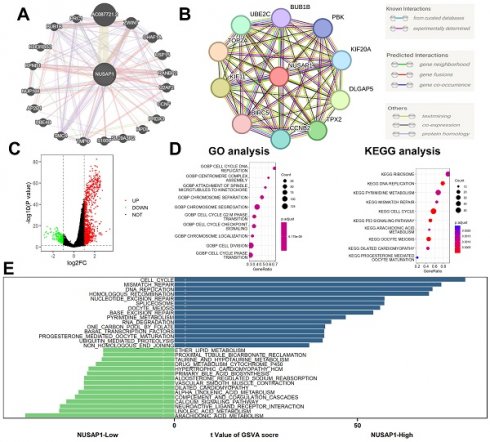
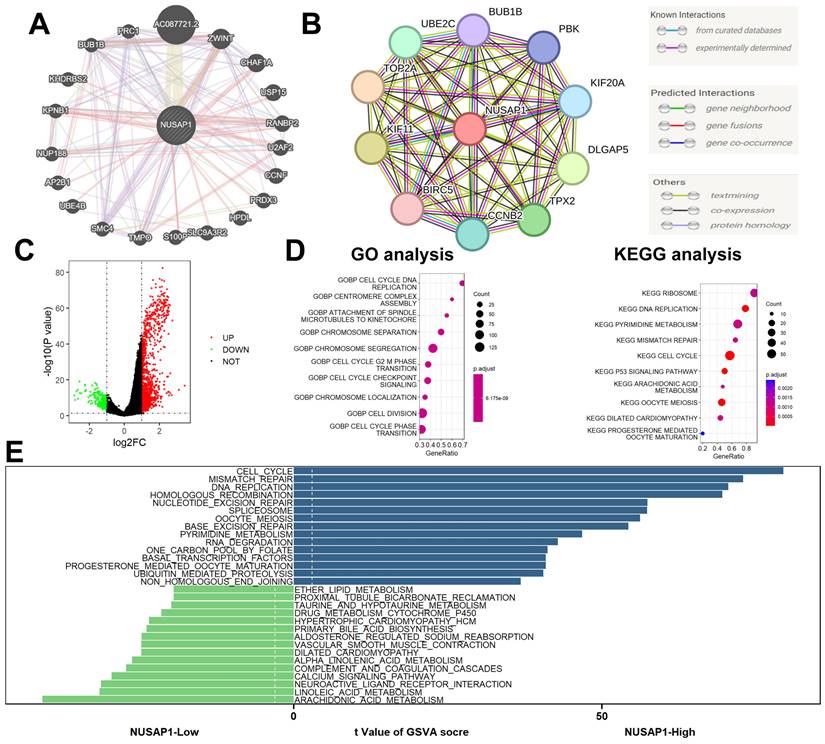
We constructed gene-gene interaction networks (Figure 8A) and protein-protein interaction networks (Figure 8B) utilizing the GeneMANIA (http://genemania.org/) [20] and STRING (https://string-db.org/) [21] databases, respectively. Subsequently, differential expression gene (DEG) analysis was conducted between NUSAP1 high- and low-expression samples, employing thresholds of |logFC| > 1 and p-value < 0.05. This analysis revealed a total of 1,378 DEGs, comprising 1,145 up-regulated genes and 233 down-regulated genes (Figure 8C). Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses unveiled that these DEGs were predominantly enriched in cell cycle-related biological processes (Figure 8D). Additionally, Gene Set Variation Analysis (GSVA) highlighted enrichment in cell cycle and lipid metabolism-related biological processes among these DEGs (Figure 8E).
Discussion
LUAD stands out as one of the most prevalent histological subtypes of non-small cell lung cancer (NSCLC), presenting consistently high morbidity and mortality rates worldwide [22]. Despite significant strides in precision medicine, which have unveiled a spectrum of biomarkers and facilitated the adoption of personalized therapeutic approaches, challenges persist in targeted therapies and immunotherapies for LUAD patients [23, 24]. As such, LUAD remains a significant global public health concern. Thus, the quest for biomarkers capable of accurately prognosticating outcomes and guiding immunotherapeutic efficacy holds paramount importance in shaping clinical interventions for LUAD patients. In our study, we established a significant correlation between elevated NUSAP1 expression and unfavorable prognosis in LUAD using multi-omics analysis. Additionally, we investigated disparities in the TME, mutation profiles, and response to immunotherapy across LUAD samples with varying NUSAP1 expression levels. This comprehensive analysis not only enhances comprehension of LUAD tumor immunology but also offers valuable insights for refining therapeutic strategies and ultimately improving LUAD patient outcomes.
Prediction of potential tumor-sensitive drugs targeting NUSAP1. (A) The eight tumor-sensitive drugs. (B) Only the IC50 values of AMONAFIDE and KPT-9274 displayed differentiation between samples with high or low NUSAP1 expression. ns, not significant; *p < 0.05.
Multiple immune cell populations within the TME play pivotal roles in the onset, progression, and treatment outcomes of LUAD patients [25]. A comprehensive understanding of TME characteristics is paramount for devising novel immunotherapeutic strategies. Tumor-associated macrophages (M2-type) and IL-10-secreting B cells exhibit pro-tumorigenic traits and immunosuppressive functions [26, 27]. Regulatory T cells facilitate tumor growth by suppressing anti-tumor responses [28]. CD8+ T cells induce direct cytotoxic effects on cancer cells through the secretion of granzymes and perforins [29]. Dendritic cells capture tumor antigens, instigating CD8+ or CD4+ T cell-mediated immune responses [30]. Conversely, tumor-associated macrophages (M1-type) secrete tumor necrosis factor-alpha and nitric oxide to eliminate tumor cells and bolster T cell-mediated immune responses [31]. In this study, we utilized various algorithms, including ESTIMATE, CIBERSORT, xCELL, TIMER, and ssGSEA, to assess the TME of LUAD samples. Our findings revealed a robust correlation between NUSAP1 expression and immune cell infiltration. Notably, we observed diverse NUSAP1 expression patterns, with high NUSAP1 expression associated with reduced immune cell infiltration compared to samples exhibiting low NUSAP1 expression. Subsequent analysis of multiple single-cell RNA sequencing datasets unveiled predominant NUSAP1 expression on Tprolif cells, supported by gene enrichment analyses. GSEA and GSVA results highlighted the association between NUSAP1 and lipid metabolism, a process pivotal in cancer cell tumorigenesis, disease progression, immune cell recruitment, and modulation of the immune microenvironment response [32]. Importantly, lipid metabolism may impact the biological activity of Tprolif cells [33].
Relationship between NUSAP1 expression and m6A-related genes. (A) The significant positive correlation between NUSAP1 expression and the expression of these m6A-related genes (except FTO). (B) The notable elevation in the expression of these m6A-related genes in samples. **p <0.01; ***p < 0.001.
The TME harboring multiple immunotherapy-associated targets such as PDCD1, PD-L1, LAG3, and CTLA4 can significantly influence the efficacy of immunotherapy in LUAD patients [34]. Consequently, immunotherapeutic approaches targeting the TME to impede tumor growth have garnered considerable attention in recent years. However, resistance to immunotherapy in LUAD stems from the intricate nature of the immune system. One of the foremost unresolved challenges in immunotherapy research is the absence of specific biomarkers to accurately predict the response to immunotherapy in LUAD [35]. In our study, we observed that LUAD samples exhibiting high NUSAP1 expression displayed increased somatic mutation frequency, elevated TMB values, and heightened expression of ICI genes compared to samples with low NUSAP1 expression. Furthermore, analysis of the IMvigor210 cohort revealed that patients with high NUSAP1 expression exhibited superior survival rates and treatment response rates in comparison to those with low NUSAP1 expression. These findings suggest that NUSAP1 holds promise as a predictive biomarker for the efficacy of anti-PD-L1 therapy in LUAD patients. Consequently, our study advocates for the inclusion of LUAD patients with high NUSAP1 expression in future multicenter trials investigating anti-PD-L1 immunotherapy. Furthermore, given that m6A-modified genes often exhibit oncogenic functions in cancer, m6A-related therapies, such as modulation or inhibition of m6A modifications, hold potential as promising cancer treatments [36]. In this study, we identified a positive correlation between m6A modification genes and NUSAP1 expression. Notably, there was a significant increase in the expression of these m6A modifier genes in LUAD samples with high NUSAP1 expression compared to those with low NUSAP1 expression. These findings suggest that NUSAP1 could play a role in predicting the efficacy of m6A-related therapies for managing LUAD.
Function enrichment analyses. (A) Gene-gene interaction networks. (B) Protein-protein interaction networks. (C) A total of 1,378 DEGs, comprising 1,145 up-regulated genes and 233 down-regulated genes. (D) GO and KEGG analyses. (E) GSVA analysis.
In our study, utilizing the CellMiner database, we discovered that amonafide and KPT-9274 show increased effectiveness in anti-tumor therapy for patients exhibiting low NUSAP1 expression. Amonafide, currently under clinical development for cancer treatment, acts as a DNA intercalator, which contains a 5-position amine that undergoes variable acetylation to produce a toxic metabolite in humans, leading to heightened adverse effects and complicating the dosing of amonafide [37]. Amonafide demonstrates significant inhibition of tumor growth, reduction in tumor size, and promising activity in treating advanced breast cancer [38] and acting as a second-line option for acute myeloid leukemia [39]. Unfortunately, in a Phase II clinical study, 18 recruited NSCLC patients were unable to benefit from Amonafide treatment [40]. Additionally, KPT-9274, a dual-specific inhibitor targeting PAK4 and nicotinamide phosphoribosyltransferase, effectively suppressed the growth, survival, and migration of triple-negative breast cancer [41, 42], kidney cancer [43], acute myeloid leukemia [44], and ovarian cancer cells [45]. It could also increase the therapeutic sensitivity of pancreatic neuroendocrine tumors to everolimus [46]. Regrettably, there is currently no study reporting whether treatment with KPT-9274 benefits LUAD patients. This necessitates the recruitment of a specific number of patients for clinical drug trials in the future to validate its efficacy.
Conclusion
In conclusion, the present study identified NUSAP1 as a potential prognostic and immunotherapeutic biomarker for LUAD patients, which warrants further investigation.
Acknowledgements
Thanks to all researchers who participated in this work.
Funding
This work is supported by the National Natural Science Foundation of China (NSFC, No. 22204146), the Joint Funds of Research and Development Program for the Science and Technology Supported by Henan Province (No. 232301420057), PhD research startup foundation of the Third Affiliated Hospital of Zhengzhou University (No. 2022016), and Medical Science and Technology Research Program of Henan Province (No. SBGJ202302081).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found within the article.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committees of Zhengzhou University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
JM and WZ contributed equally to this work. JM: Designing, Project administration, Writing - review and editing, Writing - original draft, Funding acquisition. WZ: Data collection, Data analysis, Performed experiments, Writing - original draft. YW: Data collection, Data analysis, Writing - original draft. HD: Performed experiments related to independent IHC tests, Writing - original draft. LM: Performed experiments related to qRT-PCR, Writing - original draft. LL: Data analysis, Writing - original draft. CN: Data collection, Writing - original draft. SL: Writing - review and editing. KZ: Designing, Writing - review and editing, Conceptualization. EY: Project administration, Writing - review and editing, Funding acquisition. All authors contributed to the article and approved the submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ribbeck K, Raemaekers T, Carmeliet G, Mattaj IW. A role for NuSAP in linking microtubules to mitotic chromosomes. Curr Biol. 2007;17:230-6
2. Matsuda K, Miyoshi H, Hiraoka K, Yokoyama S, Haraguchi T, Hashiguchi T. et al. Clinicopathological and prognostic value of transforming acidic coiled-coil-containing protein 3 (TACC3) expression in soft tissue sarcomas. PLoS One. 2017;12:e0188096
3. Wen X, Hou J, Chu Y, Liao G, Wu G, Fang S. et al. Immunotherapeutic value of NUSAP1 associated with bladder cancer through a comprehensive analysis of 33 human cancer cases. Am J Cancer Res. 2024;14:959-78
4. Liu Y, Tang R, Meng QC, Shi S, Xu J, Yu XJ. et al. NUSAP1 promotes pancreatic ductal adenocarcinoma progression by drives the epithelial-mesenchymal transition and reduces AMPK phosphorylation. BMC Cancer. 2024;24:87
5. Kong J, Xu S, Zhang P, Wang Y. Transcription Factor E2F8 Promotes Cisplatin Resistance in Hepatocellular Carcinoma by Regulating DNA Damage via NUSAP1. Int J Toxicol. 2023;42:420-9
6. Meng J, Yang Z, Jiang X, Zou J. Unveiling NUSAP1 as a common gene signature linking chronic HBV infection and HBV-related HCC. Discov Oncol. 2024;15:61
7. Zhu Y, Liu Y, Zhang L, Zeng S, Xu W. NUSAP1 regulates basal cell carcinoma migration, invasion and DNA damage through activation of the Hedgehog signaling pathway. Physiol Int. 2023;110:160-72
8. Hu Y, Xue Z, Qiu C, Feng Z, Qi Q, Wang J. et al. Knockdown of NUSAP1 inhibits cell proliferation and invasion through downregulation of TOP2A in human glioblastoma. Cell Cycle. 2022;21:1842-55
9. Gou R, Zheng M, Hu Y, Gao L, Wang S, Liu O. et al. Identification and clinical validation of NUSAP1 as a novel prognostic biomarker in ovarian cancer. BMC Cancer. 2022;22:690
10. Yadav DK, Sharma A, Dube P, Shaikh S, Vaghasia H, Rawal RM. Identification of crucial hub genes and potential molecular mechanisms in breast cancer by integrated bioinformatics analysis and experimental validation. Comput Biol Med. 2022;149:106036
11. Luo B, Feng S, Li T, Wang J, Qi Z, Zhao Y. et al. Transcription factor HOXB2 upregulates NUSAP1 to promote the proliferation, invasion and migration of nephroblastoma cells via the PI3K/Akt signaling pathway. Mol Med Rep. 2022;25:205
12. Zhang G. Regulatory T-cells-related signature for identifying a prognostic subtype of hepatocellular carcinoma with an exhausted tumor microenvironment. Front Immunol. 2022;13:975762
13. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612
14. Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D. et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nature medicine. 2015;21:938-45
15. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome biology. 2017;18:220
16. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer research. 2017;77:e108-e10
17. Han Y, Wang Y, Dong X, Sun D, Liu Z, Yue J. et al. TISCH2: expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment. Nucleic Acids Res. 2023;51:D1425-d31
18. Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J. et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012;72:3499-511
19. Tian L, Wang Y, Tian J, Song W, Li L, Che G. Prognostic Value and Genome Signature of m6A/m5C Regulated Genes in Early-Stage Lung Adenocarcinoma. Int J Mol Sci. 2023;24:6520
20. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214-20
21. Aran D, Camarda R, Odegaard J, Paik H, Oskotsky B, Krings G. et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun. 2017;8:1077
22. Cao M, Li H, Sun D, Chen W. Cancer burden of major cancers in China: A need for sustainable actions. Cancer Commun (Lond). 2020;40:205-10
23. Blumenschein GR Jr, Smit EF, Planchard D, Kim DW, Cadranel J, De Pas T. et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)†. Ann Oncol. 2015;26:894-901
24. Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP. et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018-28
25. Zhang Y, Yang M, Ng DM, Haleem M, Yi T, Hu S. et al. Multi-omics Data Analyses Construct TME and Identify the Immune-Related Prognosis Signatures in Human LUAD. Mol Ther Nucleic Acids. 2020;21:860-73
26. Chen D, Zhang X, Li Z, Zhu B. Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics. 2021;11:1016-30
27. Wang H, Meng D, Guo H, Sun C, Chen P, Jiang M. et al. Single-Cell Sequencing, an Advanced Technology in Lung Cancer Research. Onco Targets Ther. 2021;14:1895-909
28. Fan XX, Wu Q. Decoding Lung Cancer at Single-Cell Level. Front Immunol. 2022;13:883758
29. Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu J. et al. Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol. 2021;14:91
30. Genova C, Dellepiane C, Carrega P, Sommariva S, Ferlazzo G, Pronzato P. et al. Therapeutic Implications of Tumor Microenvironment in Lung Cancer: Focus on Immune Checkpoint Blockade. Front Immunol. 2021;12:799455
31. Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79:4557-66
32. Wang Z, Zhang Z, Zhang K, Zhou Q, Chen S, Zheng H. et al. Multi-Omics Characterization of a Glycerolipid Metabolism-Related Gene Enrichment Score in Colon Cancer. Front Oncol. 2022;12:881953
33. Lin L, Ding Y, Wang Y, Wang Z, Yin X, Yan G. et al. Functional lipidomics: Palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism. Hepatology. 2017;66:432-48
34. Yang L, He YT, Dong S, Wei XW, Chen ZH, Zhang B. et al. Single-cell transcriptome analysis revealed a suppressive tumor immune microenvironment in EGFR mutant lung adenocarcinoma. J Immunother Cancer. 2022;10:e003534
35. Tang L, Chen Z, Yang J, Li Q, Wang S, Mo T. et al. Single-cell and Bulk RNA-Seq reveal angiogenic heterogeneity and microenvironmental features to evaluate prognosis and therapeutic response in lung adenocarcinoma. Front Immunol. 2024;15:1352893
36. Li D, Li K, Zhang W, Yang KW, Mu DA, Jiang GJ. et al. The m6A/m5C/m1A Regulated Gene Signature Predicts the Prognosis and Correlates With the Immune Status of Hepatocellular Carcinoma. Frontiers in immunology. 2022;13:918140
37. Liu Y, Norton JT, Witschi MA, Xu Q, Lou G, Wang C. et al. Methoxyethylamino-numonafide is an efficacious and minimally toxic amonafide derivative in murine models of human cancer. Neoplasia. 2011;13:453-60
38. Costanza ME, Berry D, Henderson IC, Ratain MJ, Wu K, Shapiro C. et al. Amonafide: An active agent in the treatment of previously untreated advanced breast cancer-a cancer and leukemia group B study (CALGB 8642). Clin Cancer Res. 1995;1:699-704
39. Ratain MJ, Rosner G, Allen SL, Costanza M, Van Echo DA, Henderson IC. et al. Population pharmacodynamic study of amonafide: a Cancer and Leukemia Group B study. J Clin Oncol. 1995;13:741-7
40. Chang AY, Tu ZN, Smith JL, Bonomi P, Smith TJ, Wiernik PH. et al. Phase II trial of gallium nitrate, amonafide and teniposide in metastatic non-small cell lung cancer. An Eastern Cooperative Oncology Group study (E2588). Invest New Drugs. 1995;13:137-41
41. Cordover E, Wei J, Patel C, Shan NL, Gionco J, Sargsyan D. et al. KPT-9274, an Inhibitor of PAK4 and NAMPT, Leads to Downregulation of mTORC2 in Triple Negative Breast Cancer Cells. Chem Res Toxicol. 2020;33:482-91
42. Rane C, Senapedis W, Baloglu E, Landesman Y, Crochiere M, Das-Gupta S. et al. A novel orally bioavailable compound KPT-9274 inhibits PAK4, and blocks triple negative breast cancer tumor growth. Sci Rep. 2017;7:42555
43. Abu Aboud O, Chen CH, Senapedis W, Baloglu E, Argueta C, Weiss RH. Dual and Specific Inhibition of NAMPT and PAK4 By KPT-9274 Decreases Kidney Cancer Growth. Mol Cancer Ther. 2016;15:2119-29
44. Mitchell SR, Larkin K, Grieselhuber NR, Lai TH, Cannon M, Orwick S. et al. Selective targeting of NAMPT by KPT-9274 in acute myeloid leukemia. Blood Adv. 2019;3:242-55
45. Kudo K, Greer YE, Yoshida T, Harrington BS, Korrapati S, Shibuya Y. et al. Dual-inhibition of NAMPT and PAK4 induces anti-tumor effects in 3D-spheroids model of platinum-resistant ovarian cancer. Cancer Gene Ther. 2024;31:721-35
46. Mpilla GB, Uddin MH, Al-Hallak MN, Aboukameel A, Li Y, Kim SH. et al. PAK4-NAMPT Dual Inhibition Sensitizes Pancreatic Neuroendocrine Tumors to Everolimus. Mol Cancer Ther. 2021;20:1836-45
Author contact
![]() Corresponding author: Corresponding Author: Jun Ma: majun89505edu.cn; Kai Zhang: zhangkai0163com; Enwu Yuan: yuanenwucom.
Corresponding author: Corresponding Author: Jun Ma: majun89505edu.cn; Kai Zhang: zhangkai0163com; Enwu Yuan: yuanenwucom.

 Global reach, higher impact
Global reach, higher impact