Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(14):2851-2861. doi:10.7150/ijms.103186 This issue Cite
Research Paper
Effect of SDF-1 and CXCR4 gene variants on the development of diabetic kidney disease
1. Division of Cardiology, Department of Internal Medicine, Changhua Christian Hospital, Yunlin Branch, Yunlin, Taiwan.
2. Department of Nursing, Hungkuang University, Taichung, Taiwan.
3. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan.
4. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
5. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
6. Department of Family and Community Medicine, Chung Shan Medical University Hospital, Taichung, Taiwan.
7. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan.
8. Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan.
9. Division of Nephrology, Department of Internal Medicine, Chung Shan Medical University Hospital, Taichung, Taiwan.
10. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
Received 2024-9-3; Accepted 2024-10-21; Published 2024-10-28
Abstract
Diabetic kidney disease (DKD) is the gradual loss of renal function occurring in patients with diabetes. Stromal cell-derived factor-1 (SDF-1, encoded by SDF-1 gene) is a chemokine that binds to its receptor, CXCR4, to mediate many aspects of renal biology. To test the potential impact of SDF-1/CXCR4 gene variations on the risk for DKD, single-nucleotide polymorphisms (SNPs) of SDF-1/CXCR4 genes were genotyped in 388 DKD patients and 335 DKD-free diabetic controls. Among 6 SNPs examined, we demonstrated that rs1801157 of SDF-1 gene was associated with an increased risk for DKD (GA vs GG, AOR=2.252, p=0.035; GA+AA vs GG, AOR=2.156, p=0.036). Further stratification revealed that the correlation of rs1801157 with DKD was particularly detected in diabetic patients with early CKD but not in those with severe renal impairment. Instead, another SNP of SDF-1 gene, rs266085, was found in association with the advanced form of DKD (TC vs TT, AOR=2.106, p=0.027; TC+CC vs TT, AOR=2.130, p=0.019), indicating differential impacts of SDF-1 gene polymorphisms on the progressive loss of renal function in diabetic patients. Moreover, preliminary survey of public gene expression datasets showed that rs1801157 and rs266085 modulated SDF-1 expression in many human tissues, and SDF-1/CXCR4 levels were elevated in renal tissues of DKD patients. These data suggest that allele-specific expression of SDF-1 gene may influence DKD progression.
Keywords: SDF-1, CXCR4, gene polymorphism, diabetic kidney disease
Introduction
As the microvascular damages to the kidney frequently arise in diabetes, diabetic kidney disease (DKD), a hallmark comorbidity of diabetes that occurs in roughly 20-50% of patients with type 2 diabetes mellitus (T2DM) [1], has been believed to stem from a complex interplay of metabolic, inflammatory, hemodynamic, and fibrotic abnormalities [2]. These dysfunctions, comprising massive elimination of metabolic products derived from distorted glucose catabolism [3], perturbation of the renin-angiotensin-aldosterone system (RAAS) [4], and modulation of various signaling pathways associated with renal fibrosis [5, 6], toxicity of reactive oxygen species [7, 8], complement cascade [9], and inflammatory activity [10, 11], to a large extent account for the pathophysiology of DKD, resulting in renal impairment. Diverse risk factors for DKD are known and conceptually classified as susceptibility (e.g., age, gender, and genetic inheritance), initiation (e.g., hyperglycemia and acute kidney injury), and progression factors (e.g., obesity, hypertension, and diet) [12]. Among these established risks, hyperglycemia, obesity, and hypertension are conceivably modifiable via appropriate diabetes management [13]. This complex nature of disease etiology contributes to a high variation of DKD epidemiology and clinical outcomes, thereby urging us for the identification of new targets or manipulable parameters to ameliorate the prevention and treatment of DKD.
Cumulative evidence has clearly linked genetic inheritance to the development of both diabetes and its complications [14]. Moreover, a family history of cardiovascular conditions in diabetic individuals was found to be associated with an increased risk for DKD [15, 16], highlighting an impact of inherited factors on developing DKD in DM cases. Until now, genome-wide screening has uncovered a myriad of genetic variants that confer the susceptibility to diabetes and DKD [17-21]. These genes represent the genetic landscape of diabetes and shed light on the understanding of DKD pathogenesis, notably in the glycemic control, albuminuria, and renal function decline in distinct racial groups [22, 23]. Yet, the architecture of DKD predisposing factors is of great heterogeneity and only partly explains why some individuals suffer from renal diseases and some preserve kidney function [24]. Thus, further dissecting genetic background of DKD may provide additional biomarkers for the improvement of disease diagnosis and management.
Stromal cell-derived factor 1 (SDF-1), also named CXCL12, was initially identified as a member of CXC chemokine family secreted by bone marrow stromal cells [25, 26]. It is known that SDF-1 binds to its cognate receptor, CXC chemokine receptor 4 (CXCR4), to mediate diverse biological effects, such as the induction of cell adhesion, proliferation, motility, chemotactic activity, and angiogenic responses [27, 28]. In the kidney, SDF-1 was shown to be expressed in the stromal cells and podocytes of mature glomeruli to control nephrogenesis and the development of renal vasculature [29, 30]. As renal SDF-1 has been demonstrated to play a key role in kidney repair [31], a crucial involvement of SDF-1 signal in the pathogenesis of many kidney diseases or renal complications of diabetes was proposed [32]. Moreover, blockage of SDF-1/CXCR4 axis in the rodent models of diabetes caused podocyte loss and promoted mesangial expansion and tubular epithelial cell death, thereby leading to albuminuria and glomerulosclerosis [33, 34]. These observations indicate a connection between SDF-1/CXCR4 axis and renal health in diabetic patients. Additionally, there have been significant efforts to explore whether single nucleotide polymorphisms (SNPs) of SDF-1 gene (encoding SDF-1) influence the susceptibility to acquired immunodeficiency syndrome (AIDS) [35], chronic myeloproliferative disease [36] and many forms of malignancies [37]. To date, the association of SDF-1 gene polymorphisms with the development of DKD remains elusive, as an effect of SDF-1 gene variations on an ocular complication of diabetes, diabetic retinopathy, has been reported [38]. In this case-control study, we attempted to test the potential impact of SDF-1/CXCR4 gene polymorphisms on the risk for DKD.
Materials and methods
Study cohorts
In this study, 388 DKD cases and 335 diabetic patients with normal kidney function [estimated by glomerular filtration rate (GFR)] were enrolled and approval by the institutional review board (CSMUH No: CS2-22190) in Chung Shan Medical University Hospital, Taichung, Taiwan. Kidney disease was diagnosed as the sign of albuminuria or an estimated GFR (eGFR) of less than 60 mL/min/1.73 m2 from two individual visits. To exploring the disease severity, we stratified DKD cases into early CKD (n=308, CKD stage 1-3; with an eGFR ≥ 30) and pre-ESRD (n=80, CKD stage 4-5; with an eGFR < 30). Demographic and laboratory data regarding age, gender, diabetic condition, hyperlipidemic status, and renal function were obtained.
Genotyping
A total of six SNPs, including four from the SDF-1 gene (rs1801157, rs2297630, rs2839693, and rs266085) and two from the CXCR4 gene (rs2228014 and rs6430612) were examined based on their connection with the susceptibility to various diseases [37-41]. Genomic DNA of the whole blood samples was isolated by using QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA, USA). Biallelic discrimination for rs1801157 (assay ID: C_3223115_10, rs2297630 (assay ID: C_3223122_1), rs2839693 (assay ID: C_31777299_10), rs266085 (assay ID: C_1033724_30), rs2228014 (assay ID: C_27378716_10) and rs6430612 (assay ID: C_30721949_10) SNPs was carried out through the TaqMan assay (Applied Biosystems, Foster City, CA, USA), and genotypes were determined by SDS version 3.0 software.
Statistical analysis
Hardy-Weinberg equilibrium for six selected SNPs was assessed by using a χ2 goodness-of-fit approach. Demographic and laboratory data between cases and controls were compared with the Mann-Whitney U test. Association of polymorphic alleles with the risk and severity of DKD was evaluated by multiple logistic regression analyses combined with the adjustment for potential confounders (age, the duration of diabetes, HbA1c, systolic blood pressure, serum creatinine levels, glomerular filtration rate, HDL cholesterol levels, LDL cholesterol levels, triglycerides levels, TC/HDL ratio, microalbumin and UACR). Differences in SDF-1 expression among genotypic groups from the Genotype-Tissue Expression (GTEx) database [42] were calculated with one-way ANOVA. Gene expression of SDF-1 and CXCR4 was retrieved from the Gene Expression Omnibus repository (GSE30122) [43] and compared by t-test. A p value of <0.05 was considered statistically significant.
Results
Baseline characteristics of study cohorts
To examine the potential effect of SDF-1/CXCR4 gene variants on the risk for DKD, 388 DKD cases were recruited and compared with 335 CKD-free controls. The baseline characteristics of two study groups were evaluated (Table 1). Differences in the age but not the gender were detected between cases and controls. The mean of age in DKD cases and controls was 62.74 and 59.13 (years), respectively. Besides common indications of kidney function decline (impaired glomerular filtration rate and elevation of urinal albumin and serum creatinine), an increase in the duration of diabetic conditions and severity of hyperglycemia (determined by HbA1c values) was seen in the case group, as compared with controls. Furthermore, several indications of cardiovascular diseases, such as systolic blood pressure and ratio of total cholesterol to high-density lipoprotein cholesterol, were elevated in DKD patients in comparison with CKD-free controls.
Effect of SDF-1/CXCR4 gene polymorphisms on the risk for DKD
To test the influence of SDF-1/CXCR4 gene variations on the susceptibility to DKD, genotypes of four SNPs from the SDF-1 gene (rs1801157, rs2297630, rs2839693, and rs266085) and of two SNPs from the CXCR4 gene (rs2228014 and rs6430612) were surveyed in our cohorts. For all six SNPs tested, no deviation (p>0.05) from Hardy-Weinberg equilibrium in both study cohorts was seen. Of note, we observed that diabetic individuals who are heterozygous for the minor allele (A) of SDF-1 rs1801157 (GA; AOR, 2.252; 95% CI, 1.060-4.741; p=0.035) are more frequently to develop renal complications (Table 2), while diabetic patients homozygous for the major allele (G) of rs1801157 (GG) are used as the reference in additive model. In addition, DM patients who carry at least one minor allele (A) of SDF-1 rs1801157 (GA and AA; AOR, 2.156; 95% CI, 1.051-4.424; p=0.036) are more commonly suffering from CKD than are those homozygotes for the major allele (GG) in dominant model. Yet, for two SNPs of the CXCR4 gene tested, no interaction with the risk for DKD was detected from our study cohorts (Table 2). However, there are no significant correlations noted in allele model (Table 3). These data indicate that SDF-1 rs1801157 genotypes confer the predisposition to renal complications in diabetic patients.
Clinical and laboratory characteristics of diabetic patients with/without kidney diseases.
| Variable | Non-diabetic kidney disease (N=335) | Diabetic kidney disease (N=388) | p value |
|---|---|---|---|
| Age (years) | 59.13 ± 11.01 | 62.74 ± 10.96 | <0.001 |
| Male gender [n (%)] | 173 (51.6%) | 217 (55.9%) | 0.249 |
| Duration of diabetes (years) | 9.10 ± 6.60 | 11.55 ± 8.04 | <0.001 |
| HbA1c [% (mmol/mol)] | 6.95 ± 0.96 | 7.40 ± 1.35 | <0.001 |
| Body mass index [kg/m2] | 25.78 ± 4.13 | 26.38 ± 4.43 | 0.064 |
| Systolic blood pressure [mmHg] | 133.16 ± 15.58 | 138.85 ± 16.27 | <0.001 |
| Diastolic blood pressure [mm Hg] | 75.76 ± 11.19 | 76.62 ± 11.59 | 0.311 |
| Serum creatinine [mg/dL] | 0.79 ± 0.19 | 1.43 ± 1.56 | <0.001 |
| Glomerular filtration rate [ml/min] | 85.94 ± 26.43 | 61.23 ± 30.38 | <0.001 |
| Total cholesterol [mmol/L] | 164.02 ± 36.97 | 160.72 ± 50.54 | 0.327 |
| HDL cholesterol [μmol/L] | 47.12 ± 12.77 | 43.94 ± 13.09 | 0.001 |
| LDL cholesterol [μmol/L] | 89.58 ± 26.73 | 83.87 ± 32.30 | 0.011 |
| Triglycerides, [μmol/L] | 134.86 ± 115.90 | 156.01 ± 173.10 | 0.060 |
| TC/HDL ratio | 3.66 ± 1.12 | 3.96 ± 2.37 | 0.040 |
| Microalbumin (mg/dL) | 1.12 ± 1.07 | 48.68 ± 103.33 | <0.001 |
| UACR (mg/g) | 9.97 ± 7.09 | 613.81 ± 1457.35 | <0.001 |
Differential effects of SDF-1/CXCR4 gene polymorphisms on the disease progression of DKD
Since a genetic predisposition of DKD was observed in our cohorts, we subsequently performed stratification analyses to explore whether specific genotypes of SDF-1/CXCR4 genes are associated with the progression of renal impairment. By stratifying DKD cases into two severity groups (early CKD and pre-ESRD), we found that the correlation of SDF-1 rs1801157 with DKD was particularly detected in diabetic patients with early CKD (GA vs GG, AOR, 2.198; 95% CI, 1.036-4.663, p=0.040; GA+AA vs GG, AOR, 2.116; 95% CI, 1.029-4.353, p=0.042) in additive model and dominant model (Table 4). However, this genetic association was not observed in diabetic subjects with advanced CKD (the pre-ESRD group) (Table 5). Instead, specific genotypes of another SNP of SDF-1 gene, rs266085, were associated with advanced DKD (the pre-ESRD) (TC vs TT, AOR, 2.106; 95% CI, 1.090-4.069, p=0.027; TC+CC vs TT, AOR, 2.130; 95% CI, 1.130-4.014, p=0.019) in additive model and dominant model (Table 5). These results indicate differential impacts of SDF-1 gene polymorphisms on the progressive loss of kidney function in DM patients.
Association between SDF-1/CXCR4 genotypes and diabetic kidney disease.
| Variable | Non-diabetic kidney disease (N=335) | diabetic kidney disease (N=388) | AOR (95% CI) | p value |
|---|---|---|---|---|
| SDF-1 rs1801157 | ||||
| Additive model | ||||
| GG | 175 (52.2%) | 176 (45.4%) | 1.000 (reference) | |
| GA | 139 (41.5%) | 180 (46.4%) | 2.242 (1.060-4.741) | p=0.035 |
| AA | 21 (6.3%) | 32 (8.2%) | 1.741 (0.441-6.875) | p=0.429 |
| Dominant model | ||||
| GG | 175 (52.2%) | 176 (45.4%) | 1.000 (reference) | |
| GA+AA | 160 (47.8%) | 212 (54.6%) | 2.156 (1.051-4.424) | p=0.036 |
| SDF-1 rs2297630 | ||||
| Additive model | ||||
| GG | 267 (79.7%) | 303 (78.1%) | 1.000 (reference) | |
| GA | 60 (17.9%) | 75 (19.3%) | 0.583 (0.211-1.613) | p=0.299 |
| AA | 8 (2.4%) | 10 (2.6%) | 0.571 (0.052-6.274) | p=0.647 |
| Dominant model | ||||
| GG | 267 (79.7%) | 303 (78.1%) | 1.000 (reference) | |
| GA+AA | 68 (20.3%) | 85 (21.9%) | 0.581 (0.224-1.509) | p=0.265 |
| SDF-1 rs2839693 | ||||
| Additive model | ||||
| CC | 263 (78.5%) | 296 (76.3%) | 1.000 (reference) | |
| CT | 68 (20.3%) | 87 (22.4%) | 1.832 (0.769-4.367) | p=0.172 |
| TT | 4 (1.2%) | 5 (1.3%) | 0.597 (0.029-12.144) | p=0.737 |
| Dominant model | ||||
| CC | 263 (78.5%) | 296 (76.3%) | 1.000 (reference) | |
| CT+TT | 72 (21.5%) | 92 (23.7%) | 1.712 (0.726-4.036) | p=0.219 |
| SDF-1 rs266085 | ||||
| Additive model | ||||
| TT | 119 (35.5%) | 123 (31.7%) | 1.000 (reference) | |
| TC | 166 (49.6%) | 193 (49.7%) | 0.689 (0.319-1.488) | p=0.343 |
| CC | 50 (14.9%) | 72 (18.6%) | 1.150 (0.402-3.285) | p=0.794 |
| Dominant model | ||||
| TT | 119 (35.5%) | 123 (31.7%) | 1.000 (reference) | |
| TC+CC | 216 (64.5%) | 265 (68.3%) | 0.786 (0.384-1.608) | p=0.509 |
| CXCR4 rs2228014 | ||||
| Additive model | ||||
| CC | 257 (76.7%) | 295 (76.0%) | 1.000 (reference) | |
| CT | 70 (20.9%) | 84 (21.6%) | 1.581 (0.667-3.745) | p=0.298 |
| TT | 8 (2.4%) | 9 (2.4%) | 1.845 (0.186-18.342) | p=0.601 |
| Dominant model | ||||
| CC | 257 (76.7%) | 295 (76.0%) | 1.000 (reference) | |
| CT+TT | 78 (23.3%) | 93 (24.0%) | 1.606 (0.703-3.671) | p=0.261 |
| CXCR4 rs6430612 | ||||
| Additive model | ||||
| CC | 310 (92.5%) | 353 (91.0%) | 1.000 (reference) | |
| CT | 25 (7.5%) | 35 (9.0%) | 1.233 (0.370-4.110) | p=0.734 |
| TT | 0 (0.0%) | 0 (0.0%) | --- | --- |
| Dominant model | ||||
| CC | 310 (92.5%) | 353 (91.0%) | 1.000 (reference) | |
| CT+TT | 25 (7.5%) | 35 (9.0%) | 1.233 (0.370-4.110) | p=0.734 |
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models.
Association between SDF-1/CXCR4 genotypes and diabetic kidney disease with genetic allele model.
| Variable | Non-diabetic kidney disease (N=670) | diabetic kidney disease (N=776) | OR (95% CI) | p value |
|---|---|---|---|---|
| SDF-1 rs1801157 | ||||
| Allele model | ||||
| G | 489 (73.0%) | 532 (68.6%) | 1.000 (reference) | |
| A | 181 (27.0%) | 244 (31.4%) | 1.239 (0.986-1.557) | p=0.065 |
| SDF-1 rs2297630 | ||||
| Allele model | ||||
| G | 594 (88.7%) | 681 (87.8%) | 1.000 (reference) | |
| A | 76 (11.3%) | 95 (12.2%) | 1.090 (0.791-1.503) | p=0.598 |
| SDF-1 rs2839693 | ||||
| Allele model | ||||
| C | 594 (88.7%) | 679 (87.5%) | 1.000 (reference) | |
| T | 76 (11.3%) | 97 (12.5%) | 1.117 (0.811-1.537) | p=0.499 |
| SDF-1 rs266085 | ||||
| Allele model | ||||
| T | 404 (60.3%) | 439 (56.6%) | 1.000 (reference) | |
| C | 266 (39.7%) | 337 (43.4%) | 1.166 (0.945-1.438) | p=0.152 |
| CXCR4 rs2228014 | ||||
| Allele model | ||||
| C | 584 (87.2%) | 674 (86.9%) | 1.000 (reference) | |
| T | 86 (12.8%) | 102 (13.1%) | 1.028 (0.756-1.398) | p=0.862 |
| CXCR4 rs6430612 | ||||
| Allele model | ||||
| C | 645 (96.3%) | 741 (95.5%) | 1.000 (reference) | |
| T | 25 (3.7%) | 35 (4.5%) | 1.219 (0.722-2.058) | p=0.459 |
Functional insight of rs1801157 and rs266085 in DKD
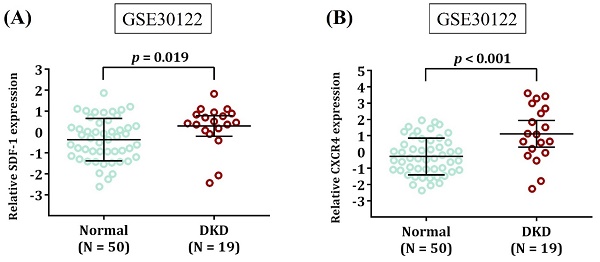
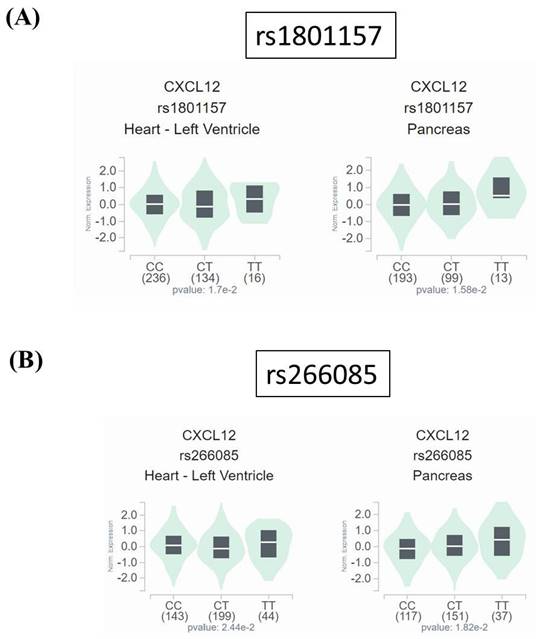
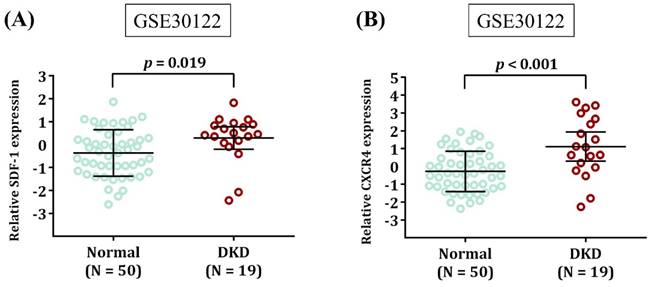
Since neither rs1801157 nor rs266085 is located on the coding region of SDF-1 gene, we also surveyed public datasets to obtain a possible clue for the function of these two DKD-associated alleles. We demonstrated fluctuations of SDF-1 expression in the heart and pancreas among different genotypic groups of rs1801157 and rs266085 in the Genotype-Tissue Expression (GTEx) database (Figure 1). In addition, through analyzing a transcriptomic dataset in Gene Expression Omnibus repository (GSE30122) [43], a significant increase in expression levels of SDF-1 and its receptor, CXCR4, was noted in renal tissues of DKD patients in comparison with that of healthy donors (Figure 2). There data imply that alteration in allele-specific SDF-1 expression might influence the disease progression of DKD.
Discussion
A body of emerging evidence has pointed out that the complex etiology of DKD is controlled by an interplay of genetic and acquired parameters. Here, by using a candidate gene approach, we demonstrated an association of SDF-1 rs1801157 with the initiation of DKD. In addition, genotypes of SDF-1 rs266085 were correlated with the progression into the advanced form of DKD, unveiling a differential effect of SDF-1 gene variations on orchestrating the disease course of DKD.
Genetic variations in SDF-1 gene have configured intricate patterns of disease susceptibility. Among these disease-associated alleles of SDF-1 gene, one SNP located in the 3' untranslated region (3'UTR), rs1801157, is the best-studied polymorphism. It has been demonstrated that rs1801157 is correlated with many clinical manifestations of human immunodeficiency virus (HIV) infection, outcomes of liver transplantation, age-at-onset of diabetes, and risks for hematologic malignancies and solid tumors [44]. In this study, we extend the list of its involvements in pathological conditions to include renal complications of diabetes. Moreover, rs1801157 gene polymorphism is associated with susceptibility to adverse long-term allograft outcomes in non-diabetic kidney transplant recipients [45]. The functional relevance of rs1801157 variants was originally accounted for by induction of SDF-1 levels [35]. However, opposite findings that carriers homozygous for the minor allele (A) of rs1801157 was associated with low plasma SDF-1 levels have been reported [46, 47], while other studies documented that there was no difference in the SDF-1 production by the presence of rs1801157 polymorphisms [48, 49]. Such controversies might be attributed to the frequency and structure of unique haplotypes in different ethnic groups, as additional SNPs in high linkage disequilibrium with rs1801157, instead of rs1801157 itself, were found to mediate differential transcription levels [50]. These observations collectively indicate that rs1801157 likely contributes to allele-specific expression of SDF-1 gene to render a functional impact.
Association between SDF-1/CXCR4 genotypes and early diabetic kidney disease.
| Variable | Non-diabetic kidney disease (N=335) | Early CKD (N=308) | AOR (95% CI) | p value |
|---|---|---|---|---|
| SDF-1 rs1801157 | ||||
| Additive model | ||||
| GG | 175 (52.2%) | 137 (44.5%) | 1.000 (reference) | |
| GA | 139 (41.5%) | 143 (46.4%) | 2.198 (1.036-4.663) | p=0.040 |
| AA | 21 (6.3%) | 28 (9.1%) | 1.721 (0.438-6.758) | p=0.436 |
| Dominant model | ||||
| GG | 175 (52.2%) | 137 (44.5%) | 1.000 (reference) | |
| GA+AA | 160 (47.8%) | 171 (55.5%) | 2.116 (1.029-4.353) | p=0.042 |
| SDF-1 rs2297630 | ||||
| Additive model | ||||
| GG | 267 (79.7%) | 238 (77.3%) | 1.000 (reference) | |
| GA | 60 (17.9%) | 62 (20.1%) | 0.593 (0.215-1.633) | p=0.312 |
| AA | 8 (2.4%) | 8 (2.6%) | 0.614 (0.056-6.715) | p=0.689 |
| Dominant model | ||||
| GG | 267 (79.7%) | 238 (77.3%) | 1.000 (reference) | |
| GA+AA | 68 (20.3%) | 70 (22.7%) | 0.595 (0.230-1.544) | p=0.286 |
| SDF-1 rs2839693 | ||||
| Additive model | ||||
| CC | 263 (78.5%) | 238 (77.3%) | 1.000 (reference) | |
| CT | 68 (20.3%) | 65 (21.1%) | 1.846 (0.774-4.402) | p=0.167 |
| TT | 4 (1.2%) | 5 (1.6%) | 0.613 (0.032-11.899) | p=0.746 |
| Dominant model | ||||
| CC | 263 (78.5%) | 238 (77.3%) | 1.000 (reference) | |
| CT+TT | 72 (21.5%) | 70 (22.7%) | 1.724 (0.731-4.066) | p=0.213 |
| SDF-1 rs266085 | ||||
| Additive model | ||||
| TT | 119 (35.5%) | 105 (34.1%) | 1.000 (reference) | |
| TC | 166 (49.6%) | 146 (47.4%) | 0.693 (0.321-1.497) | p=0.350 |
| CC | 50 (14.9%) | 57 (18.5%) | 1.174 (0.410-3.362) | p=0.764 |
| Dominant model | ||||
| TT | 119 (35.5%) | 105 (34.1%) | 1.000 (reference) | |
| TC+CC | 216 (64.5%) | 203 (65.9%) | 0.793 (0.387-1.622) | p=0.525 |
| CXCR4 rs2228014 | ||||
| Additive model | ||||
| CC | 257 (76.7%) | 232 (75.3%) | 1.000 (reference) | |
| CT | 70 (20.9%) | 68 (22.1%) | 1.591 (0.674-3.758) | p=0.289 |
| TT | 8 (2.4%) | 8 (2.6%) | 1.858 (0.196-17.595) | p=0.589 |
| Dominant model | ||||
| CC | 257 (76.7%) | 232 (75.3%) | 1.000 (reference) | |
| CT+TT | 78 (23.3%) | 76 (24.7%) | 1.618 (0.711-3.682) | p=0.252 |
| CXCR4 rs6430612 | ||||
| Additive model | ||||
| CC | 310 (92.5%) | 276 (89.6%) | 1.000 (reference) | |
| CT | 25 (7.5%) | 32 (10.4%) | 1.173 (0.344-4.001) | p=0.799 |
| TT | 0 (0.0%) | 0 (0.0%) | --- | --- |
| Dominant model | ||||
| CC | 310 (92.5%) | 276 (89.6%) | 1.000 (reference) | |
| CT+TT | 25 (7.5%) | 32 (10.4%) | 1.173 (0.344-4.001) | p=0.799 |
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models.
Association between SDF-1/CXCR4 genotypes and Pre-ESRD disease.
| Variable | Non-diabetic kidney disease (N=335) | Pre-ESRD (N=80) | AOR (95% CI) | p value |
|---|---|---|---|---|
| SDF-1 rs1801157 | ||||
| Additive model | ||||
| GG | 175 (52.2%) | 39 (48.7%) | 1.000 (reference) | |
| GA | 139 (41.5%) | 37 (46.3%) | 1.163 (0.328-4.123) | p=0.815 |
| AA | 21 (6.3%) | 4 (5.0%) | 0.715 (0.193-2.640) | p=0.614 |
| Dominant model | ||||
| GG | 175 (52.2%) | 39 (48.7%) | 1.000 (reference) | |
| GA+AA | 160 (47.8%) | 41 (51.3%) | 0.964 (0.276-3.364) | p=0.954 |
| SDF-1 rs2297630 | ||||
| Additive model | ||||
| GG | 267 (79.7%) | 65 (81.3%) | 1.000 (reference) | |
| GA | 60 (17.9%) | 13 (16.3%) | 0.463 (0.054-3.987) | p=0.483 |
| AA | 8 (2.4%) | 2 (2.5%) | 0.540 (0.085-3.428) | p=0.514 |
| Dominant model | ||||
| GG | 267 (79.7%) | 65 (81.3%) | 1.000 (reference) | |
| GA+AA | 68 (20.3%) | 15 (18.8%) | 0.440 (0.051-3.772) | p=0.454 |
| SDF-1 rs2839693 | ||||
| Additive model | ||||
| CC | 263 (78.5%) | 58 (72.5%) | 1.000 (reference) | |
| CT | 68 (20.3%) | 22 (27.5%) | 1.324 (0.345-5.083) | p=0.682 |
| TT | 4 (1.2%) | 0 (0.0%) | --- | --- |
| Dominant model | ||||
| CC | 263 (78.5%) | 58 (72.5%) | 1.000 (reference) | |
| CT+TT | 72 (21.5%) | 22 (27.5%) | 1.311 (0.342-5.024) | p=0.693 |
| SDF-1 rs266085 | ||||
| Additive model | ||||
| TT | 119 (35.5%) | 18 (22.5%) | 1.000 (reference) | |
| TC | 166 (49.6%) | 47 (58.8%) | 2.106 (1.090-4.069) | p=0.027 |
| CC | 50 (14.9%) | 15 (18.7%) | 2.208 (0.937-5.204) | p=0.070 |
| Dominant model | ||||
| TT | 119 (35.5%) | 18 (22.5%) | 1.000 (reference) | |
| TC+CC | 216 (64.5%) | 62 (77.5%) | 2.130 (1.130-4.014) | p=0.019 |
| CXCR4 rs2228014 | ||||
| Additive model | ||||
| CC | 257 (76.7%) | 63 (78.8%) | 1.000 (reference) | |
| CT | 70 (20.9%) | 16 (20.0%) | 1.103 (0.223-5.456) | p=0.905 |
| TT | 8 (2.4%) | 1 (1.3%) | 0.284 (0.029-2.786) | p=0.280 |
| Dominant model | ||||
| CC | 257 (76.7%) | 63 (78.8%) | 1.000 (reference) | |
| CT+TT | 78 (23.3%) | 17 (21.3%) | 0.875 (0.184-4.165) | p=0.867 |
| CXCR4 rs6430612 | ||||
| Additive model | ||||
| CC | 310 (92.5%) | 77 (96.3%) | 1.000 (reference) | |
| CT | 25 (7.5%) | 3 (3.7%) | 1.462 (0.132-16.189) | p=0.757 |
| TT | 0 (0.0%) | 0 (0.0%) | --- | --- |
| Dominant model | ||||
| CC | 310 (92.5%) | 77 (96.3%) | 1.000 (reference) | |
| CT+TT | 25 (7.5%) | 3 (3.7%) | 1.462 (0.132-16.189) | p=0.757 |
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models.
As a matter of fact, expression levels of SDF-1 are crucial for renal development and exert modulatory effects on the course of kidney diseases under different etiologic settings by binding to its cognate receptor, CXCR4 [32]. Expression of SDF-1 by stromal cells or podocytes acts on endothelial cells to regulate vascular and glomerular development in the kidney [30]. In a mouse model of diabetes, aberrant expression of SDF-1 by glomerular podocytes augmented proteinuria and glomerulosclerosis, while the use of a specific inhibitor of SDF-1, NOX-A12, corrected glomerulosclerosis, enhanced the number of podocytes, maintained the peritubular vasculature and delayed the onset of albuminuria [33]. On the contrary, reduction of endothelial SDF-1 was accompanied by proteinuria, elevated oxidative stress, podocyte foot process effacement and augmented glomerular size in a rat model of obesity [51]. Also, SDF-1 was proposed to participate in promotion of renal fibrosis, which is believed to be the final common pathogenic mechanism leading to CKD and ESRD [52]. In addition to the involvement in the progressive loss of renal function, robust upregulation of SDF-1 after ischemia/reperfusion-induced acute kidney injury contributed to homing and migration of CXCR4-positive cells toward the injured kidney, governing renal regeneration and repair [31]. These results, together with our findings, support a conjecture that fluctuations in SDF-1 levels derived from gene polymorphisms affect the susceptibility to DKD in diabetic patients.
Intriguingly, we exhibited an association of an intronic SNP of SDF-1 gene, rs266085, with the risk for the advanced form of DKD. It has been reported that rs266085 was correlated with the susceptibility to cervical carcinoma, likely via production of distinct SDF-1 splice variants [53]. To date, at least six human SDF-1 isoforms derived from alternative splicing events have been identified (SDF-1α, SDF-1β, SDF-1γ, SDF-1δ, SDF-1ε, and SDF-1Φ) [54, 55] and subjected to different proteolytic processing [56], thus explaining functional diversity. All these SDF-1 splice variants share the same first three exons but contain different fourth exons [55]. Although rs266085 is located in the third intron between exon 2 and 3 of the SDF-1 gene, another intronic SNP located between exon 3 and 4 of the SDF-1 gene, rs266087, was in perfect linkage disequilibrium with rs266085 [53]. It is possible that rs266087 (or others variants on the same common haplotype) affected the relative accumulation of different SDF-1 isoforms [50]. In addition to generation of different SDF-1 isoforms, the same study indicated that specific haplotypes containing the minor allele of rs266085 were associated with strong SDF-1 induction, implicating a potential role of rs266085 in allele-specific expression of different SDF-1 splice variants.
Impact of rs1801157 and rs266085 genotypes on SDF-1 expression. Comparisons of SDF-1 (CXCL12) expression among (A) rs1801157 and (B) rs266085 genotypic groups in representative normal tissues based on data from the GTEx portal. p values were calculated among groups by one-way ANOVA.
Expression levels of SDF-1 and CXCR4 are increased in DKD. Comparison of SDF-1 (A) and CXCR4 expression (B) in the renal tissues between DKD patients and healthy donors (Normal). Expression data were retrieved from Gene Expression Omnibus repository (GSE30122). The total number of samples is given in parentheses. p values are calculated with Student's t test.
As the exclusive receptor for SDF-1, CXCR4 has been implicated as a crucial mediator of renal regeneration and kidney diseases [57]. However, for two SNPs (rs2228014 and rs6430612) of the CXCR4 gene tested, no significant association with the risk for DKD was observed from our cohorts. As a relatively low frequency (<10%) for the alternative allele of rs6430612 was detected in our study groups, rs2228014 polymorphism is more common in our survey. To date, many case-control studies aiming to investigate the association of CXCR4 rs2228014 with disease susceptibility have yielded conflicting results [58-64], which may be accounted for by insufficient sample size of individual study, different distributions of cases or controls, different disease pathology, and various methodologies. Determining the relationship between CXCR4 variants and DKD risks will require further investigation with a greater sample size and subgroup analyses.
In our survey, we detected an association between SDF-1 gene variants and the development of DKD. However, there are several limitations to this investigation. One potential issue is that the diverse comorbidities of diabetes (e.g. ocular, cutaneous, neurological, cardiovascular, and muscular conditions) and their inherent genetic components likely result in a different finding concerning the impact of SDF-1/CXCR4 gene variations with DKD. Moreover, causal expression quantitative trait loci were highly enriched in 3'UTR [65], yet we did not test whether rs1801157 variants orchestrate SDF-1 transcription in our own cohort or related kidney cell types, such as podocytes, renal stromal, and endothelial cells. Also unavailable are the data to support a link of rs266085 to alternative splicing, as well as to dissect an involvement of SDF-1 splice variants in DKD pathogenesis through functional validation. Additionally, recent multi-ethnic investigations indicated an ethnicity-specific involvement of SDF-1 variants in disease susceptibility [37, 66], suggesting the presence of variations in the frequency of particular SDF-1 alleles among different ethnic cohorts. Thus, the genetic impact observed in our study may be constrained to unique populations unless a replication cohort with other ethnicities was investigated.
In conclusion, our data unveiled an impact of SDF-1 gene variations on acting as a gatekeeper during the disease course of DKD. This genetic association likely connects allele-specific expression of SDF-1 splice variants to the aggravation of renal impairment in diabetic subjects.
Acknowledgements
We are grateful to the Human Biobank of Chung Shan Medical University Hospital, Taichung, Taiwan for sample preparation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Reutens AT. Epidemiology of diabetic kidney disease. Med Clin North Am. 2013;97:1-18
2. Sinha SK, Nicholas SB. Pathomechanisms of Diabetic Kidney Disease. J Clin Med. 2023;12:7349
3. Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest. 2014;124:2333-40
4. Roscioni SS, Heerspink HJ, de Zeeuw D. The effect of RAAS blockade on the progression of diabetic nephropathy. Nat Rev Nephrol. 2014;10:77-87
5. Wang L, Wang HL, Liu TT, Lan HY. TGF-Beta as a Master Regulator of Diabetic Nephropathy. Int J Mol Sci. 2021;22:7881
6. Zhang Y, Jin D, Kang X, Zhou R, Sun Y, Lian F. et al. Signaling Pathways Involved in Diabetic Renal Fibrosis. Front Cell Dev Biol. 2021;9:696542
7. Schaffer SW, Jong CJ, Mozaffari M. Role of oxidative stress in diabetes-mediated vascular dysfunction: unifying hypothesis of diabetes revisited. Vascul Pharmacol. 2012;57:139-49
8. Susztak K, Raff AC, Schiffer M, Bottinger EP. Glucose-induced reactive oxygen species cause apoptosis of podocytes and podocyte depletion at the onset of diabetic nephropathy. Diabetes. 2006;55:225-33
9. Flyvbjerg A. The role of the complement system in diabetic nephropathy. Nat Rev Nephrol. 2017;13:311-8
10. Matoba K, Takeda Y, Nagai Y, Kawanami D, Utsunomiya K, Nishimura R. Unraveling the Role of Inflammation in the Pathogenesis of Diabetic Kidney Disease. Int J Mol Sci. 2019;20:3393
11. Navarro-Gonzalez JF, Mora-Fernandez C, Muros de Fuentes M, Garcia-Perez J. Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat Rev Nephrol. 2011;7:327-40
12. Annual facility survey of providers of ESRD therapy. USRDS. United States Renal Data System. Am J Kidney Dis. 1997;30:S178-86
13. Alicic RZ, Rooney MT, Tuttle KR. Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clin J Am Soc Nephrol. 2017;12:2032-45
14. Cole JB, Florez JC. Genetics of diabetes mellitus and diabetes complications. Nat Rev Nephrol. 2020;16:377-90
15. Tedla FM, Brar A, Browne R, Brown C. Hypertension in chronic kidney disease: navigating the evidence. Int J Hypertens. 2011;2011:132405
16. Yeung EH, Pankow JS, Astor BC, Powe NR, Saudek CD, Kao WH. Increased risk of type 2 diabetes from a family history of coronary heart disease and type 2 diabetes. Diabetes Care. 2007;30:154-6
17. Mahajan A, Spracklen CN, Zhang W, Ng MCY, Petty LE, Kitajima H. et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat Genet. 2022;54:560-72
18. Xue A, Wu Y, Zhu Z, Zhang F, Kemper KE, Zheng Z. et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018;9:2941
19. Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ. et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41-7
20. Kwak SH, Srinivasan S, Chen L, Todd J, Mercader JM, Jensen ET. et al. Genetic architecture and biology of youth-onset type 2 diabetes. Nat Metab. 2024;6:226-37
21. van Zuydam NR, Ahlqvist E, Sandholm N, Deshmukh H, Rayner NW, Abdalla M. et al. A Genome-Wide Association Study of Diabetic Kidney Disease in Subjects With Type 2 Diabetes. Diabetes. 2018;67:1414-27
22. Sandholm N, Dahlstrom EH, Groop PH. Genetic and epigenetic background of diabetic kidney disease. Front Endocrinol (Lausanne). 2023;14:1163001
23. Sheng X, Qiu C, Liu H, Gluck C, Hsu JY, He J. et al. Systematic integrated analysis of genetic and epigenetic variation in diabetic kidney disease. Proc Natl Acad Sci U S A. 2020;117:29013-24
24. Thomas MC, Brownlee M, Susztak K, Sharma K, Jandeleit-Dahm KA, Zoungas S. et al. Diabetic kidney disease. Nat Rev Dis Primers. 2015;1:15018
25. Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J Exp Med. 1996;184:1101-9
26. Nagasawa T, Kikutani H, Kishimoto T. Molecular cloning and structure of a pre-B-cell growth-stimulating factor. Proc Natl Acad Sci U S A. 1994;91:2305-9
27. Petit I, Jin D, Rafii S. The SDF-1-CXCR4 signaling pathway: a molecular hub modulating neo-angiogenesis. Trends Immunol. 2007;28:299-307
28. Sadri F, Rezaei Z, Fereidouni M. The significance of the SDF-1/CXCR4 signaling pathway in the normal development. Mol Biol Rep. 2022;49:3307-20
29. Grone HJ, Cohen CD, Grone E, Schmidt C, Kretzler M, Schlondorff D. et al. Spatial and temporally restricted expression of chemokines and chemokine receptors in the developing human kidney. J Am Soc Nephrol. 2002;13:957-67
30. Takabatake Y, Sugiyama T, Kohara H, Matsusaka T, Kurihara H, Koni PA. et al. The CXCL12 (SDF-1)/CXCR4 axis is essential for the development of renal vasculature. J Am Soc Nephrol. 2009;20:1714-23
31. Togel F, Isaac J, Hu Z, Weiss K, Westenfelder C. Renal SDF-1 signals mobilization and homing of CXCR4-positive cells to the kidney after ischemic injury. Kidney Int. 2005;67:1772-84
32. Song A, Jiang A, Xiong W, Zhang C. The Role of CXCL12 in Kidney Diseases: A Friend or Foe? Kidney Dis (Basel). 2021;7:176-85
33. Sayyed SG, Hagele H, Kulkarni OP, Endlich K, Segerer S, Eulberg D. et al. Podocytes produce homeostatic chemokine stromal cell-derived factor-1/CXCL12, which contributes to glomerulosclerosis, podocyte loss and albuminuria in a mouse model of type 2 diabetes. Diabetologia. 2009;52:2445-54
34. Takashima S, Fujita H, Fujishima H, Shimizu T, Sato T, Morii T. et al. Stromal cell-derived factor-1 is upregulated by dipeptidyl peptidase-4 inhibition and has protective roles in progressive diabetic nephropathy. Kidney Int. 2016;90:783-96
35. Winkler C, Modi W, Smith MW, Nelson GW, Wu X, Carrington M. et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science. 1998;279:389-93
36. Gerli G, Vanelli C, Turri O, Erario M, Gardellini A, Pugliano M. et al. SDF1-3'A gene polymorphism is associated with chronic myeloproliferative disease and thrombotic events. Clin Chem. 2005;51:2411-4
37. Tong X, Ma Y, Deng H, Wang X, Liu S, Yan Z. et al. The SDF-1 rs1801157 Polymorphism is Associated with Cancer Risk: An Update Pooled Analysis and FPRP Test of 17,876 Participants. Sci Rep. 2016;6:27466
38. Peng SY, Chuang CC, Hwang YS, Yen CH, Lee CY, Yang SF. Association of SDF-1 and its receptor CXCR4 polymorphisms on the susceptibility of diabetic retinopathy in the Taiwanese population. Front Genet. 2023;14:1296773
39. Alonso N, Albagha OME, Azfer A, Larraz-Prieto B, Berg K, Riches PL. et al. Genome-wide association study identifies genetic variants which predict the response of bone mineral density to teriparatide therapy. Ann Rheum Dis. 2023;82:985-91
40. Wu Y, Zhang C, Xu W, Zhang J, Zheng Y, Lu Z. et al. CXC motif chemokine receptor 4 gene polymorphism and cancer risk. Medicine (Baltimore). 2016;95:e5317
41. Yin G, Zhu T, Li J, Wu A, Liang J, Zhi Y. CXCL12 rs266085 and TNF-alpha rs1799724 polymorphisms and susceptibility to cervical cancer in a Chinese population. Int J Clin Exp Pathol. 2015;8:5768-74
42. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580-5
43. Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60:2354-69
44. Colobran R, Pujol-Borrell R, Armengol MP, Juan M. The chemokine network. II. On how polymorphisms and alternative splicing increase the number of molecular species and configure intricate patterns of disease susceptibility. Clin Exp Immunol. 2007;150:1-12
45. Wang CJ, Tsai JP, Yang SF, Lian JD, Chang HR. Stromal cell-derived factor 1 gene polymorphism is associated with susceptibility to adverse long-term allograft outcomes in non-diabetic kidney transplant recipients. Int J Mol Sci. 2014;15:12495-506
46. Soriano A, Martinez C, Garcia F, Plana M, Palou E, Lejeune M. et al. Plasma stromal cell-derived factor (SDF)-1 levels, SDF1-3'A genotype, and expression of CXCR4 on T lymphocytes: their impact on resistance to human immunodeficiency virus type 1 infection and its progression. J Infect Dis. 2002;186:922-31
47. de Oliveira KB, Guembarovski RL, Oda JM, Mantovani MS, Carrera CM, Reiche EM. et al. CXCL12 rs1801157 polymorphism and expression in peripheral blood from breast cancer patients. Cytokine. 2011;55:260-5
48. Kimura R, Nishioka T, Ishida T. The SDF1-G801A polymorphism is not associated with SDF1 gene expression in Epstein-Barr virus-transformed lymphoblastoid cells. Genes Immun. 2003;4:356-61
49. Arya SK, Ginsberg CC, Davis-Warren A, D'Costa J. In vitro phenotype of SDF1 gene mutant that delays the onset of human immunodeficiency virus disease in vivo. J Hum Virol. 1999;2:133-8
50. Kimura R, Nishioka T, Soemantri A, Ishida T. Allele-specific transcript quantification detects haplotypic variation in the levels of the SDF-1 transcripts. Hum Mol Genet. 2005;14:1579-85
51. Nistala R, Habibi J, Aroor A, Sowers JR, Hayden MR, Meuth A. et al. DPP4 inhibition attenuates filtration barrier injury and oxidant stress in the zucker obese rat. Obesity (Silver Spring). 2014;22:2172-9
52. Wu X, Qian L, Zhao H, Lei W, Liu Y, Xu X. et al. CXCL12/CXCR4: An amazing challenge and opportunity in the fight against fibrosis. Ageing Res Rev. 2023;83:101809
53. Maley SN, Schwartz SM, Johnson LG, Malkki M, Du Q, Daling JR. et al. Genetic variation in CXCL12 and risk of cervical carcinoma: a population-based case-control study. Int J Immunogenet. 2009;36:367-75
54. Shirozu M, Nakano T, Inazawa J, Tashiro K, Tada H, Shinohara T. et al. Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics. 1995;28:495-500
55. Yu L, Cecil J, Peng SB, Schrementi J, Kovacevic S, Paul D. et al. Identification and expression of novel isoforms of human stromal cell-derived factor 1. Gene. 2006;374:174-9
56. De La Luz Sierra M, Yang F, Narazaki M, Salvucci O, Davis D, Yarchoan R. et al. Differential processing of stromal-derived factor-1alpha and stromal-derived factor-1beta explains functional diversity. Blood. 2004;103:2452-9
57. Togel FE, Westenfelder C. Role of SDF-1 as a regulatory chemokine in renal regeneration after acute kidney injury. Kidney Int Suppl (2011). 2011;1:87-9
58. Cai C, Wang LH, Dong Q, Wu ZJ, Li MY, Sun YH. Association of CXCL12 and CXCR4 gene polymorphisms with the susceptibility and prognosis of renal cell carcinoma. Tissue Antigens. 2013;82:165-70
59. Chang CC, Chen SC, Hsieh YH, Chen YC, Chen TY, Chu YH. et al. Stromal cell-derived factor-1 but not its receptor, CXCR4, gene variants increase susceptibility and pathological development of hepatocellular carcinoma. Clin Chem Lab Med. 2009;47:412-8
60. Zheng Q, Shuai X, Ye Y, Jin Y, Jiang N, Chen X. et al. The role of polymorphisms of stromal-derived factor-1 and CXC receptor 4 in acute myeloid leukemia and leukemia cell dissemination. Gene. 2016;588:103-8
61. Cacina C, Bulgurcuoglu-Kuran S, Iyibozkurt AC, Yaylim-Eraltan I, Cakmakoglu B. Genetic variants of SDF-1 and CXCR4 genes in endometrial carcinoma. Mol Biol Rep. 2012;39:1225-9
62. Crowther-Swanepoel D, Qureshi M, Dyer MJ, Matutes E, Dearden C, Catovsky D. et al. Genetic variation in CXCR4 and risk of chronic lymphocytic leukemia. Blood. 2009;114:4843-6
63. Bodelon C, Malone KE, Johnson LG, Malkki M, Petersdorf EW, McKnight B. et al. Common sequence variants in chemokine-related genes and risk of breast cancer in post-menopausal women. Int J Mol Epidemiol Genet. 2013;4:218-27
64. Isman FK, Kucukgergin C, Dasdemir S, Cakmakoglu B, Sanli O, Seckin S. Association between SDF1-3'A or CXCR4 gene polymorphisms with predisposition to and clinicopathological characteristics of prostate cancer with or without metastases. Mol Biol Rep. 2012;39:11073-9
65. Consortium GT. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318-30
66. Alijanpour A, Golshan A, Vakili-Ojarood M, Shirinzadeh-Dastgiri A, Naseri A, Karimi-Zarchi M. et al. Association of the CXCL12 rs1801157 Polymorphism with Breast Cancer Risk: A Meta-Analysis. Asian Pac J Cancer Prev. 2024;25:767-76
Author contact
![]() Corresponding author: Shun-Fa Yang, Ph.D. Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Tel.: +886-4-24739595 ext. 34253; Fax: +886-4-24723229; E-mail: ysfedu.tw.
Corresponding author: Shun-Fa Yang, Ph.D. Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Tel.: +886-4-24739595 ext. 34253; Fax: +886-4-24723229; E-mail: ysfedu.tw.

 Global reach, higher impact
Global reach, higher impact