Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(15):2934-2942. doi:10.7150/ijms.103179 This issue Cite
Research Paper
Genetic variants of ADAM9 as potential predictors for biochemical recurrence in prostate cancer patients after receiving a radical prostatectomy
1. International Master/PhD Program in Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan.
2. Department of Urology, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan.
3. Department of Urology, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan.
4. Division of Urology, Department of Surgery, Taichung Veterans General Hospital, Taichung, Taiwan.
5. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
6. School of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan.
7. Graduate Institute of Clinical Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan.
8. Department of Mathematical Sciences, Florida Atlantic University, Boca Raton, FL, USA.
9. Department of Applied Chemistry, National Chi Nan University, Nantou, Taiwan.
10. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
11. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
12. Pulmonary Research Center, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan.
13. Traditional Herbal Medicine Research Center, Taipei Medical University Hospital, Taipei, Taiwan.
14. TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei, Taiwan.
#Equal contribution.
Received 2024-9-3; Accepted 2024-10-30; Published 2024-11-4
Abstract
A disintegrin and metalloproteinase domain-containing protein 9 (ADAM9) functions as a membranous bridge, forming cell-cell and cell-matrix connections that regulate tumor aggressiveness in various cancer types, including prostate cancer (PCa). Elevated ADAM9 levels in PCa were identified as a prognostic marker for biochemical recurrence (BCR) in patients who had undergone a radical prostatectomy (RP). However, impacts of genetic variants of ADAM9 on clinicopathological development and BCR remain unclear. Herein, we recruited 702 patients with PCa to evaluate associations of single-nucleotide polymorphisms (SNPs) of ADAM9 with the risk of BCR and clinicopathological development. We genotyped four loci of ADAM9 SNPs located in the promoter and intron regions using a TaqMan allelic discrimination assay, including rs10105311 (C/T), rs7006414 (T/C), rs6474526 (T/G), and rs78451751 (T/C) in 702 Taiwanese PCa patients. Our results showed that the risk of postoperative BCR was 1.508-fold higher in patients carrying the T/C genotype in ADAM9 rs7006414 compared to those with the homozygous T/T genotype, a phenomenon more pronounced in younger PCa patients (aged ≤ 65 years). Furthermore, patients with at least one polymorphic G allele in ADAM9 rs6474526 had a 2.016-fold increased risk of developing an advanced clinical primary tumor stage, particularly in a subpopulation without BCR. Clinical observations from the Genotype-Tissue Expression (GTEx) database showed increased ADAM9 expression in whole blood tissues among individuals carrying the polymorphic C allele of rs7006414 and the G allele of rs6474526. Additionally, data from The Cancer Genome Atlas indicated that elevated ADAM9 levels were observed in PCa tissues compared to corresponding matched normal tissues. Our findings suggest that the rs7006414 and rs6474526 genetic variants of ADAM9 may influence ADAM9 expression and are associated with BCR and clinicopathological development in PCa patients after an RP.
Keywords: A disintegrin and metalloprotease 9, Single-nucleotide polymorphism, Biochemical recurrence, Clinicopathologic progression, Prostate cancer
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related mortality in males, particularly in Western countries [1]. Recently, there has been increasing trends in PCa incidences in developed countries, including Taiwan, likely due to more-advanced medical care facilities and early-stage prostate-specific antigen (PSA) screening [2]. While early-stage PCa can be effectively managed through a radical prostatectomy (RP) and radiotherapy, 20%-50% of PCa patients will experience biochemical recurrence (BCR) within 10 years after initial definitive therapy, characterized by rising serum PSA levels [3]. Although PSA is used as a biomarker for PCa screening, detection, and postoperative tumor recurrence diagnosis, its accuracy is limited. For instance, elevated serum PSA levels can occur in PCa and also in other non-malignant conditions such as prostatic hyperplasia, prostatitis, an older age, and an increased prostate volume [4]. Moreover, the PSA threshold indicative of BCR after an RP has long been debated, with no consensus on the best PSA threshold for defining BCR [5-7]. Therefore, given the importance of early PCa diagnoses for preventing progression and facilitating timely treatment, it is crucial to conduct research aimed at identifying novel and efficient predictive biomarkers.
Recent epidemiological studies suggested that PCa is a highly heritable malignancy, indicating a strong causal association between genetic factors and PCa development [8]. Single-nucleotide polymorphisms (SNPs), which are variations in a genome's base pairs in DNA sequences, cause variations in genes that alter the protein and enzymatic machinery of cells [9,10]. Genome-wide association studies and fine-mapping efforts have identified more than 100 well-recognized SNPs associated with PCa, which constitute major risk factors for susceptibility to, development of, and BCR of PCa [11,12]. For instance, SNPs (rs12422149, rs1789693, and rs1077858) in androgen transporter genes, such as solute carrier organic anion transporter family member 2B1, may serve as pharmacogenomic determinants of resistance to androgen deprivation therapy (ADT) in PCa [13]. Y-box-binding protein-1, a critical regulator of androgen receptor variants involved in resistance to ADT for PCa, has an intronic SNP (rs1203072) that affects gene expression and is linked to PCa metastasis [14]. Additionally, a genetic interaction between insulin-like growth factor 1 (IGF1) rs2946834 and IGF1 receptor (IGF1R) rs2016347 was reported to be a predictor of BCR in PCa patients after an RP [15]. Furthermore, the rs10895304 polymorphism of matrix metalloproteinase-7 is associated with an increased risk of BCR in PCa patients who have undergone an RP [16].
A disintegrin and metalloproteinase 9 (ADAM9) is a type I transmembrane glycoprotein and a significant member of the ADAM family. Recent studies demonstrated a relationship between elevated ADAM9 levels and progression of various cancer cells [17,18]. ADAM9 is overexpressed in PCa and was reported to promote the transition from castration-sensitive PCa to castration-resistant PCa (CRPC) [19]. Elevated ADAM9 was also identified as an independent prognostic marker for PSA relapse-free survival following an RP [20]. Functionally, ADAM9 was reported to promote growth, metastasis, and therapeutic resistance in PCa through various mechanisms. For example, Tang et al. indicated that the ADAM9/WNT1-inducible signaling pathway protein 1 axis cooperates with osteoblasts to induce PCa growth and metastasis [21]. Lin et al. reported that N-α-acetyltransferase 10 stabilizes ADAM9 to promote metastasis in CRPC [22]. Additionally, Josson et al. showed that ADAM9 enhances radio- and chemoresistance in PCa by altering E-cadherin and integrin expressions [23]. Although several studies investigated the clinical significance and functional roles of ADAM9 in PCa, impacts of genetic variants of ADAM9 on PCa have not been explored. In this study, we aimed to examine associations of SNPs within the ADAM9 gene with the risk of BCR and clinicopathological development in Taiwanese PCa patients who had received an RP.
Materials and Methods
Study populations and ethics
In this study, we collected blood samples from 702 PCa patients who had undergone a robotic-assisted laparoscopic RP at Taichung Veterans General Hospital (Taichung, Taiwan) between 2012 and 2018. Prior to venous blood collection, written informed consent was obtained from each participant, and the study protocol was approved by the Institutional Review Board (IRB no. CE19062A-2) of Taichung Veterans General Hospital. Medical data for the recruited patients, including PSA values, pathologic Gleason grades, clinical and pathologic T (tumor) and N (node) staging, cancer cell invasion areas (perineural and lymphovascular regions), and D'Amico classification, were extracted from their medical records at the time of diagnosis. In our study, BCR among the recruited PCa patients was defined by the detection of two consecutive PSA measurements, each exceeding a threshold of 0.2 ng/mL. This criterion was used to indicate a potential recurrence of cancer following initial treatment.
Genomic DNA extraction
Whole-blood samples from PCa patients were collected in tubes containing ethylenediaminetetraacetic acid. After centrifugation, genomic DNA was extracted from buffy coats using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol. The quality of the isolated DNA was evaluated by a Nanodrop-2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) before it was used as a template for a polymerase chain reaction (PCR).
Genotyping of ADAM9 SNPs
In this study, we selected four SNPs in the ADAM9 gene: two in the promoter region (rs7006414 and rs10105311) and two in the intron region (rs6474526 and rs78451751). Alleles of ADAM9 SNPs of rs7006414 (assay ID: C_28949418_10), rs10105311 (assay ID: C_26007162_10), rs6474526 (assay ID: C_449039_10), and rs78451751 (assay ID: C_105349195_10) were discriminated using a TaqMan SNP Genotyping Assay on an ABI StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Final results were analyzed with SDS vers. 3.0 software (Applied Biosystems). Detailed procedures of DNA genotyping were outlined in our previous study [24].
Bioinformatics analysis
Clinical data and messenger (m)RNA sequencing for prostate adenocarcinoma (PRAD) samples from The Cancer Genome Atlas (TCGA) were retrieved from the UCSC Xena database (https://xena.ucsc.edu/). ADAM9 gene expression levels were examined in relation to various clinical features, including Gleason scores, clinical stages, pathological tumor sizes, and lymph node involvement. The Wilcoxon signed-rank test was used for comparisons between two groups, while the Kruskal-Wallis test followed by Dunn's post-hoc test was applied for features with more than two groups.
Statistical analysis
Between-group differences in demographic characteristics were analyzed using a Chi-square test and Student's t-tests. Multivariate logistic regression models were employed to estimate odds ratios (ORs) and adjusted ORs (AORs) with 95% confidence intervals (CIs) of associations between genotypic frequencies and clinicopathologic features. All analyses were conducted using SAS software (vers. 9.1, 2005, for Windows; SAS Institute, Cary, NC, USA), with statistical significance set at p < 0.05.
Results
Demographic characteristics of PCa patients
In Table 1, comparisons are presented of demographic characteristics of PCa patients with postoperative BCR (222 patients) to those without BCR (480 patients). Patients with BCR had a significantly higher incidence of advanced clinical T stages (T3+T4) and elevated PSA levels (> 10 ng/mL) at diagnosis compared to those without BCR. Surgical pathological findings also revealed that patients with BCR more frequently exhibited high pathologic Gleason grades (3+4+5), advanced pathologic T (T3+T4) and N (N1) stages, and evidence of seminal vesicle, perineural, and lymphovascular invasion by tumor cells. According to the D'Amico risk classification for PCa, it was observed that a higher proportion of patients with BCR fell into the high-risk group. Overall, demographic characteristics of our recruited PCa subjects, both with and without BCR, were consistent with those reported in previous studies [25].
Distributions of demographic characteristics among 702 prostate cancer patients
| Variable | Biochemical recurrence (BCR) | ||
|---|---|---|---|
| No (N=480) | Yes (N=222) | p value | |
| Age at diagnosis (years) | |||
| ≤ 65 | 206 (42.9%) | 91 (41.0%) | 0.631 |
| > 65 | 274 (57.1%) | 131 (59.0%) | |
| PSA at diagnosis (ng/mL) | |||
| ≤ 10 | 267 (55.6%) | 67 (30.2%) | < 0.001* |
| > 10 | 213 (44.4%) | 155 (69.8%) | |
| Pathologic Gleason grade group | |||
| 1+2 | 351 (73.1%) | 70 (31.5%) | < 0.001* |
| 3+4+5 | 129 (26.9%) | 152 (68.5%) | |
| Clinical T (tumor) stage | |||
| 1+2 | 438 (91.2%) | 167 (75.2%) | < 0.001* |
| 3+4 | 42 (8.8%) | 55 (24.8%) | |
| Clinical N (node) stage | |||
| N0 | 473 (98.5%) | 215 (96.8%) | 0.135 |
| N1 | 7 (1.5%) | 7 (3.2%) | |
| Pathologic T stage | |||
| 2 | 319 (66.5%) | 52 (23.4%) | < 0.001* |
| 3+4 | 161 (33.5%) | 170 (76.6%) | |
| Pathologic N stage | |||
| N0 | 468 (97.5%) | 174 (78.4%) | < 0.001* |
| N1 | 12 (2.5%) | 48 (21.6%) | |
| Seminal vesicle invasion | |||
| No | 435 (90.6%) | 117 (52.7%) | < 0.001* |
| Yes | 45 (9.4%) | 105 (47.3%) | |
| Perineural invasion | |||
| No | 168 (35.0%) | 18 (8.1%) | < 0.001* |
| Yes | 312 (65.0%) | 204 (91.9%) | |
| Lymphovascular invasion | |||
| No | 446 (92.9%) | 144 (64.9%) | < 0.001* |
| Yes | 34 (7.1%) | 78 (35.1%) | |
| D'Amico classification | |||
| Low risk/Intermediate risk | 273 (56.9%) | 75 (33.8%) | < 0.001* |
| High risk | 207 (43.1%) | 147 (66.2%) | |
* A p value of < 0.05 indicates statistical significance.
Associations of ADAM9 SNPs with postoperative BCR in PCa patients
We then explored the potential effects of four selected SNPs (rs6474526 (T/G), rs7006414 (T/C), rs10105311 (C/T), and rs78451751 (T/C)) in the ADAM9 gene on BCR in PCa patients after an RP. Genotype frequencies of these SNPs were initially analyzed in our cohort of 702 PCa patients. The most common alleles were homozygous T/T for rs6474526, rs7006414, and rs78451751, while homozygous C/C was most prevalent for rs10105311 (Table 2). After adjusting for all potential confounders, only the ADAM9 rs7006414 T/C genotype showed a significant association with a higher risk of postoperative BCR (AOR = 1.508, 95% CI = 1.011-2.250) compared to the rs7006414 T/T genotype (Table 2). No significant trends were observed in polymorphic frequencies of rs6474526, rs10105311, or rs78451751.
Distribution frequencies of ADAM9 genotypes in 702 prostate cancer patients with or without biochemical recurrence (BCR)
| Variable | BCR | |||
|---|---|---|---|---|
| No (N=480) | Yes (N=222) | AOR (95% CI) | p value | |
| rs6474526 | ||||
| TT | 437 (91.0%) | 195 (87.8%) | 1.000 | |
| TG | 41 (8.5%) | 26 (11.7%) | 1.620 (0.866~3.032) | 0.131 |
| GG | 2 (0.5%) | 1 (0.5%) | 0.827 (0.022~30.603) | 0.918 |
| TG+GG | 43 (9.0%) | 27 (12.2%) | 1.262 (0.925~1.720) | 0.142 |
| rs7006414 | ||||
| TT | 276 (57.5%) | 120 (54.1%) | 1.000 | |
| TC | 171 (35.6%) | 93 (41.9%) | 1.508 (1.011~2.250) | 0.044* |
| CC | 33 (6.9%) | 9 (4.0%) | 0.738 (0.283~1.927) | 0.535 |
| TC+CC | 204 (42.5%) | 102 (45.9%) | 1.181 (0.973~1.434) | 0.092 |
| rs10105311 | ||||
| CC | 309 (64.4%) | 143 (64.4%) | 1.000 | |
| CT | 151 (31.5%) | 72 (32.4%) | 1.230 (0.815~1.856) | 0.324 |
| TT | 20 (4.1%) | 7 (3.2%) | 0.804 (0.256~2.525) | 0.709 |
| CT+TT | 171 (35.6%) | 79 (35.6%) | 1.088 (0.891~1.329) | 0.406 |
| rs78451751 | ||||
| TT | 473 (98.5%) | 220 (99.1%) | 1.000 | |
| TC | 7 (1.5%) | 2 (0.9%) | 0.539 (0.183~1.589) | 0.263 |
| CC | 0 (0.0%) | 0 (0.0%) | --- | --- |
| TC+CC | 7 (1.5%) | 2 (0.9%) | 0.539 (0.183~1.589) | 0.263 |
Odds ratios (ORs) and with their 95% confidence intervals (CIs) were estimated by logistic regression models. Adjusted ORs (AORs) with their 95% CIs were estimated by multiple logistic regression models after controlling for age at diagnosis, prostate-specific antigen, pathologic Gleason grade group, clinical T stage, clinical N stage, clinical M stage, pathologic T stage, seminal vesicle invasion, perineural invasion, lymphovascular invasion, and D'Amico classification. * A p value of < 0.05 indicates statistical significance.
In patients aged under 65 years, those carrying the ADAM9 rs7006414 polymorphism also had an increased risk of BCR under both the codominant model (TC vs. TT: AOR = 2.105, 95% CI = 1.111-3.988) and dominant model (TC + CC vs. TT: AOR = 1.432, 95% CI = 1.050-1.953) (Table 3). However, no associations were observed in elderly PCa patients (over 65 years) (data not shown). Additionally, our findings revealed that younger PCa patients carrying at least one minor allele of rs6474526 (TG + GG) had an increased risk of BCR (Table 3). These results provide insights into potential associations between ADAM9 genetic variations and BCR, particularly in younger PCa patients after an RP.
Distribution frequencies of ADAM9 genotypes in 297 younger prostate cancer patients (aged ≤ 65 years) with or without biochemical recurrence (BCR)
| Variable | BCR | |||
|---|---|---|---|---|
| No (N=206) | Yes (N=91) | AOR (95% CI) | p value | |
| rs6474526 | ||||
| TT | 193 (93.7%) | 78 (85.7%) | 1.000 | |
| TG | 13 (6.3%) | 13 (14.3%) | 1.769 (1.085~2.884) | 0.022* |
| GG | 0 (0.0%) | 0 (0.0%) | --- | --- |
| TG+GG | 13 (6.3%) | 13 (14.3%) | 1.769 (1.085~2.884) | 0.022* |
| rs7006414 | ||||
| TT | 117 (56.8%) | 47 (51.6%) | 1.000 | |
| TC | 75 (36.4%) | 39 (42.9%) | 2.105 (1.111~3.988) | 0.025* |
| CC | 14 (6.8%) | 5 (5.5%) | 1.680 (0.432~6.529) | 0.454 |
| TC+CC | 89 (43.2%) | 44 (48.4%) | 1.432 (1.050~1.953) | 0.023* |
| rs10105311 | ||||
| CC | 128 (62.1%) | 60 (65.9%) | 1.000 | |
| CT | 67 (32.5%) | 26 (28.6%) | 1.221 (0.636~2.343) | 0.549 |
| TT | 11 (5.3%) | 5 (5.5%) | 1.767 (0.433~7.207) | 0.428 |
| CT+TT | 78 (37.9%) | 31 (34.1%) | 1.131 (0.829~1.544) | 0.437 |
| rs78451751 | ||||
| TT | 202 (98.1%) | 91 (100.0%) | 1.000 | |
| TC | 4 (1.9%) | 0 (0.0%) | --- | --- |
| CC | 0 (0.0%) | 0 (0.0%) | --- | --- |
| TC+CC | 4 (1.9%) | 0 (0.0%) | --- | --- |
Odds ratios (ORs) with their 95% confidence intervals (CIs) were estimated by logistic regression models. Adjusted odds ratios (AORs) with their 95% CIs were estimated by multiple logistic regression models after controlling for prostate-specific antigen, pathologic Gleason grade group, clinical T stage, clinical N stage, pathologic T stage, pathologic N stage, seminal vesicle invasion, perineural invasion, lymphovascular invasion, and D'Amico classification. * A p value of < 0.05 indicates statistical significance.
Relationships between clinicopathological features and ADAM9 SNPs in PCa patients with and those without BCR
Next, to clarify the effects of ADAM9 genetic polymorphisms on the clinicopathological status of PCa, we examined factors such as pathologic staging, pathologic Gleason grade groups, clinical staging, tumor invasion statuses, and D'Amico classification. Among the four ADAM9 loci analyzed, we found that PCa patients carrying at least one minor allele (TG+GG) of rs6474526 had a significantly higher risk of developing advanced clinical T stages (cT3+4) (OR = 2.016, 95% CI = 1.101-3.690; p = 0.021) compared to those with the wild-type (WT) homozygote (TT), as shown in Table 4. None of the other three ADAM9 SNPs—rs7006414, rs10105311, or rs78451751—showed significant associations with the clinicopathological features mentioned above (see Table 4 and data not shown). We further stratified PCa patients into subgroups with or without BCR and investigated relationships between ADAM9 SNPs and PCa clinicopathological statuses within both groups. Notably, among the 480 PCa patients without BCR, those carrying the rs6474526 G-allele had a significantly higher risk of developing advanced clinical T stages (OR = 3.241, 95% CI = 1.433-7.328; p = 0.003) compared to patients with the WT T-allele (Table 5).
Potential impacts of ADAM9 genetic polymorphisms on ADAM9 expression
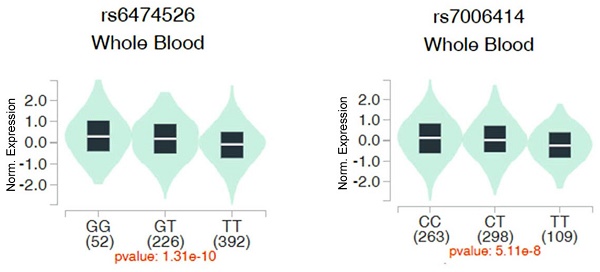
We then evaluated associations between ADAM9 polymorphisms and ADAM9 gene expression in whole blood samples from healthy individuals using data from the Genotype-Tissue Expression (GTEx) database. Individuals with the WT homozygous TT genotype for rs6474526 and rs7006414 all showed the lowest ADAM9 expression levels compared to those carrying at least one minor allele (Figure 1).
Correlations of ADAM9 expression levels with clinicopathologic features and prognoses of PCa patients
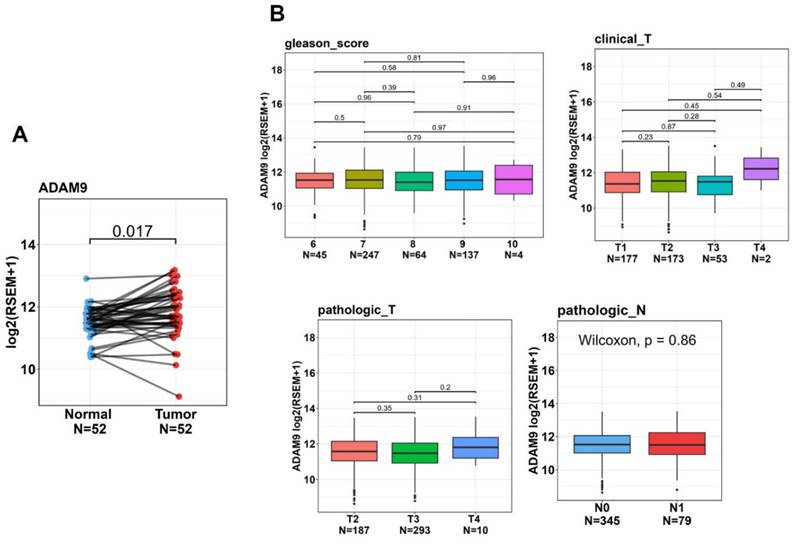
To further examine ADAM9 expression levels in normal and PCa tissues and explore correlations between ADAM9 levels and disease progression, we utilized the TCGA-PRAD dataset. Our analysis revealed that ADAM9 expression was significantly higher in tumor tissues compared to corresponding matched normal tissues (Figure 2A). Although we found no significant correlations between elevated ADAM9 expression levels and various clinicopathological features, such as Gleason scores, clinical T stages, pathological T stages, and lymph node metastasis, we observed a trend where tumor tissues with a clinical T4 stage exhibited higher ADAM9 expression compared to those with clinical T1-T3 stages (Figure 2B).
Discussion
Given the significant role of ADAM9 as a protease with oncogenic effects in PCa progression [17,18], we identified polymorphisms in the promoter and intron regions of the ADAM9 gene that displayed different distributions in PCa patients with and those without BCR. Our findings showed that patients with the mutant TC genotype of rs7006414 had a significantly higher risk of developing postoperative BCR, with stronger associations observed in younger PCa patients (aged ≤ 65 years).
Odds ratios (ORs) and 95% confidence intervals (CIs) for the clinical status and genotypic frequencies of ADAM9 rs6474526 and rs7006414 in 702 prostate cancer patients
| Variable | rs6474526 | rs7006414 | ||||||
|---|---|---|---|---|---|---|---|---|
| TT (N=632) | TG+GG (N=70) | OR (95% CI) | p value | TT (N=396) | TC+CC (N=306) | OR (95% CI) | p value | |
| Pathologic Gleason grade group | ||||||||
| 1+2 | 380 (60.1%) | 41 (58.6%) | 1.000 | 0.801 | 235 (59.3%) | 186 (60.8%) | 1.000 | 0.699 |
| 3+4+5 | 252 (39.9%) | 29 (41.4%) | 1.067 (0.646~1.761) | 161 (40.7%) | 120 (39.2%) | 0.942 (0.694~1.277) | ||
| Clinical T (tumor) stage | ||||||||
| 1+2 | 551 (87.2%) | 54 (77.1%) | 1.000 | 0.021* | 343 (86.6%) | 262 (85.6%) | 1.000 | 0.705 |
| 3+4 | 81 (12.8%) | 16 (22.9%) | 2.016 (1.101~3.690) | 53 (13.4%) | 44 (14.4%) | 1.087 (0.706~1.672) | ||
| Clinical N (node) stage | ||||||||
| N0 | 619 (97.9%) | 69 (98.6%) | 1.000 | 0.721 | 387 (97.7%) | 301 (98.4%) | 1.000 | 0.548 |
| N1 | 13 (2.1%) | 1 (1.4%) | 0.690 (0.089~5.356) | 9 (2.3%) | 5 (1.6%) | 0.714 (0.237~2.153) | ||
| Pathologic T stage | ||||||||
| 2 | 338 (53.5%) | 33 (47.1%) | 1.000 | 0.313 | 205 (51.8%) | 166 (54.2%) | 1.000 | 0.514 |
| 3+4 | 294 (46.5%) | 37 (52.9%) | 1.289 (0.786~2.114) | 191 (48.2%) | 140 (45.8%) | 0.905 (0.671~1.221) | ||
| Pathologic N stage | ||||||||
| N0 | 580 (91.8%) | 62 (88.6%) | 1.000 | 0.363 | 361 (91.2%) | 281 (91.8%) | 1.000 | 0.753 |
| N1 | 52 (8.2%) | 8 (11.4%) | 1.439 (0.654~3.168) | 35 (8.8%) | 25 (8.2%) | 0.918 (0.537~1.569) | ||
| Seminal vesicle invasion | ||||||||
| No | 500 (79.1%) | 52 (74.3%) | 1.000 | 0.350 | 308 (77.8%) | 244 (79.7%) | 1.000 | 0.530 |
| Yes | 132 (20.9%) | 18 (25.7%) | 1.311 (0.742~2.317) | 88 (22.2%) | 62 (20.3%) | 0.889 (0.617~1.282) | ||
| Perineural invasion | ||||||||
| No | 171 (27.1%) | 15 (21.4%) | 1.000 | 0.311 | 104 (26.4%) | 82 (26.8%) | 1.000 | 0.874 |
| Yes | 461 (72.9%) | 55 (78.6%) | 1.360 (0.748~2.471) | 292 (73.7%) | 224 (73.2%) | 0.973 (0.694~1.364) | ||
| Lymphovascular invasion | ||||||||
| No | 529 (83.7%) | 61 (87.1%) | 1.000 | 0.456 | 334 (84.3%) | 256 (83.7%) | 1.000 | 0.806 |
| Yes | 103 (16.3%) | 9 (12.9%) | 0.758 (0.365~1.574) | 62 (15.7%) | 50 (16.3%) | 1.052 (0.701~1.580) | ||
| D'Amico classification | ||||||||
| Low risk/Intermediate risk | 312 (49.4%) | 36 (51.4%) | 1.000 | 0.743 | 185 (46.7%) | 163 (53.3%) | 1.000 | 0.085 |
| High risk | 320 (50.6%) | 34 (48.6%) | 0.921 (0.562~1.509) | 211 (53.3%) | 143 (46.7%) | 0.769 (0.570~1.037) | ||
ORs with their 95% CIs were estimated by logistic regression models.
* p < 0.05 indicates statistical significance.
Odds ratios (ORs) and 95% confidence intervals (CIs) for the clinical status and ADAM9 rs6474526 genotypic frequencies in 702 prostate cancer patients with and those without biochemical recurrence
| Variable | No biochemical recurrence (N=480) | With biochemical recurrence (N=222) | ||||||
|---|---|---|---|---|---|---|---|---|
| TT (N=437) | TG+GG (N=43) | OR (95% CI) | p value | TT (N=195) | TG+GG (N=27) | OR (95% CI) | p value | |
| Pathologic Gleason grade group | ||||||||
| 1+2 | 319 (73.0%) | 32 (74.4%) | 1.000 | 0.841 | 61 (31.3%) | 9 (33.3%) | 1.000 | 0.830 |
| 3+4+5 | 118 (27.0%) | 11 (25.6%) | 0.929 (0.454~1.903) | 134 (68.7%) | 18 (66.7%) | 0.91 (0.387~2.142) | ||
| Clinical T (tumor) stage | ||||||||
| 1+2 | 404 (92.4%) | 34 (79.1%) | 1.000 | 0.003* | 147 (75.4%) | 20 (74.1%) | 1.000 | 0.882 |
| 3+4 | 33 (7.6%) | 9 (20.9%) | 3.241 (1.433~7.328) | 48 (24.6%) | 7 (25.9%) | 1.072 (0.427~2.691) | ||
| Clinical N (node) stage | ||||||||
| N0 | 431 (98.6%) | 69 (97.7%) | 1.000 | 0.619 | 188 (96.4%) | 27 (100.0%) | 1.000 | --- |
| N1 | 6 (1.4%) | 1 (2.3%) | 1.710 (0.201~14.545) | 7 (3.6%) | 0 (0.0%) | --- | ||
| Pathologic T stage | ||||||||
| 2 | 292 (66.8%) | 27 (62.8%) | 1.000 | 0.593 | 46 (23.6%) | 6 (22.2%) | 1.000 | 0.875 |
| 3+4 | 145 (33.2%) | 16 (37.2%) | 1.193 (0.623~2.285) | 149 (76.4%) | 21 (77.8%) | 1.081 (0.411~2.838) | ||
| Pathologic N stage | ||||||||
| N0 | 428 (97.9%) | 40 (93.0%) | 1.000 | 0.083 | 152 (77.9%) | 22 (81.5%) | 1.000 | 0.676 |
| N1 | 9 (2.1%) | 3 (7.0%) | 3.567 (0.928~13.706) | 43 (22.1%) | 5 (18.5%) | 0.803 (0.287~2.247) | ||
| Seminal vesicle invasion | ||||||||
| No | 398 (91.1%) | 37 (86.0%) | 1.000 | 0.280 | 102 (52.3%) | 15 (56.6%) | 1.000 | 0.751 |
| Yes | 39 (8.9%) | 6 (14.0%) | 1.655 (0.657~4.166) | 93 (47.7%) | 12 (44.4%) | 0.877 (0.391~1.971) | ||
| Perineural invasion | ||||||||
| No | 156 (35.7%) | 12 (27.9%) | 1.000 | 0.307 | 15 (7.7%) | 3 (11.1%) | 1.000 | 0.542 |
| Yes | 281 (64.3%) | 31 (72.1%) | 1.434 (0.716~2.872) | 180 (92.3%) | 24 (88.9%) | 0.667 (0.180~2.473) | ||
| Lymphovascular invasion | ||||||||
| No | 405 (92.7%) | 41 (95.3%) | 1.000 | 0.515 | 124 (63.6%) | 20 (74.1%) | 1.000 | 0.285 |
| Yes | 32 (7.3%) | 2 (4.7%) | 0.617 (0.143~2.670) | 71 (36.4%) | 7 (25.9%) | 0.611 (0.246~1.517) | ||
| D'Amico classification | ||||||||
| Low risk/Intermediate risk | 248 (56.8%) | 25 (58.1%) | 1.000 | 0.861 | 64 (32.8%) | 11 (40.7%) | 1.000 | 0.415 |
| High risk | 189 (43.2%) | 18 (41.9%) | 0.945 (0.501~1.782) | 131 (67.2%) | 16 (59.3%) | 0.711 (0.312~1.620) | ||
ORs with their 95% CIs were estimated by logistic regression models.
* p < 0.05 indicates statistical significance.
Impacts of ADAM9 rs6474526 and rs7006414 polymorphisms on ADAM9 expression were assessed using validated data from the Genotype Tissue Expression (GTEx) portal (https://www.gtexportal.org/home/). Violin plots illustrate that the G allele of rs6474526 (left panel) and the C allele of rs7006414 (right panel) were associated with higher ADAM9 expression levels in whole blood.
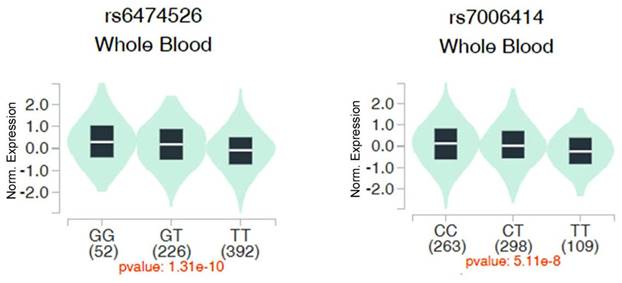
In this subgroup, we also found that those carrying at least one minor G allele of rs6474526 had an increased risk of BCR. These results highlight the potential influence of specific genetic variations of ADAM9 on BCR, particularly in younger PCa patients after an RP. Additionally, PCa patients with a mutated G variant of ADAM9 rs6474526 exhibited a notably increased risk of developing advanced clinical T stages (3 or 4). More-robust associations between the rs6474526 SNP and advanced clinical T stages were observed in PCa patients without BCR, suggesting that the ADAM9 rs6474526 SNP may influence tumor growth, especially in patients without BCR.
The clinical relevance of ADAM9 levels in prostate cancer (PCa) patients was analyzed using data from the TCGA-prostate adenocarcinoma (PRAD) dataset. (A) ADAM9 gene expressions were evaluated in paired normal and tumor tissues from PCa patients. (B) ADAM9 gene expression levels in PCa were compared from the TCGA-PRAD dataset based on Gleason scores, clinical T stages, pathological T stages, and lymph node metastasis.
The rs7006414 SNP is located at position -1314 in the promoter region of the ADAM9 gene. Results of an ADAM9 promoter reporter assay demonstrated that the rs7006414 C allele exhibited higher transcriptional activity compared to the rs7006414 T allele in both neural and non-neural cell lines, such as the LNCaP PCa cell line [26], thereby increasing ADAM9 expression. This difference in activity may be due to variations in the affinity of DNA-binding proteins for the two alleles of the ADAM9 promoter. The same study indicated that the rs7006414 C allele enhances DNA/protein interactions, as shown by electrophoretic mobility shift assay results, suggesting that the rs7006414 C polymorphism may more effectively bind with transcription factors, leading to increased ADAM9 expression [26]. Consistent with these findings, we also observed elevated ADAM9 expression in whole blood tissues of individuals carrying the polymorphic C allele of ADAM9 rs7006414, as indicated by data from the GTEx database. Indeed, elevated ADAM9 levels were identified as a prognostic marker for BCR in PCa patients [20]. Collectively, these observations suggest that the polymorphic C allele of ADAM9 rs7006414 may upregulate ADAM9 expression, thereby increasing the likelihood of BCR in PCa patients after an RP.
ADT is the primary treatment strategy for PCa as it reduces circulating androgen levels to castration levels [27]. ADAM9 upregulation was reported to play a critical role in resistance to ADT in PCa patients [19]. In addition to androgens, estrogens—particularly estradiol, the most potent endogenous estrogen—also influence PCa cell biology. For instance, plasma estradiol levels are a significant predictor of high-grade PCa in patients undergoing an RP [28] and in patients undergoing ADT who progress to CRPC [29]. Cong et al. showed estrogen to be an inducer of ADAM9 promoter activity in LNCaP cells, especially in cases where the ADAM9 gene carries the rs7006414 C polymorphism [20]. We propose that age-related estrogen declines may lead to lower ADAM9 levels in older PCa patients, which could explain why we observed stronger associations between the rs7006414 SNP and BCR in younger PCa patients after an RP.
Beyond rs7006414, our findings indicated that the G polymorphism of ADAM9 rs6474526 also affected BCR and advanced clinical T stages in PCa patients. Notably, previous research demonstrated that elevated ADAM9 expression promoted tumor growth in PCa [21]. The rs6474526 SNP resides within an intron, and data from the GTEx database revealed that individuals with a polymorphic G allele of rs6474526 showed higher ADAM9 expression in whole blood tissues. Although intronic polymorphisms generally do not alter protein sequences, emerging evidence suggests that these variations can cause splicing abnormalities, potentially affecting transcription and contributing to various human diseases, including cancers. For example, the rs12203592 intronic polymorphism in the interferon regulatory factor 4 (IRF4) gene was linked to a higher risk of acute lymphoblastic leukemia, where a C to T substitution increased IRF4 gene expression by altering the binding affinity of the activator protein 2α transcription factor (TF) [30]. Additionally, intronic regions often contain numerous cis-regulatory elements, such as TF-binding sites, enhancers, and silencers, which can either upregulate or downregulate gene expressions. For instance, three independent variants (rs2981578, rs35054928, and rs45631563) in fibroblast growth factor receptor 2 (FGFR2) were mapped to transcriptional silencer elements, enhancing silencer activity, reducing FGFR2 expression, and increasing breast cancer risk due to greater estrogen responsiveness [31]. Moreover, intronic regions also encompass many non-coding RNA motifs, including long non-coding (lnc)RNAs. Intronic lncRNAs were shown to play crucial roles in regulating expressions of their host genes [32]. Interestingly, lncADAM9 was reported to downregulate ADAM9 mRNA [33]. However, whether the rs6474526 SNP is located within lncADAM9 or another intronic lncRNA region remains unclear. The transcriptional potential of the rs6474526 T>G SNP within the ADAM9 intron will be further investigated in future studies.
Nevertheless, this study still has some limitations that need to be discussed. First, due to the relatively small sample size, the frequencies of certain homozygous variants (e.g., rs7006414 CC or rs6474526 GG genotypes) were low in subgroups, which may have limited the statistical power and precision of our results. Therefore, conducting studies with larger independent cohorts from different medical centers would provide more robust and reliable findings on the impact of ADAM9 SNPs on BCR and the development of PCa. Additionally, all PCa patients in this SNP study were Taiwanese (of Chinese ethnicity), whereas the correlations between ADAM9 expression and clinicopathologic features were analyzed using the TCGA-PRAD dataset, which predominantly includes White and African American individuals. It remains unclear whether similar results would be observed across different racial/ethnic groups. Further investigations of these SNPs in diverse racial populations of PCa patients are needed to confirm our findings. Finally, our current study only demonstrated the effects of ADAM9 rs6474526 and rs7006414 SNPs on ADAM9 gene expression in whole blood tissues of healthy individuals, based on the GTEx database. To validate the influence of ADAM9 SNPs on ADAM9 expression in PCa patients, future studies should simultaneously collect mRNA and DNA from the same samples of PCa patients.
In summary, ours is the first study to explore distinct allelic effects of ADAM9 SNPs (rs7006414 and rs6474526) in a Taiwanese population, highlighting their impacts on the incidence of BCR and tumor growth in PCa. Our findings suggest that the rs7006414 promoter SNP and the rs6474526 intronic SNP may influence ADAM9 gene expression, thereby contributing to promoting BCR and tumor growth in PCa. These genetic variants could potentially serve as valuable markers for predicting recurrence in PCa patients undergoing an RP.
Acknowledgements
This study was supported by the TMU Research Center of Cancer Translational Medicine from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (to M.-H. Chien) and by grant no. 113-wf-swf-05 from Taipei Medical University-Wan Fang Hospital (to Y.-C. Wen).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48
2. Chang HJ, Pong YH, Chiang CY, Huang PC, Wang MH, Chan YJ. et al. A matched case-control study in Taiwan to evaluate potential risk factors for prostate cancer. Sci Rep. 2023;13:4382
3. Kupelian PA, Buchsbaum JC, Elshaikh M, Reddy CA, Zippe C, Klein EA. Factors affecting recurrence rates after prostatectomy or radiotherapy in localized prostate carcinoma patients with biopsy Gleason score 8 or above. Cancer. 2002;95:2302-2307
4. Bo M, Ventura M, Marinello R, Capello S, Casetta G, Fabris F. Relationship between Prostatic Specific Antigen (PSA) and volume of the prostate in the Benign Prostatic Hyperplasia in the elderly. Crit Rev Oncol Hematol. 2003;47:207-211
5. Cookson MS, Aus G, Burnett AL, Canby-Hagino ED, D'Amico AV, Dmochowski RR. et al. Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: the American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel report and recommendations for a standard in the reporting of surgical outcomes. J Urol. 2007;177:540-545
6. Toussi A, Stewart-Merrill SB, Boorjian SA, Psutka SP, Thompson RH, Frank I. et al. Standardizing the Definition of Biochemical Recurrence after Radical Prostatectomy-What Prostate Specific Antigen Cut Point Best Predicts a Durable Increase and Subsequent Systemic Progression? J Urol. 2016;195:1754-1759
7. Tourinho-Barbosa R, Srougi V, Nunes-Silva I, Baghdadi M, Rembeyo G, Eiffel SS. et al. Biochemical recurrence after radical prostatectomy: what does it mean? Int Braz J Urol. 2018;44:14-21
8. Pernar CH, Ebot EM, Wilson KM, Mucci LA. The Epidemiology of Prostate Cancer. Cold Spring Harb Perspect Med. 2018;8:a030361
9. Chan EY. Next-generation sequencing methods: impact of sequencing accuracy on SNP discovery. Methods Mol Biol. 2009;578:95-111
10. Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009;578:23-39
11. Benafif S, Kote-Jarai Z, Eeles RA. A Review of Prostate Cancer Genome-Wide Association Studies (GWAS). Cancer Epidemiol Biomarkers Prev. 2018;27:845-857
12. Allemailem KS, Almatroudi A, Alrumaihi F, Makki Almansour N, Aldakheel FM, Rather RA. et al. Single nucleotide polymorphisms (SNPs) in prostate cancer: its implications in diagnostics and therapeutics. Am J Transl Res. 2021;13:3868-3889
13. Yang M, Xie W, Mostaghel E, Nakabayashi M, Werner L, Sun T. et al. SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J Clin Oncol. 2011;29:2565-2573
14. Shiota M, Fujimoto N, Imada K, Yokomizo A, Itsumi M, Takeuchi A. et al. Potential Role for YB-1 in Castration-Resistant Prostate Cancer and Resistance to Enzalutamide Through the Androgen Receptor V7. J Natl Cancer Inst. 2016;108:djw005
15. Chang CF, Pao JB, Yu CC, Huang CY, Huang SP, Yang YP. et al. Common variants in IGF1 pathway genes and clinical outcomes after radical prostatectomy. Ann Surg Oncol. 2013;20:2446-2452
16. Jaboin JJ, Hwang M, Lopater Z, Chen H, Ray GL, Perez C. et al. The matrix metalloproteinase-7 polymorphism rs10895304 is associated with increased recurrence risk in patients with clinically localized prostate cancer. Int J Radiat Oncol Biol Phys. 2011;79:1330-1335
17. Haoyuan MA, Yanshu LI. Structure, regulatory factors and cancer-related physiological effects of ADAM9. Cell Adh Migr. 2020;14:165-181
18. Chou CW, Huang YK, Kuo TT, Liu JP, Sher YP. An Overview of ADAM9: Structure, Activation, and Regulation in Human Diseases. Int J Mol Sci. 2020;21:7790
19. Le TT, Hsieh CL, Lin IH, Chu CY, Do AD, Chen SH. et al. The ADAM9/UBN2/AKR1C3 axis promotes resistance to androgen-deprivation in prostate cancer. Am J Cancer Res. 2022;12:176-197
20. Fritzsche FR, Jung M, Tölle A, Wild P, Hartmann A, Wassermann K. et al. ADAM9 expression is a significant and independent prognostic marker of PSA relapse in prostate cancer. Eur Urol. 2008;54:1097-1106
21. Chang AC, Lin LW, Chen YC, Chen PC, Liu SC, Tai HC. et al. The ADAM9/WISP-1 axis cooperates with osteoblasts to stimulate primary prostate tumor growth and metastasis. Int J Biol Sci. 2023;19:760-771
22. Lin YW, Wen YC, Chu CY, Tung MC, Yang YC, Hua KT. et al. Stabilization of ADAM9 by N-α-acetyltransferase 10 protein contributes to promoting progression of androgen-independent prostate cancer. Cell Death Dis. 2020;11:591
23. Josson S, Anderson CS, Sung SY, Johnstone PA, Kubo H, Hsieh CL. et al. Inhibition of ADAM9 expression induces epithelial phenotypic alterations and sensitizes human prostate cancer cells to radiation and chemotherapy. Prostate. 2011;71:232-240
24. Lin YW, Wang SS, Wen YC, Tung MC, Lee LM, Yang SF. et al. Genetic Variations of Melatonin Receptor Type 1A are Associated with the Clinicopathologic Development of Urothelial Cell Carcinoma. Int J Med Sci. 2017;14:1130-1135
25. Koupparis AJ, Grummet JP, Hurtado-Coll A, Bell RH, Buchan N, Goldenberg SL. et al. Radical prostatectomy for high-risk clinically localized prostate cancer: a prospective single institution series. Can Urol Assoc J. 2011;5:E156-161
26. Cong L, Jia J. Promoter polymorphisms which regulate ADAM9 transcription are protective against sporadic Alzheimer's disease. Neurobiol Aging. 2011;32:54-62
27. Polotti CF, Kim CJ, Chuchvara N, Polotti AB, Singer EA, Elsamra S. Androgen deprivation therapy for the treatment of prostate cancer: a focus on pharmacokinetics. Expert Opin Drug Metab Toxicol. 2017;13:1265-1273
28. Salonia A, Gallina A, Briganti A, Suardi N, Capitanio U, Abdollah F. et al. Circulating estradiol, but not testosterone, is a significant predictor of high-grade prostate cancer in patients undergoing radical prostatectomy. Cancer. 2011;117:5029-5038
29. Toren P, Hoffman A, Ding K, Joncas FH, Turcotte V, Caron P. et al. Serum Sex Steroids as Prognostic Biomarkers in Patients Receiving Androgen Deprivation Therapy for Recurrent Prostate Cancer: A Post Hoc Analysis of the PR.7 Trial. Clin Cancer Res. 2018;24:5305-5312
30. Do TN, Ucisik-Akkaya E, Davis CF, Morrison BA, Dorak MT. An intronic polymorphism of IRF4 gene influences gene transcription in vitro and shows a risk association with childhood acute lymphoblastic leukemia in males. Biochim Biophys Acta. 2010;1802:292-300
31. Campbell TM, Castro MAA, de Santiago I, Fletcher MNC, Halim S, Prathalingam R. et al. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis. 2016;37:741-750
32. Wu H, Yang L, Chen LL. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017;33:540-552
33. Wang X, Liu A, Hou D, Dong X, Chu C, Ju W. et al. Epigenetic impact of long non-coding RNA lnc-ADAM9 on extracellular matrix pathway in preterm syndrome through down-regulation of mRNA-ADAM9. J Transl Genet Genom. 2023;7:126-140
Author contact
![]() Corresponding authors: Ming-Hsien Chien, PhD; E-mail: d002089002edu.tw or Shun-Fa Yang, PhD; E-mail: ysfedu.tw.
Corresponding authors: Ming-Hsien Chien, PhD; E-mail: d002089002edu.tw or Shun-Fa Yang, PhD; E-mail: ysfedu.tw.

 Global reach, higher impact
Global reach, higher impact