Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(15):2870-2882. doi:10.7150/ijms.100957 This issue Cite
Research Paper
Co-inhibition of PGF and VEGFA enhances the effectiveness of immunotherapy in bladder cancer
1. Institute of Urology, The Third Affiliated Hospital of Shenzhen University (Luohu Hospital Group), Shenzhen 518000, China.
2. Department of Urology, South China Hospital of Shenzhen University, Shenzhen 518116, China.
3. Shantou University Medical College, Shantou University, Shantou, China.
4. Medical School, Anhui University of Science and Technology, Huainan 232001, China.
5. Guangzhou Laboratory, Guangzhou International BioIsland, Guangzhou 510005, China.
6. BioSyngen Pte Ltd, Taiseng Exchange, 5 Tai Seng Avenue, 536671, Singapore.
† These authors contributed equally to this work.
Received 2024-7-14; Accepted 2024-10-9; Published 2024-10-28
Abstract
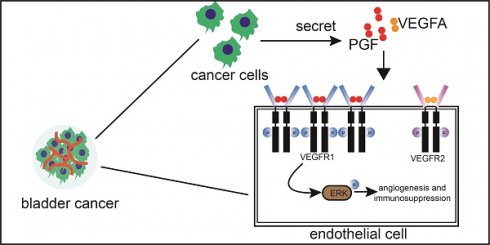
Background: Anti-angiogenic inhibitors and immune checkpoint blockade combination therapy offers a novel approach to circumvent the challenges associated with limited responsiveness to checkpoint inhibitors in bladder cancer. However, the effective strategies for inhibiting angiogenesis in bladder cancer need further elucidation.
Objective: This work aims to identify key targets for the effective inhibition of angiogenesis in bladder cancer and to explore the potential benefits of combining anti-angiogenic therapies with immune checkpoint blockade strategies in the treatment of this disease.
Methods: Cell-cell interaction analysis was performed using bladder cancer single-cell transcriptome datasets downloaded from the Gene Expression Omnibus (GEO) database to determine the regulatory network driving angiogenesis in bladder cancer. The bladder cancer cell line MBT2 was orthotopically transplanted into mice to investigate the impact of pro-angiogenic molecules on angiogenesis and tumor growth, and to evaluate the synergistic therapeutic potential of a combination therapy targeting angiogenesis and Programmed Cell Death Protein 1 (PD-1). Proliferation and tube formation assays with Human Umbilical Vein Endothelial Cells (HUVECs) were used to explore the regulatory functions of pro-angiogenic molecules in angiogenesis.
Results: Placental growth factor (PGF) is a pro-angiogenic factor in bladder cancer, in addition to vascular endothelial growth factor A (VEGFA). Suppression of PGF reduced the tumor size and angiogenesis in bladder cancer. The expression level of vascular endothelial growth factor receptor 1 (VEGFR1) is higher than that of vascular endothelial growth factor receptor2 (VEGFR2) in the endothelial cells of bladder cancer. The pro-angiogenic activity of PGF is dependent on the expression level of VEGFR1 in endothelial cells. The combined inhibition of PGF and VEGFA exerts a synergistic effect on suppressing tumor growth and angiogenesis. The concurrent inhibition of PGF and VEGFA stands out as the only intervention capable of significantly enhancing the infiltration of CD8+ cytotoxic T cells within the bladder cancer microenvironment. In the bladder cancer mouse model, the introduction of an anti- programmed cell death protein 1 (PD-1) therapeutic regimen combined with the targeted inhibition of PGF and VEGFA, led to a significantly elevated survival rate compared to the outcome observed with anti-PD-1 monotherapy.
Conclusion: PGF is a pro-angiogenic molecule in bladder cancer that requires significant expression levels of VEGFR1 in endothelial cells. Notably, the concurrent inhibition of PGF and VEGFA amplifies the therapeutic impact of anti-PD-1 treatment in bladder cancer. These findings provide further insights into the role of PGF in angiogenesis regulation and have conceptual implications for combining anti-angiogenic therapy with immune therapy in bladder cancer treatment.
Keywords: anti-angiogenesis, anti-PD1, PGF, bladder cancer
Introduction
Immune checkpoint inhibitors (ICIs) have shown survival advantages in some bladder cancer patients [1-4]. Pembrolizumab has been approved by the U.S. Food and Drug administration (FDA) for non-muscle invasive bladder cancer (NMIBC) patients who exhibit refractoriness to Bacillus Calmette Guerin (BCG) [5, 6] and for patients with advanced urothelial carcinoma who are ineligible for cisplatin-containing chemotherapy [5, 7]. However, this therapy has a limitation due to its low response rates, with only about 25% of the tumors demonstrating responsiveness [8-10]. A common strategy to overcome this limitation is to develop combination therapies [11]. For example, the National Comprehensive Cancer Network (NCCN) guidelines have incorporated the combination of pembrolizumab with the antibody-drug conjugate enfortumab vedotin as the preferred regimen for patients with locally advanced or metastatic urothelial cancer [12].
Numerous investigations demonstrate that combining an anti-angiogenic therapy with ICIs can improve the efficiency of ICIs [13, 14]. The combination of the checkpoint inhibitor atezolizumab (A) with the anti-angiogenic drug bevacizumab (B) and chemotherapy (C) resulted in improved overall survival compared to AC and BC in chemotherapy-naive patients with metastatic non-squamous non-small lung cancer (NSCLC), However, this survival advantage was not observed in the AC group, suggesting a potential synergistic effect between bevacizumab and atezolizumab [15, 16]. Tumor vessels lacking a basement membrane and pericytes prevent effector T cells from accessing deeper regions of tumors. Additionally, the tumor vasculature expresses high levels of immunosuppressive molecules, including programmed cell death 1 protein ligand 1 (PD-L1), resulting in the inactivation of T cells within the vascular lumen [13, 17, 18]. Consequently, targeting tumor vasculature can enhance the infiltration and activation of cytotoxic T cells [19, 20]. VEGFA is the critical driver of tumor angiogenesis and is the most extensively investigated target of tumor vasculature [21]. However, VEGFA exhibits higher expression in pT1 bladder cancers than muscle-invasive (≥pT2) bladder cancers [22-24]. This suggests tumors more likely to exhibit areas of hypoxia (e.g., Stage pT2-4) display reduced VEGFA expression [23]. This finding contradicts prevailing views, as hypoxia is always associated with more angiogenesis and higher VEGFA expression [25]. The clinical trials evaluating the efficacy of VEGFA inhibitors in treating bladder cancer have also shown modest benefit [26, 27]. Therefore, another pro-angiogenic mechanism may be activated in bladder cancer. A better understanding of the molecular network regulating angiogenesis in bladder cancer may pave the way for future integration of checkpoint immunotherapy with anti-angiogenic therapy in the treatment of this malignancy.
In this study, we analyzed bladder cancer single-cell transcriptome datasets from the GEO database. We found that PGF is an additional pro-angiogenic factor in bladder cancer, complementing the well-known role of VEGFA. To reconcile the seemingly contradictory functions of PGF in angiogenesis, our research unveiled that the expression level of VEGFR1 in endothelial cells is a pivotal determinant of PGF's pro-angiogenic activity. Additionally, the concurrent inhibition of PGF and VEGFA, when coupled with anti-PD-1 therapy, provides a survival benefit exceeding the beneficial effect of anti-PD-1 monotherapy. These findings highlight the potential of this combinatorial approach in overcoming the limitations of checkpoint immunotherapy in bladder cancer.
Materials and Methods
Single-cell transcriptome datasets analysis
The GSE135337 bladder cancer single-cell transcriptome datasets, which include NMIBC and muscle invasive bladder cancer (MIBC), were downloaded from the GEO database [28]. These datasets were analyzed using the Seurat package (R package, version 5.1.1). As these datasets were already normalized in the GEO database, the identification of highly variable genes and dataset integration were carried out without normalization. Principal component analysis (PCA) was performed using the RunPCA function. Significantly enriched principal components (PCs) were used for cell clustering and uniform manifold approximation and projection (UMAP) dimensional reduction. Cell clustering was performed by the FindClusters function. Clusters were annotated based on the expression of well-established marker genes for each cell type: urothelial cells (EPCAM, KRT8, and KRT18), myeloid/macrophage (CD14, CSF1R, and AIF1), T cells (CD2, CD3D, and CD3E), fibroblasts (DCN, PDPN, and TAGLN) and endothelial cells (PECAM1, VWF, and CLDN5), as reported previously [28]. These cells were clustered into 21 clusters; cells of clusters 0-10, 12, 14, 16, and 20 were defined as cells of urothelial origin (bladder cancer cells); clusters 11, 13, and 18 were defined as fibroblasts; cluster 17 as T cells, cluster 15 as myeloid/macrophage, and cluster 19 as endothelial cells (Figure S1A). To explore the interaction between endothelial cells and other cell types, the Python package CellPhoneDB (version 2.1.7) was used for ligand-receptor analysis with default parameters [29]. The endothelial cell subset data was then separated and an expression analysis of VEGFR1 and VEGFR2 in these endothelial cells was performed.
The GSE163558 gastric cancer (GC) single-cell transcriptome datasets were downloaded from the GEO database [30]. Three tumor samples were included in our study. These datasets were also analyzed by the Seurat package. Data normalization, identification of highly variable genes, and data scaling were performed by the SCTransform function. Datasets were integrated according to the integration workflow for SCTransform in Seurat. PCA, UMAP dimensional reduction, and cell clustering analyses were done as described for the bladder cancer datasets. Cell clusters were identified using known lineage markers, such as those for epithelial cells (EPCAM, KRT18, and CLDN4), endothelial cells (PECAM1, and VWF), proliferation (STMN1, and PCNA), T cells (CD3D, CD3E, CD2), B cells (CD79A, IGHG1, and MS4A1), for natural killer (KLRD1, GNLY, and KLRF1), and myeloid (CSF1R, CSF3R, CD68), according to published data [30]. Sixteen clusters were identified; cluster 8 was defined as endothelial cells (Figure S1B). The endothelial cell subset data was then separated, and an expression analysis of VEGFR1 and VEGFR2 was performed.
The GSE149614 hepatocellular carcinoma (HCC) single-cell transcriptome datasets were downloaded from the GEO database [31]. Because the datasets contain a normalized expression matrix and a cell annotation matrix, the expression profiles of endothelial cells were acquired directly from these matrices. Then, the expression levels of VEGFR1 and VEGFR2 in HCC endothelial cells were analyzed.
Immunohistochemistry
Bladder cancer tissue microarray slides were purchased from Shanghai Outdo Biotech Company (HBlaU060CS02). The tissue microarray slides were deparaffinized in xylene and then rehydrated in a series of 100%, 90%, and 70% ethanol. Antigen retrieval was performed by incubating the tissue sections with citric acid buffer (Solarbio, C1032) under a combination of high temperature and pressure within a pressure cooker for 5 minutes. Endogenous peroxidase was inactivated by incubating the sections with 3% hydrogen peroxide for 15 minutes. Then the sections were blocked for 30 min in 10% goat serum. After blocking, the sections were incubated with an anti-PGF antibody (Proteintech, 10642-1-AP, 1:200 dilution) at 4°C overnight. The next day, the tissue sections were washed with PBS three times for 5 minutes each, then incubated with an HRP-linked secondary antibody for 1 hour at room temperature, followed by incubation with Diaminobenzidine (DAB) and counterstaining with hematoxylin.
Immunofluorescence
The multiplex immunohistochemical kit (Panovue, 10001100020) was used for immunofluorescence labeling. The procedures for deparaffinization, hydration, and antigen retrieval were the same as those for immunohistochemistry described above. Antigen detection was carried out according to the instruction manual. Tissue samples were initially treated with a blocking solution for 30 minutes, followed by an overnight incubation with the primary antibody at 4°C. Subsequently, they were incubated with the secondary antibody for 1 hour and then stained with a chromogenic agent for 10 minutes. This process was repeated for a second round of antigen retrieval, antibody incubation, and staining. Lastly, the samples were counterstained with DAPI for nuclear visualization.
Cell lines and culture conditions
The MBT2 (RRID: CVCL_4660) mouse bladder cancer cell line was purchased from OriCell, and the human umbilical vein endothelial cells (HUVECs, (RRID: CVCL_2959)) was purchased from Biospecies. All cell lines have been authenticated using STR profiling and confirmed without mycoplasma pollution. MBT2 cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. HUVECs cell lines were cultured in complete endothelial cell medium (elGbio, EP1001). All cell lines were maintained at 37°C under 5% CO2.
Generation of stable cell lines
The coding sequence of VEGFR1 was cloned into the pCDH vector to generate the VEGFR1-overexpression plasmid. Pgf- and Vegfa-specific shRNA oligonucleotides were inserted into the pLKO.1 to generate Pgf- and Vegfa- knockdown plasmids. Then, these plasmids were transfected into the human embryonic kidney-293 (HEK293) cells using polyethyleneimine (PEI) to produce lentiviruses. The lentiviral supernatant was harvested at 48 hours. HUVECs was infected with a VEGFR1 lentivirus using polybrene (8μg/ml) to generate a VEGFR1-overexpressing cell line. MBT2 was infected with Pgf shRNA or/and Vegfa shRNA lentiviruses, resulting in the development of MBT2 sublines with diminished expression of Pgf, Vegfa, or both. To simultaneously knockdown Pgf and Vegfa in MBT2, the pLKO vectors for Pgf shRNA and Vegfa shRNA were designed to confer distinct antibiotic resistance. Following virus infection, positive cells were selected by culturing in media containing specific antibiotics. Blasticidin was used for VEGFR1-overexpressing cells (6μg/ml, 7days), puromycin for Pgf-knockdown cells (8μg/ml, 2days), hygromycin for Vegfa-knockdown cells (800μg/ml, 4days). The knockdown of Pgf and Vegfa, as well as the overexpression of VEGFR1, were validated by western blot analysis probing these proteins. The sequences of primes are provided in supplementary materials (Table S1,2).
Tumor model and treatment regimens
Cells of the MBT2 sublines, suspended in DMEM devoid of FBS and antibiotics, were mixed with Matrigel (Corning, 354262) at a ratio of 100: 15, then orthotopically implanted into the bladder wall of C57BL/6 mice (2.5x105 cells/mouse). The C57BL/6 mice purchased from Gempharmatech were administered intraperitoneal injections of phosphate-buffered saline (PBS) and the anti-mouse PD-1 antibody (200μg/mouse, SELLECK, A2122).
Western blot
Western blotting was performed according to standard procedures. Cells were washed with ice-cold PBS and then lysed using 1×SDS lysis buffer. The samples were boiled, resolved in 12% Tris-glycine gels, and transferred to a polyvinylidene difluoride membrane using wet transfer. The polyvinylidene difluoride membranes were blocked with 5% milk in Tris-buffered saline with 0.02% Tween20 (TBS-T) for 1 hour. They were then probed with primary antibodies overnight at 4°C, followed by incubation with a secondary antibody conjugated to horseradish peroxidase for 1-2 hours at room temperature. The secondary antibody was diluted in 2.5% milk in TBS-T. The antibodies used for western blot were as follows: anti-PGF (1:500, Proteintech, 10642-1-AP), anti-VEGFA (1:500, Proteintech, 19003-1-AP), anti-VEGFR1 (1:1000, HuaBio, ET1605-11), Anti-phosphorylated ERK1/2 (1:500, Servicebio, GB11004), and anti-ERK1/2 (1:1000, Servicebio, GB11560). The specific proteins were detected using a ChemiDocTMMP Imaging System (Biorad) after incubation with the western HRP substrate (Beyotime, P0018S).
Proliferation assay
HUVECs cell lines were seeded into a 96-well plate (2000 cells/well) and cultured with complete endothelial cell medium. To explore the effect of PGF on endothelial cell proliferation, the complete endothelial cell medium was replaced with either a standard endothelial cell medium depleted of growth factors or a medium containing 100 ng/ml of PGF (MCE, HY-P70749). Cell numbers were determined using the CCK8 assay (Abbkine, BMU106-CN) according to the standard procedure. In brief, 10 μl CCK8 solution was added to each well. Then, the cells were incubated at 37°C for 2 hours. The absorbance was detected using the iMarkTM Microplate Reader (Biorad).
Tube formation assay
To assess the effects of PGF on the tube formation ability of HUVECs cell lines, 96-well angiogenesis plates (Ibidi, 89646) were coated with 10μl Matrigel (Corning, 356231). Then, the plates were incubated at 37°C for at least 30 minutes to allow matrigel polymerization. The HUVECs cell lines cultured in distinct conditions described above for 3 days were seeded into the plates (8000 cells/well) and incubated at 37°C under 5% CO2. 8 hours later, these HUVECs cell lines were imaged using an Incucyte S3 system (ESSEN Bioscience).
Bulk RNA seq and analysis
Total RNA was extracted using the TRIZOL reagent according to the standard procedure. The mRNA was purified using poly-T oligo-coupled magnetic beads, fragmented, and cDNA was synthesized and converted into a library for sequencing. Libraries were sequenced on an Illumina Novaseq platform, and 150bp paired-end reads were generated.
Raw data of fastq format was processed. In this step, clean data was obtained by removing reads containing adapter, poly-N, and low-quality reads from raw data. At the same time, Q20, Q30, and GC content of the clean data were calculated. The paired-end reads were aligned to the reference genome using Hisat2 v2.0.5. FeatureCounts v1.5.0-p3 was used to count the reads numbers mapped to each gene. Then, the FPKM of each gene was calculated based on the length of the gene and the read counts mapped to this gene. Differential expression analysis of two conditions was performed using the edgeR R package (3.22.5). Gene Ontology (GO) enrichment analysis of differentially expressed genes was performed using the DAVID website (https://david.ncifcrf.gov/tools.jsp). GO terms with p-values lower than 0.05 were considered significantly enriched. Gene Set Enrichment Analysis (GSEA) was performed using GSEA_4.3.2 software.
Flow cytometry analysis
Mouse bladder tumors were cut into small pieces and incubated in a digestion buffer comprising 0.1% collagenase I, 0.25% collagenase IV, and 1mg/ml hyaluronidase at 37°C, 200rpm for 2 hours to obtain single-cell suspensions. The cell suspensions were incubated in red cell lysis buffer for 10 minutes to remove red blood cells and then they were filtered through a 40 μm cell strainer. The dissociated cells were stained with the following primary antibodies: anti-CD45 (Biolegend, 157606), anti-CD3 (Biolegend, 100204), anti-CD8a (Biolegend, 100708), anti-CD4 (Biolegend, 100407), and anti-CD25 (Biolegend, 101915). The cells were further permeabilized using True-Nuclear Transcription Factor Buffer set (Biolegend, 424401) and stained with anti-FOXP3 (Biolegend, 126419). Flow cytometry was performed using a BD LSRFortessaTM X-20 flow cytometer (BD Biosciences), and the data were analyzed using FlowJo software version 10.
Statistical analysis
All statistical analyses were performed using the GraphPad Prism 8.0 software. Differential expressions of FLT1 and KDR were assessed using a paired two-tailed T test. Differences in survival rates were calculated using the Log-rank (Mantel-Cox) test. Differences among multiple groups were initially assessed using one-way ANOVA, followed by comparisons between each pair of groups using Tukey's multiple comparisons test. Differences in all other categories were analyzed using an unpaired two-tailed T-test. Values were presented as mean±SD. Statistical significance was set at P<0.05.
Results
FLT1-PGF interaction between endothelial cells and carcinoma cells is highly expressed in both NMIBC and MIBC
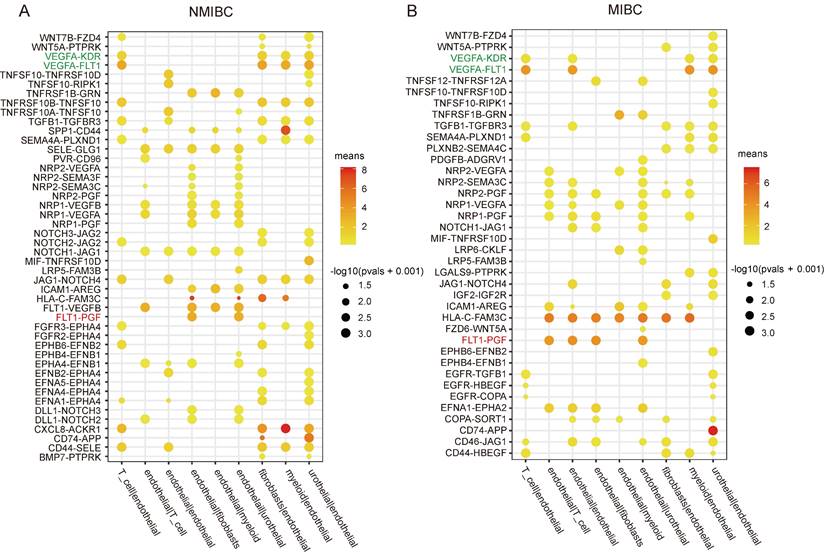
To elucidate the angiogenic regulatory mechanisms operating in bladder cancer, we analyzed single-cell transcriptome datasets of bladder cancer obtained from the GEO database. The CellphoneDB package was used to explore the ligand-receptor interactions between endothelial cells and other cell populations, based on expression levels of ligands and receptors within distinct cell clusters. Interestingly, the pro-angiogenic ligand-receptor interactions of FLT1(VEGFR1)-PGF, FLT1(VEGFR1)-VEGFB, ACKR1-CXCL8, and FLT1(VEGFR1)-VEGFA between endothelial cells and urothelial-derived carcinoma cells were enriched and displayed higher expression levels compared to other interactions in NMIBC (Figure 1A); however, only FLT1(VEGFR1)-PGF and FLT1(VEGFR1)-VEGFA exhibited higher expression in MIBC (Figure 1B). PGF belongs to the VEGF family, which also includes VEGFA, B, C and D [32]. Therefore, this finding suggests that PGF may play an important role in angiogenesis in bladder cancer, in addition to VEGFA.
VEGFA is known to bind to FLT1 (VEGFR1) and KDR (VEGFR2). However, VEGFA-FLT1 interaction surprisingly exceeded VEGFA-KDR interaction in both NMIBC and MIBC, even though KDR is recognized as the primary receptor for VEGFA-induced angiogenesis [33]. As the results derived from CellphoneDB were based on the expression levels of ligands and receptors in distinct cell types, this phenomenon could be attributed to the expression levels of FLT1 and KDR in the endothelial cells of bladder cancers (Fig. 1A.B).
PGF promotes angiogenesis in bladder cancer
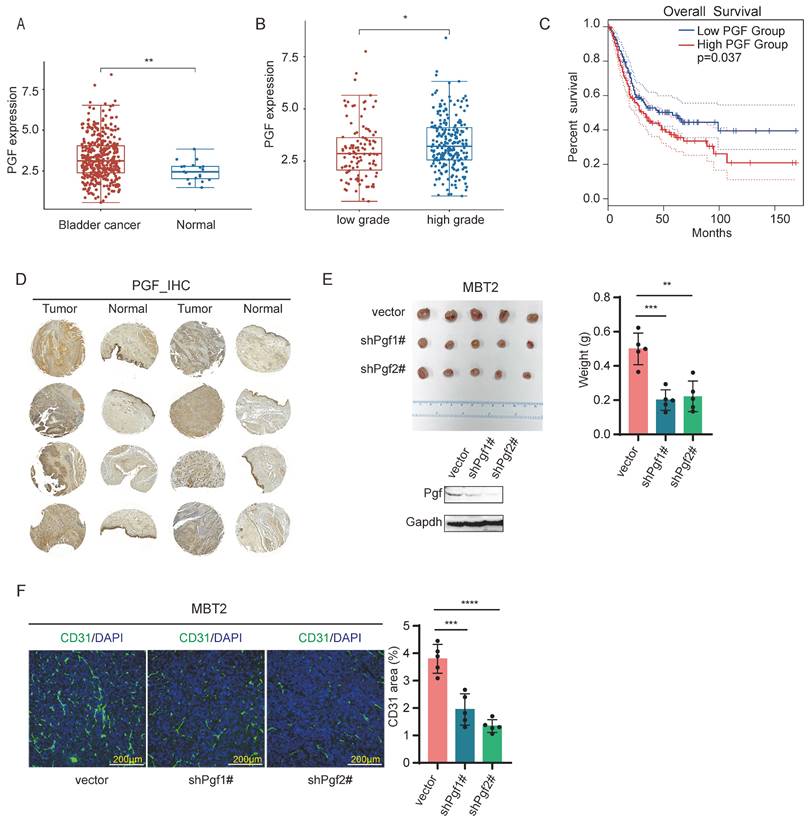
To explore whether PGF serves as a pro-angiogenic factor in bladder cancer, we analyzed its expression levels and assessed its correlation with patient survival rate through Gene Expression Profiling Interactive Analysis 2 (GEPIA2) web platform (http://gepia2.cancer-pku.cn/#index) [34] and Assistant of Clinical Bioinformatics web platform (https://www.aclbi.com/static/index.html#/). These analyses were conducted using bladder cancer clinical datasets from The Cancer Genome Atlas Program (TCGA) database. The expression levels of PGF are elevated in tumor samples relative to normal tissue counterparts (Figure 2A), and in bladder cancer, high-grade tumors (>pT2) present higher PGF expression compared to low-grade tumors (<=pT2) (Figure 2B). A negative correlation between PGF expression levels and overall survival was observed in bladder cancer patients (Figure 2C). These findings underscore the significance of PGF in the context of bladder cancer pathology. To further validate this, immunohistochemical staining of PGF expression was conducted using a tissue microarray of clinical bladder cancer specimens. The expression levels of PGF were elevated in tumor samples when compared to matched normal bladder tissues (Figure 2D). Additionally, in vivo studies using a mouse model orthotopically transplanted with MBT2 demonstrated that knockdown of Pgf led to a reduction in tumor size (Figure 2E). Histological analysis of bladder tissues from these mice, using immunofluorescence labeling of CD31, revealed a decrease in vessel density following the silencing of Pgf (Figure 2F).
The pro-angiogenic activity of PGF depends on the expression levels of VEGFR1 in endothelial cells
The role of PGF in angiogenesis remains unclear. Previous studies have shown that PGF can positively or negatively modulate angiogenesis. The overexpression of PGF has been reported to lead to the inhibition of angiogenesis and the normalization of blood vessels, which in turn suppresses tumor growth [35]. However, other studies have found that PGF contributes to pathological angiogenesis with little impact on normal vasculature [36, 37]. PGF exclusively activates FLT1, also known as VEGFR1. VEGFR1 is a receptor tyrosine kinase (RTK) exhibiting lower kinase activity than VEGFR2 [38]. It was thought to act as an angiogenic antagonist by competing with VEGFR2 for VEGFA binding [39, 40]. However, VEGFA, a well-established pro-angiogenic factor, can activate both FLT1 and KDR, also known as VEGFR2. VEGFR2 is considered the predominant receptor mediating the pro-angiogenic activity of VEGFA [41]. The expression level of VEGFR2 is usually higher than that of VEGFR1 in normal blood vessels [42]. Surprisingly, the expression level of VEGFA-FLT1(VEGFR1) interaction between bladder cancer cells and endothelial cells is higher than that of VEGFA-KDR(VEGFR2) interaction (Figure 1). As the results derived from CellphoneDB are based on the expression profiles of ligands and receptors [29], the expression level of FLT1 in the endothelium of bladder cancer might be higher than that of KDR. The higher expression of FLT1 might contribute to the pro-angiogenic activity of PGF in bladder cancer. This could explain why PGF does not affect normal vessels or even inhibits angiogenesis in some studies but activates angiogenesis in bladder cancer.
Cell-cell interaction analysis of NMIBC and MIBC. (A, B) The ligand-receptor interactions between endothelial cells and other cell populations are significantly enriched (p-value < 0.05) in NMIBC and MIBC. The intensity of dot colors corresponds to the expression level, while the size of the dots reflects the statistical significance (p-value).
To test this hypothesis, an expression analysis of FLT1 and KDR in endothelial cells was performed using single-cell transcriptome datasets. Notably, the expression level of FLT1 was significantly higher than that of KDR in both NMIBC and MIBC (Figure 3A). This finding was further validated via immunofluorescence double staining of CD31 with FLT1 or KDR in clinical bladder cancer tissues. The results confirmed a greater degree of co-localization between CD31 and FLT1 compared to that between CD31 and KDR (Figure 3B). The high expression of FLT1 coupled with low expression of KDR in endothelial cells is also observed in gastric cancer (GC) and hepatocellular carcinoma (HCC), where the expression level of PGF inversely correlates with patient overall survival rates (Figure 3C, D). To further evaluate the role of VEGFR1, it was genetically overexpressed in HUVECs (Figure 3E). PGF did not activate proliferation and tubule formation in wild-type HUVECs, but significantly stimulated these processes in HUVECs overexpressing VEGFR1 (Figure 3F, G, H). These results support the contention that the expression level of VEGFR1 plays a crucial role in mediating the pro-angiogenic activity of PGF in bladder cancer.
PGF is a pro-angiogenic factor in bladder cancer. (A) Statistical analysis of mRNA levels of PGF in bladder cancer patients using TCGA datasets (Wilcox test). (B) Comparison of mRNA levels of PGF between high-grade and low-grade bladder cancer using TCGA datasets (Wilcox test). (C) Survival analysis of bladder cancer patients stratified by PGF expression levels using data from TCGA. (D) Immunohistochemical analysis of PGF expression in a clinically representative tissue microarray of bladder cancer samples. (E) Tumor size analysis of MBT2 with targeted knockdown of Pgf. (F) Immunofluorescence analysis of the vessel density (CD31). The histogram represents quantitative statistics of the fluorescent area. * p< 0.05, ** p <0.01, *** p <0.001, **** p <0.0001.
ERK1/2 are robustly activated upon PGF engagement with VEGFR1
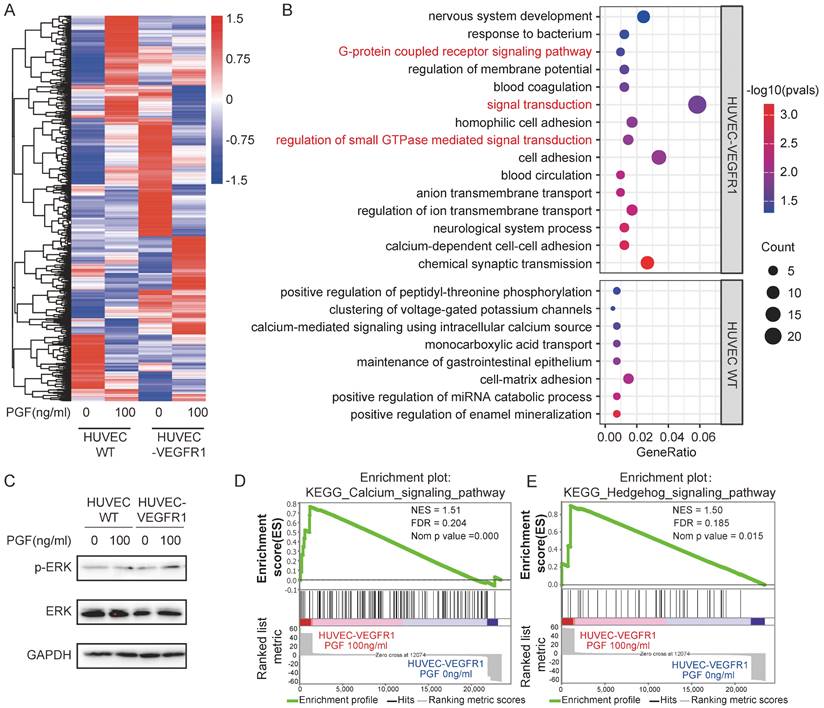
To delineate the impact of PGF stimulation on endothelial cells, an RNA-seq analysis was carried out to map the comprehensive changes in their transcriptomic landscape. The differential gene expression profiles following PGF stimulation diverged markedly between VEGFR1-overexpressing HUVECs and its wild-type counterpart (Figure 4A). According to a Gene Ontology (GO) enrichment analysis focusing on the differently expressed genes in response to PGF stimulation in VEGFR1-overexpressing HUVECs and wild-type HUVECs, a notable enrichment of several signaling transduction pathways was exclusively found in the VEGFR1-overexpressing HUVECs. These pathways included the regulation of small GTPase-mediated signal transduction, signal transduction, and the G-protein coupled receptor signaling pathway, all of which are related to vascular endothelial growth factor receptor (VEGFR) activation (Figure 4B) [43-45]. Therefore, the high expression of VEGFR1 is essential for efficient PGF signaling, consistent with previous findings.
The pro-angiogenic activity of PGF is dependent on the expression level of VEGFR1 in endothelial cells. (A) The expression levels of FLT1 (VEGFR1) and KDR (VEGFR2) in endothelial cells of NMIBC and MIBC. (B) Immunofluorescence analysis of CD31, VEGFR1, and VGFR2. (C, D) The expression of FLT1 and KDR in GC and HCC in which high expression of PGF is associated with poor survival rate. (E) VEGFR1 overexpression validation in HUVECs. (F) Proliferation curves of VEGFR1-overexpressing HUVECs (HUVEC-VEGFR1) and wild-type HUVECs (HUVEC WT) incubated with PGF protein or not. (G, H) Images of distinct tube formation patterns and quantitative analysis using ImageJ software revealed significant differences in loop numbers across three independent replicates. ns means no significant difference. ** p < 0.01, **** p < 0.0001.

Signal transduction pathways mediate the pro-angiogenic activity of PGF. (A) Heatmap displaying the expression pattern of differently expressed genes in response to PGF in HUVEC WT and HUVEC-VEGFR1. (B) GO analysis focusing on differently expressed genes in response to PGF in HUVEC-WT and HUVEC-VEGFR1. (C) Western blot probing phosphorylated ERK1/2. (D.E) GSEA plot for “KEGG calcium signaling pathway” and “KEGG hedgehog signaling pathway” in HUVEC-VEGR1 cells.
ERK1/2 activation is an essential downstream event in the angiogenic cascade orchestrated by VEGFR2 [46, 47]. ERK1/2 has also been identified as a downstream target of VEGFR1 signaling in monocytes and human retinal endothelial cells [48, 49]. We found that in VEGFR1-overexpressing HUVECs, PGF treatment led to the activation of ERK1/2, whereas this effect was absent in wild-type HUVECs (Figure 4C). This observation implies that ERK1/2 may act as a key downstream effector of VEGFR1 in mediating the pro-angiogenic effects of PGF, bridging receptor activation to the subsequent pro-angiogenic signaling. Additionally, GSEA analysis revealed that the calcium and hedgehog pathways were enriched in VEGFR1-overexpressing HUVECs treated with PGF (Figure 4D, E). Both pathways are associated with the process of angiogenesis [50-52].
Co-inhibition of PGF and VEGFA enhances the effectiveness of anti-PD-1 antibodies
According to the results described above, PGF is shown to be another pro-angiogenic factor in bladder cancer. This discovery offers insight into the downregulation of VEGFA observed during disease progression, suggesting that PGF may compensate for the reduced VEGFA activity. Therefore, co-inhibition of PGF and VEGFA could potentially achieve synergistic anti-angiogenic effects. To validate this hypothesis, both Vegfa and Pgf were knocked down in MBT2 (Figure 5B). Co-inhibition of Vegfa and Pgf resulted in the most pronounced reduction in tumor size and the lowest density of CD31- positive blood vessels (Figure 5A, C).
The combined inhibition of PGF and VEGFA significantly augments the survival outcome when administered alongside anti-PD1 antibody therapy. (A) Tumor size analysis of MBT2 with targeted knockdown of Pgf and/or Vegfa orthotopically implanted into the mouse bladder. (B) Western blot probing the PGF and VEGFA. (C) Immunofluorescence analysis of vessel density (CD31) and infiltration of CD8+ T cell (CD8). Histograms represent quantitative analysis of fluorescent areas. (D, E) Flow cytometry analysis of the CD8+ T cell and Treg within the microenvironment of MBT2 xenografts. The histogram depicts the quantitative analysis of positive cells. (F) Calculation of the CD8+ T cell to Treg cell ratio based on the above flow cytometry data. (G, H) Survival analysis evaluating the therapeutic efficacy of anti-PD1 antibody treatment in mice bearing MBT2 tumors with targeted knockdown of Pgf and/or Vegfa, and the treatment schedule. * p<0.05, ** p<0.01, *** p <0.001, **** p<0.0001, ns means no significant difference.
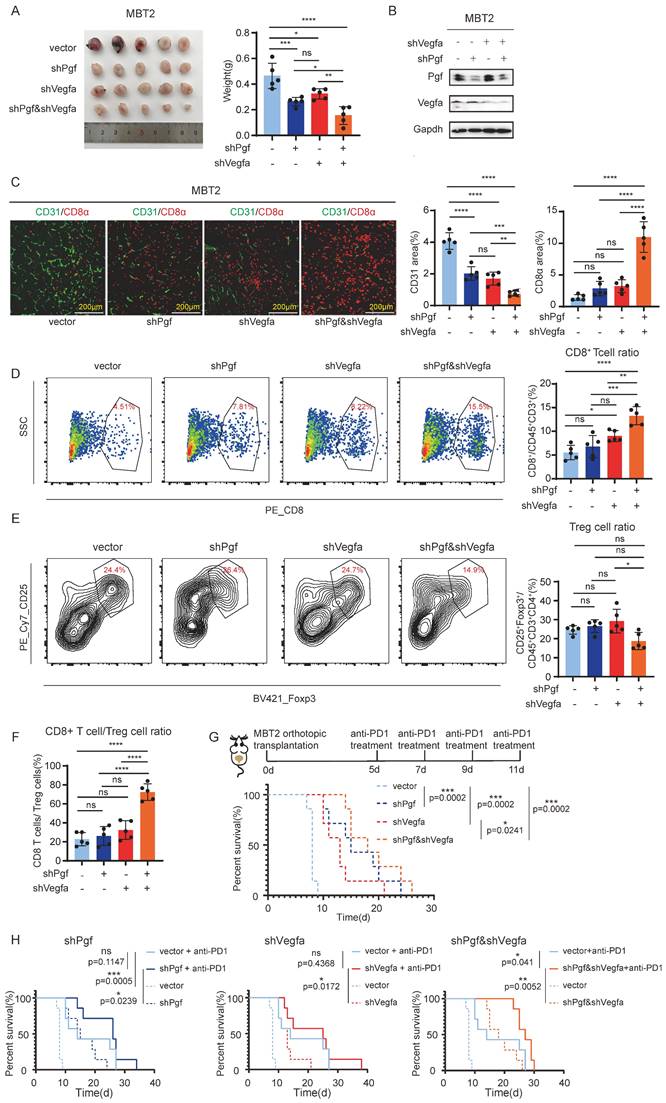
As reported in previous studies, anti-angiogenic therapies increased the infiltration of CD8+ cytotoxic T cells into tumors, potentiating the effectiveness of immunotherapy against cancer [19]. In line with these findings, we investigated the impact of concurrently targeting Vegfa and Pgf on the infiltration of immune cells within the tumor microenvironment. We found that only the co-inhibition of Vegfa and Pgf markedly increased the infiltration of CD8+ cytotoxic T cells (Figure 5C, D). Even though the infiltration of regulatory T cells (Tregs) was not significantly affected by the co-inhibition of Vegfa and Pgf (Figure 5E), the ratio of CD8+ T cell to Treg cell within the tumor was significantly elevated when Vegfa and Pgf were concurrently knocked down (Fig.5F). The individual blockade of Vegfa resulted in minimal change in CD8+ T cell infiltration, but did not affect the CD8+ T cell to Treg cell ratio within the tumor (Figure 5 D, F). Therefore, only concurrent inhibition of both Vegfa and Pgf induces significant alterations in the composition of immune cells within the tumor microenvironment. Thus, the combinatorial effect of anti-PD-1 antibody treatment and pro-angiogenic molecule silencing was evaluated. Our findings indicate that the dual inhibition of Vegfa and Pgf, as well as their individual blockades, significantly enhanced the survival rate of mice with bladder cancer. Notably, the co-inhibition of Vegfa and Pgf yielded a superior survival benefit when compared to the single inhibition of Vegfa alone. However, there was no significant difference in survival rates between the single inhibition of Pgf and the combined inhibition of Vegfa and Pgf (p-value is not shown) (Figure 5G). Only the co-inhibition of Vegfa and Pgf significantly enhances the survival benefit associated with anti-PD1 antibody therapy in the context of bladder cancer management (Figure 5H).
Discussion
Checkpoint immunotherapy has contributed to the treatment of bladder cancer, but its effectiveness still needs to be further improved. In recent years, there has been a resurgence of interest in anti-angiogenic therapy, recognizing its potential to augment the effectiveness of checkpoint immunotherapy. Combining immune checkpoint inhibitors with anti-angiogenic therapies is likely an approach to enhance the efficacy of checkpoint immunotherapy in bladder cancer treatment. To date, clinical trials using angiogenic inhibitors in the treatment of bladder cancer have not been successful. To our knowledge, almost all anti-angiogenic therapies target the VEGFA-VEGFR2 signaling pathway. However, VEGFA expression is reduced during the progression of bladder cancer, altering the use of anti-angiogenic agents that target VEGFA/VEGFR2 signaling. These findings highlight the need for further investigations into the mechanism of angiogenic regulation in bladder cancer.
We found that PGF promotes angiogenesis in bladder cancer in addition to VEGFA, and the combined inhibition of PGF and VEGFA yields synergistic effects in tumor growth inhibition and anti-tumor immunity. Anti-angiogenic therapies have been underutilized in the broader spectrum of cancer treatment due to their historically limited effectiveness. Our findings suggest that the ineffectiveness of current anti-angiogenic therapies, which mainly focus on inhibiting the VEGFA/VEGFR2 signaling axis, might result from an incomplete angiogenesis blockade. This is because the impact of VEGFA/VEGFR2 pathway inhibitors can be counteracted by activating alternative pro-angiogenic pathways.
The expression level of VEGR1 is higher than that of VEGFR2 in bladder cancer, contributing to the pro-angiogenic activity of PGF in this disease. The mechanism underlying the expression profiles of VEGFR1 and VEGFR2 is unknown. We have demonstrated that this expression pattern is not unique to bladder cancer. It is hypothesized that these distinctive expression profiles could indicate the unique characteristics of tumor blood endothelial cells, which are crucial to the formation of immature and permeable tumor vasculature. Furthermore, the hypoxic conditions prevalent within the tumor microenvironment, which are known to foster angiogenesis, may be responsible for the high VEGFR1 and low VEGFR2 expression levels. It is also plausible that these expression patterns are organ-specific, reflecting the unique physiological demands of different tissues. Exploring these hypotheses is crucial for understanding whether similar synergistic effects from the co-inhibition of PGF and VEGFA occur in other types of cancer, beyond bladder cancer, and warrants further investigation.
Unlike VEGFA, there is a lack of clinically available anti-PGF antibodies for in vivo inhibition. The effectiveness of antibody-mediated PGF inhibition relative to its genetic ablation remains unknown. Notably, PB101, a glycosylated soluble decoy receptor fusion protein, was developed to antagonize both VEGFA and PGF. PB101 demonstrated robust decoy activity against these key angiogenic factors [53]. In the tumor model, PB101 not only efficiently inhibited the tumor angiogenesis and progression [53, 54], but also enhanced antitumor immunity [55], consistent with our findings. Future works should focus on developing drugs specifically inhibiting PGF signaling or, ideally, PGF and VEGFA, and on exploring the synergistic effects of combining these drugs with immunotherapy for more effective clinical treatments of bladder cancer.
Abbreviations
FDA: the U.S. Food and drug administration; GC: gastric cancer; HCC: hepatocellular carcinoma; HUVECs: human umbilical vein endothelial cells; ICIs: immune checkpoint inhibitors; MIBC: muscle invasive bladder cancer; NCCN: the National Comprehensive Cancer Network; NMIBC: non-muscle invasive bladder cancer; PD-1: programmed cell death protein 1; PD-L1: programmed cell death 1 protein ligand 1; PGF: placental growth factor; RTK: receptor tyrosine kinase; Tregs: regulatory T cells; VEGFA: vascular endothelial growth factor A; VEGFR1: vascular endothelial growth factor receptor1; VEGFR2: vascular endothelial growth factor receptor2.
Supplementary Material
Supplementary figure and tables.
Acknowledgements
We thank Mr. Dewang Zhou for the technical assistance with bladder orthotopic transplantation. We acknowledge Guangzhou Laboratory for financial support.
Funding
This project was supported by the Shenzhen Science and Technology Innovation Commission (grant number RCJC20200714114557005), the Sanming Project of Medicine in Shenzhen (SZSM202211009), the National Natural Science Foundation of China (61931024, 92359202, 82072818).
Ethics statement
All animal procedures were approved by the Shenzhen Zhongxun Precision Medical Research Institute Experimental Animal Ethics Committee of the Third Affiliated Hospital of Shenzhen University. (approval number: ZXJZ20230501004).
Data availability statement
All data supporting the finding of this study are available within the paper and its supplementary material. Further information is available from the corresponding authors upon request.
Author contributions
Xianzhi Yang: Writing-original draft, conceptualization, formal analysis, investigation, visualization. Haoxiang Zheng: Writing-original draft, conceptualization, formal analysis, investigation, visualization. Jianxu Huang: Investigation, data curation. Yujun Liu: Investigation, data curation. Yingrui Li: Investigation, data curation, funding acquisition. Bingwen Zhang: Investigation, data curation. Chu Sun: Investigation, data curation. Yuqing Li: Writing-editing and review, conceptualization. Jean Paul Thiery: Writing-editing and review, conceptualization. Song Wu: Writing-editing and review, conceptualization, funding acquisition.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Massari F, Di Nunno V, Cubelli M. et al. Immune checkpoint inhibitors for metastatic bladder cancer. Cancer Treat Rev. 2018;64:11-20
2. Audisio A, Buttigliero C, Delcuratolo MD. et al. New perspectives in the medical treatment of non-muscle-invasive bladder cancer: immune checkpoint inhibitors and beyond. Cells. 2022;11:357
3. Lopez-Beltran A, Cookson MS, Guercio BJ. et al. Advances in diagnosis and treatment of bladder cancer. Bmj. 2024;384:e76743
4. Dyrskjøt L, Hansel DE, Efstathiou JA. et al. Bladder cancer. Nat Rev Dis Primers. 2023;9:58
5. Tran L, Xiao J, Agarwal N. et al. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104-21
6. Babjuk M, Burger M, Capoun O. et al. European association of urology guidelines on non-muscle-invasive bladder cancer (ta, t1, and carcinoma in situ). Eur Urol. 2022;81:75-94
7. Alfred Witjes J, Max Bruins H, Carrión A. et al. European association of urology guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2023 guidelines. Eur Urol. 2024;85:17-31
8. Bidnur S, Savdie R, Black PC. Inhibiting immune checkpoints for the treatment of bladder cancer. Bladder Cancer. 2016;2:15-25
9. Roviello G, Catalano M, Santi R. et al. Immune checkpoint inhibitors in urothelial bladder cancer: state of the art and future perspectives. Cancers (Basel). 2021;13:4411
10. Robertson AG, Meghani K, Cooley LF. et al. Expression-based subtypes define pathologic response to neoadjuvant immune-checkpoint inhibitors in muscle-invasive bladder cancer. Nat Commun. 2023;14:2126
11. Wu M, Huang Q, Xie Y. et al. Improvement of the anticancer efficacy of pd-1/pd-l1 blockade via combination therapy and pd-l1 regulation. J Hematol Oncol. 2022;15:24
12. Flaig TW, Spiess PE, Abern M. et al. Bladder cancer, version 3.2024. J Natl Compr Canc Netw. 2024;22:216-25
13. Lee WS, Yang H, Chon HJ. et al. Combination of anti-angiogenic therapy and immune checkpoint blockade normalizes vascular-immune crosstalk to potentiate cancer immunity. Exp Mol Med. 2020;52:1475-85
14. Brest P, Mograbi B, Pagès G. et al. Checkpoint inhibitors and anti-angiogenic agents: a winning combination. Br J Cancer. 2023;129:1367-72
15. Socinski MA, Jotte RM, Cappuzzo F. et al. Atezolizumab for first-line treatment of metastatic nonsquamous nsclc. N Engl J Med. 2018;378:2288-301
16. Popat S, Grohé C, Corral J. et al. Anti-angiogenic agents in the age of resistance to immune checkpoint inhibitors: do they have a role in non-oncogene-addicted non-small cell lung cancer? Lung Cancer. 2020;144:76-84
17. Huinen ZR, Huijbers EJM, van Beijnum JR. et al. Anti-angiogenic agents — overcoming tumour endothelial cell anergy and improving immunotherapy outcomes. Nat Rev Clin Oncol. 2021;18:527-40
18. Khan KA, Kerbel RS. Improving immunotherapy outcomes with anti-angiogenic treatments and vice versa. Nat Rev Clin Oncol. 2018;15:310-24
19. Shrimali RK, Yu Z, Theoret MR. et al. Antiangiogenic agents can increase lymphocyte infiltration into tumor and enhance the effectiveness of adoptive immunotherapy of cancer. Cancer Res. 2010;70:6171-80
20. Wallin JJ, Bendell JC, Funke R. et al. Atezolizumab in combination with bevacizumab enhances antigen-specific t-cell migration in metastatic renal cell carcinoma. Nat Commun. 2016;7:12624
21. Pérez-Gutiérrez L, Ferrara N. Biology and therapeutic targeting of vascular endothelial growth factor a. Nat Rev Mol Cell Biol. 2023;24:816-34
22. O'Brien T, Cranston D, Fuggle S. et al. Different angiogenic pathways characterize superficial and invasive bladder cancer. Cancer Res. 1995;55:510-3
23. Crew JP. Vascular endothelial growth factor: an important angiogenic mediator in bladder cancer. Eur Urol. 1999;35:2-8
24. Pignot G, Bieche I, Vacher S. et al. Large-scale real-time reverse transcription-pcr approach of angiogenic pathways in human transitional cell carcinoma of the bladder: identification of vegfa as a major independent prognostic marker. Eur Urol. 2009;56:678-89
25. de Heer EC, Jalving M, Harris AL. Hifs, angiogenesis, and metabolism: elusive enemies in breast cancer. J Clin Invest. 2020;130:5074-87
26. Pili R, Qin R, Flynn PJ. et al. A phase ii safety and efficacy study of the vascular endothelial growth factor receptor tyrosine kinase inhibitor pazopanib in patients with metastatic urothelial cancer. Clin Genitourin Cancer. 2013;11:477-83
27. Ghafouri S, Burkenroad A, Pantuck M. et al. Vegf inhibition in urothelial cancer: the past, present and future. World J Urol. 2021;39:741-9
28. Lai H, Cheng X, Liu Q. et al. Single-cell rna sequencing reveals the epithelial cell heterogeneity and invasive subpopulation in human bladder cancer. Int J Cancer. 2021;149:2099-115
29. Efremova M, Vento-Tormo M, Teichmann SA. et al. Cellphonedb: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484-506
30. Jiang H, Yu D, Yang P. et al. Revealing the transcriptional heterogeneity of organ-specific metastasis in human gastric cancer using single-cell rna sequencing. Clin Transl Med. 2022;12:e730
31. Lu Y, Yang A, Quan C. et al. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat Commun. 2022;13:4594
32. Dakowicz D, Zajkowska M, Mroczko B. Relationship between vegf family members, their receptors and cell death in the neoplastic transformation of colorectal cancer. Int J Mol Sci. 2022;23:3375
33. Peach CJ, Mignone VW, Arruda MA. et al. Molecular pharmacology of vegf-a isoforms: binding and signalling at vegfr2. Int J Mol Sci. 2018;19:1264
34. Tang Z, Kang B, Li C. et al. Gepia2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-60
35. Hedlund E, Hosaka K, Zhong Z. et al. Malignant cell-derived plgf promotes normalization and remodeling of the tumor vasculature. Proceedings of the National Academy of Sciences. 2009;106:17505-10
36. Ceci C, Atzori MG, Lacal PM. et al. Role of vegfs/vegfr-1 signaling and its inhibition in modulating tumor invasion: experimental evidence in different metastatic cancer models. International Journal of Molecular Sciences. 2020;21:1388
37. Fischer C, Jonckx B, Mazzone M. et al. Anti-plgf inhibits growth of vegf(r)-inhibitor-resistant tumors without affecting healthy vessels. Cell. 2007;131:463-75
38. Sawano A, Iwai S, Sakurai Y. et al. Flt-1, vascular endothelial growth factor receptor 1, is a novel cell surface marker for the lineage of monocyte-macrophages in humans. Blood. 2001;97:785-91
39. Kendall RL, Wang G, Thomas KA. Identification of a natural soluble form of the vascular endothelial growth factor receptor, flt-1, and its heterodimerization with kdr. Biochem Biophys Res Commun. 1996;226:324-8
40. Carmeliet P, Moons L, Luttun A. et al. Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat Med. 2001;7:575-83
41. Lacal PM, Graziani G. Therapeutic implication of vascular endothelial growth factor receptor-1 (vegfr-1) targeting in cancer cells and tumor microenvironment by competitive and non-competitive inhibitors. Pharmacol Res. 2018;136:97-107
42. Zhang Z, Neiva KG, Lingen MW. et al. Vegf-dependent tumor angiogenesis requires inverse and reciprocal regulation of vegfr1 and vegfr2. Cell Death Differ. 2010;17:499-512
43. Bruns AF, Bao L, Walker JH. et al. Vegf-a-stimulated signalling in endothelial cells via a dual receptor tyrosine kinase system is dependent on co-ordinated trafficking and proteolysis. Biochem Soc Trans. 2009;37:1193-7
44. Shimizu A, Zankov DP, Kurokawa-Seo M. et al. Vascular endothelial growth factor-a exerts diverse cellular effects via small g proteins, rho and rap. Int J Mol Sci. 2018;19:1203
45. Farzaneh BM, Zahri S, Gholami SZ. et al. Targeting signaling pathways of vegfr1 and vegfr2 as a potential target in the treatment of breast cancer. Mol Biol Rep. 2020;47:2061-71
46. Koch S, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect Med. 2012;2:a6502
47. Bazzazi H, Isenberg JS, Popel AS. Inhibition of vegfr2 activation and its downstream signaling to erk1/2 and calcium by thrombospondin-1 (tsp1): in silico investigation. Front Physiol. 2017;8:48
48. Tchaikovski V, Fellbrich G, Waltenberger J. The molecular basis of vegfr-1 signal transduction pathways in primary human monocytes. Arterioscler Thromb Vasc Biol. 2008;28:322-8
49. Jiao W, Ji JF, Xu W. et al. Distinct downstream signaling and the roles of vegf and plgf in high glucose-mediated injuries of human retinal endothelial cells in culture. Sci Rep. 2019;9:15339
50. Moccia F, Negri S, Shekha M. et al. Endothelial ca(2+) signaling, angiogenesis and vasculogenesis: just what it takes to make a blood vessel. Int J Mol Sci. 2019;20:3962
51. Bausch D, Fritz S, Bolm L. et al. Hedgehog signaling promotes angiogenesis directly and indirectly in pancreatic cancer. Angiogenesis. 2020;23:479-92
52. Mannan A, Dhiamn S, Garg N. et al. Pharmacological modulation of sonic hedgehog signaling pathways in angiogenesis: a mechanistic perspective. Dev Biol. 2023;504:58-74
53. Lee JE, Kim C, Yang H. et al. Novel glycosylated vegf decoy receptor fusion protein, vegf-grab, efficiently suppresses tumor angiogenesis and progression. Mol Cancer Ther. 2015;14:470-9
54. Hong HK, Park YJ, Kim DK. et al. Preclinical efficacy and safety of vegf-grab, a novel anti-vegf drug, and its comparison to aflibercept. Invest Ophthalmol Vis Sci. 2020;61:22
55. Go EJ, Yang H, Lee SJ. et al. Pb101, a vegf- and plgf-targeting decoy protein, enhances antitumor immunity and suppresses tumor progression and metastasis. Oncoimmunology. 2023;12:2259212
Author contact
![]() Corresponding authors: Song Wu, MD, PhD/Professor, Department of Urology, South China Hospital of Shenzhen University, Shenzhen 518116, China; E-mail: wusongedu.cn. Jean Paul Thiery, PhD/Professor, Guangzhou Laboratory, Guangzhou International BioIsland, Guangzhou 510005, China; E-mail: tjpnus.edu.sg. Yuqing Li, PhD, Department of Urology, South China Hospital of Shenzhen University, Shenzhen 518116, China; Email: liyuqingszu.edu.cn.
Corresponding authors: Song Wu, MD, PhD/Professor, Department of Urology, South China Hospital of Shenzhen University, Shenzhen 518116, China; E-mail: wusongedu.cn. Jean Paul Thiery, PhD/Professor, Guangzhou Laboratory, Guangzhou International BioIsland, Guangzhou 510005, China; E-mail: tjpnus.edu.sg. Yuqing Li, PhD, Department of Urology, South China Hospital of Shenzhen University, Shenzhen 518116, China; Email: liyuqingszu.edu.cn.

 Global reach, higher impact
Global reach, higher impact