Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(14):2837-2850. doi:10.7150/ijms.100468 This issue Cite
Research Paper
Integrating Network Pharmacology, Molecular Docking and Experimental Validation to Explore the Pharmacological Mechanisms of Quercetin Against Diabetic Wound
1. Department of General Surgery & VIP In-Patient Ward, The First Hospital of China Medical University, Shenyang, Liaoning Province, 110001, China.
2. Department of Vascular and Thyroid Surgery, The First Hospital of China Medical University, Shenyang, Liaoning Province, 110001, China.
3. Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang, Liaoning Province, 110122, China.
Received 2024-7-3; Accepted 2024-10-17; Published 2024-10-28
Abstract
The chronic non-healing diabetic wound (DW) has remained a challenge to both the society and individuals. Previous studies suggested dietary moderate consumption of quercetin (QCT) are beneficial in preventing diabetic complications, including non-healing DW. However, there were few studies that have investigated QCT-related underlying molecular mechanisms against DW. In the present study, we for the first-time combined network pharmacology with molecular docking and experimental validation to investigate QCT-related therapeutic targets and mechanisms for treating DW. Finally, 191 QCT-related targets and 1750 DW-related pathogenetic targets were obtained from online databases. After removing duplicates, a total of 90 potential therapeutic targets of quercetin for treating DW were ultimately identified. Furthermore, 7 targets with higher degree including IL-6, EGFR, SRC, TNF, AKT1, JUN and MMP9 were predicted as central therapeutic targets of QCT for treating DW. Functional enrichment analysis demonstrated that QCT exerted strong levels of multitargeting regulatory activity. In addition, the KEGG enrichment analysis indicated that several signaling pathways including AGE-RAGE signaling pathway in diabetic complications, IL-17, PI3k-AKT, TNF, HIF-1, VEGF were predicted as key regulators of QCT for treating DW. Molecular docking results suggested that QCT had strong binding activity with the predicted targets. In addition, verification experiments suggested that QCT could significantly attenuated the expression of inflammatory cytokines and the regulation of PI3K-AKT signaling pathway was probably a vital mechanism involved in the pharmacological mechanism of QCT for treating DW. Taken together, combined network pharmacological with experimental validation, we for the first time systematically investigated associated-therapeutic targets and potential pathways of QCT for DW treatment. Our study might provide theoretical basis for DW treatment.
Keywords: quercetin, diabetic wound, polyphenol, PI3K-AKT signaling, molecular docking, network pharmacology
Introduction
Diabetes mellitus (DM) is a serious, costly metabolic disorder with high morbidity. Despite improving living standards, the prevalence of DM has seen an explosive increase in the past few decades [1]. The heavy cost of diabetes and related complications have become a significant burden to society [2]. Clinically, DM patients are generally asymptomatic in the early stages. Unfortunately, without available medications, it may lead to serious damage and even cause death [3]. Diabetic foot ulcer (DFU) and refractory wounds are serious chronic complications of DM, contributing to over 84% of DM-related lower limb amputations [4]. The normal wound healing process can be divided into 3 overlapping and dynamic phases: inflammation, proliferation, and tissue remodeling [5]. However, the chronic refractory wound healing in diabetic patients is a complex and long-term process [6]. Under pathological conditions, chronic hyperglycemia negatively affects the normal healing process, triggering the activation of inflammation cascades [7]. Currently, medical therapies against refractory wounds are limited. Therefore, it is of great importance to explore novel therapies for diabetic refractory wounds.
Plant-derived natural products have gained much attention due to their affordability, safety and lower side effects [8]. Quercetin (QCT), one of the most well-studied phytochemicals, belongs to the subclass of flavonoids and is widely abundant in fruits and vegetables, such as apples, onions, peppers and berries [9]. Previous studies suggested that moderate consumption of QCT is beneficial to health. Accumulating evidence has shown that QCT has various health-prompting properties such as anti-inflammation, anti-oxidant, anti-tumor and cardioprotective capabilities [10]. The role of QCT play in wound healing is well-studied. Numerous studies suggested that QCT is a promising natural wound healing agent [11]. A recent study published by Harvinder suggested that QCT-loaded AgNPs in hydrogel matrices significantly attenuated wound gaps in an experimental DW model [12]. Furthermore, Vinay reported that QCT ointment obviously reduced expression of pro-inflammatory cytokines and improved angiogenesis in DW rats [13]. All these findings suggested that quercetin is a promising drug in accelerating DW healing.
Network pharmacology is considered the next paradigm of drug discovery and is helpful in screening potential therapeutic targets and pathways from a molecular perspective [14]. In the present study, network pharmacology methods combined with molecular docking was utilized to explore potential therapeutic targets and underlying mechanisms of QCT for treating DW. Furthermore, experimental validations were performed to testify our predictions. Our study might provide theoretical basis for DW research.
Materials & Methods
Screening for QCT-related therapeutic targets against DW
In this study, QCT-related targets were obtained from the Traditional Chinese Medicine Systems Pharmacology database [15] (TCMSP, https://old.tcmsp-e.com/index.php), the Encyclopedia of Traditional Chinese Medicine database [16] (ETCM, http://www.tcmip.cn/ETCM/index.php), and the SwissTargetPrediciton database (http://www.swisstargetprediction.ch/index.php). Furthermore, DW-related targets were obtained from the Therapeutic Target database [17] (TTD, https://db.idrblab.net/ttd/), the Online Mendelian Inheritance in Man database [18] (OMIM, https://omim.org/), the GeneCards database (https://www.gene-cards.org/) and the Disgenet database [19] (https://www.disgenet.org/). Then the drug and disease-related targets were summarized and removed the duplicates. The Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to overlap all the non-duplicates targets which were seen as potential therapeutic targets in QCT-related DW treatment.
Construction of protein-protein interaction (PPI) network and screening for potential therapeutic targets of QCT for DW treatment
To find protein-associated networks of QCT against DW, the overlapped genes were imported into the STRING Database (https://cn.string-db.org/). In this network, we set the minimum required interaction score as high confidence (0.700) and the species was set as Homo sapiens. Then, the Cytoscape software (Version 3.9.1) was used to visualize the PPI network. Then the Cytohubba plugin was used to explore core therapeutic targets of QCT against DW.
Biological enrichment analysis and construction of gene-pathway network
In this study, the Gene Ontology (GO) in biological processes (BP), molecular function (MF) and cellular components (CC) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were performed by using the Metascape database(https://metascape.org/gp/index.html). Furthermore, we used the Bioinformatic database (http://www.bioinformatics.com.cn/) to visualize identified results. Then we used Cytoscape software to construct the gene-pathway network.
Molecular docking
To better identify molecular interactions behind the ligand and macromolecules, molecular docking analysis was performed in this study. The PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was used to retrieve the structure of QCT and identified core targets. Then we used Pymol software (https://py-mol.org/2/) to remove water and unwanted atoms. The Autodock (https://autodock.scripps.edu/) was used to perform molecular docking procedures. The Pymol software was used to visualize the binding site between the ligand and macromolecules. The DiscoveryStudio 2019 software was used to visualize molecular bonds between QCT and identified core targets.
Experimental DM model establishment
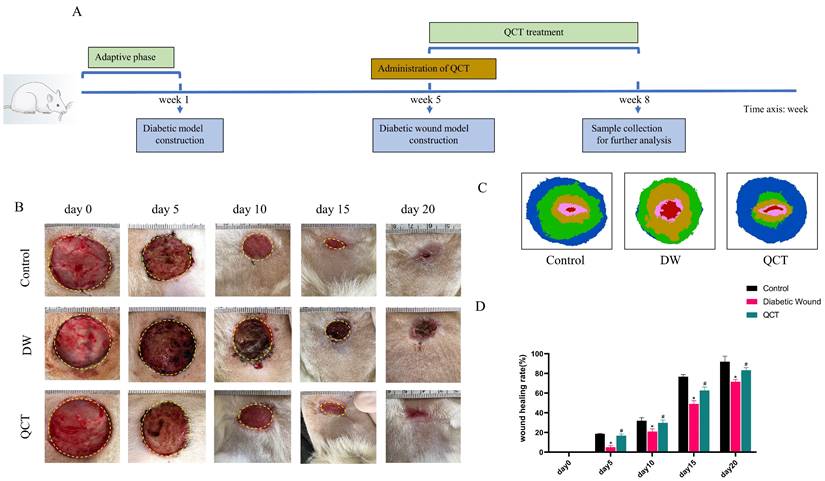
The streptozotocin (STZ) was obtained from Sigma-Aldrich (United States). The citric acid/sodium citrate buffer were obtained from Beyotime Biotechnology (Shanghai, China). 30 8-week-old, 280-300g male Sprague Dawley (SD) rats were obtained from Liaoning Changsheng Biotechnology (Liaoning, China) and were housed under controlled environment with sufficient food and water. The whole experimental animal protocols were approved by the Institutional Animal Care and Use Committee of China Medical University (Ethic number: CMU20241591). And the animal experimental procedures were performed following the guideline of China Medical University Guide for the Care and Use of Laboratory Animals. After one-week of adaptive feeding, 30 SD rats were randomly divided into 3 groups (n=10): control group, STZ-induced DW group and QCT-treated group. Rats in STZ-induced DW group and QCT-treated group were injected intraperitoneally with 65mg/kg STZ (dissolved in 0.1mol/L citric acid/sodium citrate buffer, PH=4.2) to induce diabetic model [20]. Rats in control group were injected intraperitoneally with equal amounts of 0.1mol/L citric acid/sodium citrate buffer without STZ. To prevent hypoglycaemia, 5% glucose was added to the drinking water at the day of diabetes induction. After diabetes induction, the rats' blood glucose levels were measured for consecutive 4 weeks. The rats with blood glucose level greater than 16.7mM were deemed as diabetes rats and were used for the experiment.
Experimental DW model establishment and QCT administration
QCT was purchased from Shanghai Yuanye Bio-Technology Co., Ltd (B20527, Shanghai, China). QCT was suspended in 0.5% sodium carboxymethyl cellulose. The fresh QCT solutions were prepared daily. To establish DW model, after 4 weeks of STZ injection, diabetic rats were anesthetized with 2% isoflurane and the dorsal hair were shaved. After the surgical site was disinfect with 75% alcohol, a 3*3cm full-thickness excisional wound was created on their backs. To avoid skin contraction, the wound site was stabilized with 4-0 nylon sutures. After surgery, rats in QCT-treated group were administered with 40mg/kg QCT solution by oral gavage daily and rats in other group were treated with equal amounts of solution without QCT. The dose of QCT was according to previously published work [21]. The drug treatments continued for 3 weeks. After 3-week treatment, all rats were euthanized by excessive anesthesia (5% isoflurane) and skin tissues were collected for further analysis. To record the healing process, the photos of wound area were taken at days 0, 5, 10, 15, 20 after surgery. The wound area was measured and the wound healing rate was analyzed by the imageJ software. The wound healing rate was represented as (original wound area - daily wound area) / (original wound area) * 100%.
Western blotting
In the present study, the skin tissues were collected. The total protein extraction kit was obtained from Solarbio, China to extract proteins from skin tissues. The blotting procedures were performed to identify characterization of predicted targets as previously described [22]. Furthermore, the imageJ and Graphpad Prism software was used to analyzed and visualized the image. The primary antibodies used in this study were as follows, p- indicated phosphorylation. Antibody against MMP9(No: ab76003), TNFα (No: ab34674), IL6 (No: ab259341) and β-actin (No: ab8227) was obtained from Abcam (Cambridge, MA). Antibody against mTOR (No.2983), p-mTOR (No.5536), AKT(No.4691), p-AKT (No.9271), PI3K (No.4249), p-PI3K (No.4228) was obtained from cell signaling technology (CST).
Histological study of wound sections
The wound tissues were collected and fixed in 4% paraformaldehyde and then embedded in paraffin. The tissues were sectioned into 5μm thickness slides, and stained by HE (haematoxylin and eosin), Masson and Immunohistochemical (IHC) protocols. The stained procedures were performed as previously described [22]. The level of TNF, IL6, p-PI3K, p-AKT and p-mTOR were assessed with IHC staining. The ImageJ software was used to qualify the stained area.
Statistical analysis
Quantitative data from 3 independent experiments were expressed as mean ± SEM. These data were analyzed by using Statistical Product Service Solutions (SPSS, Version 26) and visualized by GraphPad Prism software (Version 9.0.0). In addition, the two-way Analysis of Variance (ANOVA) was performed to compare the differences. P < 0.05 was considered to be statistically significant.
Results
Screening for potential therapeutic targets of QCT for DW
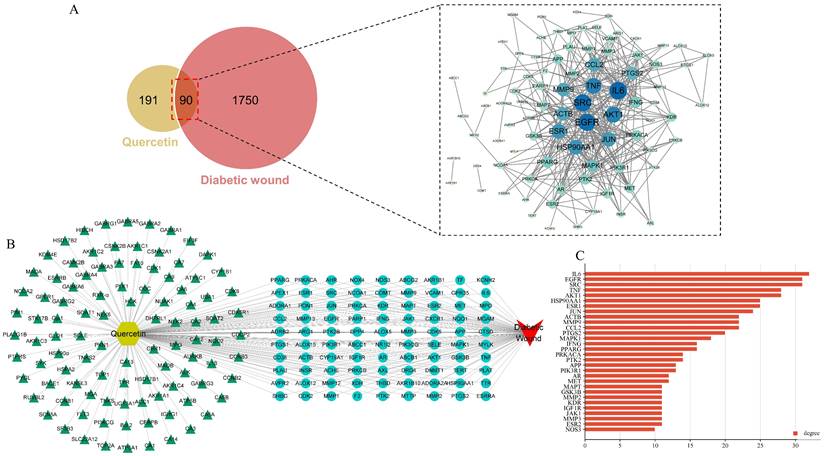
The work flow of the whole study is presented in Figure 1. The chemical structure of QCT was depicted in Figure 2. As shown in Figure 3a, a total of 191 QCT-related targets were obtained from the intersection of TCMSP, ETCM and SwissTarget database after merging and deleting duplicates. Besides, 1750 DW-related targets were obtained from the TTD, the Disgenet, the Genecards, and the OMIM database. Then, identified targets of QCT and DW were overlapped with a Venn diagram and constructed a drug-disease network. As shown in Figure 3b, a total of 90 potential therapeutic targets of QCT for treating DW were ultimately identified. The overlapped targets were shown in Table 1 and were considered potential therapeutic targets for QCT against DW.
Putative genes of QCT for DW repairing.
| No. | Gene | Uniprot ID | No. | Gene | Uniprot ID | No. | Gene | Uniprot ID |
|---|---|---|---|---|---|---|---|---|
| 1 | ABCB1 | P08183 | 31 | DPP4 | P27487 | 61 | MYLK | Q15746 |
| 2 | ABCC1 | P33527 | 32 | DRD4 | P21917 | 62 | NCOA1 | Q15788 |
| 3 | ABCG2 | Q9UNQ0 | 33 | EGFR | P00533 | 63 | NOS3 | P29474 |
| 4 | ACHE | P22303 | 34 | ESR1 | P03372 | 64 | NOX4 | Q9NPH5 |
| 5 | ACTB | P60709 | 35 | ESR2 | Q92731 | 65 | NQO1 | P15559 |
| 6 | ADORA1 | P30542 | 36 | ESRRA | P11474 | 66 | NR1I2 | O75469 |
| 7 | ADORA2A | P29274 | 37 | F2 | P00734 | 67 | PARP1 | P09874 |
| 8 | ADRB2 | P07550 | 38 | GPR35 | Q9HC97 | 68 | PIK3CG | P48736 |
| 9 | AHR | P35869 | 39 | GSK3B | P49841 | 69 | PIK3R1 | P27986 |
| 10 | AKR1B1 | P15121 | 40 | HSP90AA1 | P07900 | 70 | PLAT | P00750 |
| 11 | AKR1B10 | O60218 | 41 | IFNG | P01579 | 71 | PLAU | P00749 |
| 12 | AKT1 | P31749 | 42 | IGF1R | P08069 | 72 | PON1 | P27169 |
| 13 | ALOX12 | P18054 | 43 | IL6 | P05231 | 73 | PPARG | P37231 |
| 14 | ALOX15 | P16050 | 44 | INSR | P06213 | 74 | PRKACA | P17612 |
| 15 | ALOX5 | P09917 | 45 | JAK1 | P23458 | 75 | PRKCA | P17252 |
| 16 | APEX1 | P27695 | 46 | JUN | P05412 | 76 | PRKCB | P05771 |
| 17 | APP | P05067 | 47 | KCNH2 | Q12809 | 77 | PTGS1 | P23219 |
| 18 | AR | P10275 | 48 | KDR | P35968 | 78 | PTGS2 | P35354 |
| 19 | ARG1 | P05089 | 49 | MAPK1 | P28482 | 79 | PTK2 | Q05397 |
| 20 | AVPR2 | P30518 | 50 | MAPT | P10636 | 80 | PTK2B | Q14289 |
| 21 | AXL | P30530 | 51 | MET | P08581 | 81 | SELE | P16581 |
| 22 | CCL2 | P19875 | 52 | MGAM | O43451 | 82 | SHBG | P04278 |
| 23 | CD38 | P28907 | 53 | MMP1 | P03956 | 83 | SRC | P12931 |
| 24 | CDK2 | P24941 | 54 | MMP12 | P39900 | 84 | TERT | O14746 |
| 25 | CDK5 | Q00535 | 55 | MMP13 | P45452 | 85 | TF | P02787 |
| 26 | COMT | P21964 | 56 | MMP2 | P08253 | 86 | THBD | P07204 |
| 27 | CTSD | P07339 | 57 | MMP3 | P08254 | 87 | TNF | P01375 |
| 28 | CXCR1 | P25024 | 58 | MMP9 | P14780 | 88 | TTR | P02766 |
| 29 | CYP19A1 | P11511 | 59 | MPO | P05164 | 89 | VCAM1 | P19320 |
| 30 | DNMT1 | P26358 | 60 | MTTP | P55157 | 90 | XDH | P47989 |
Identification of core targets of QCT against DW
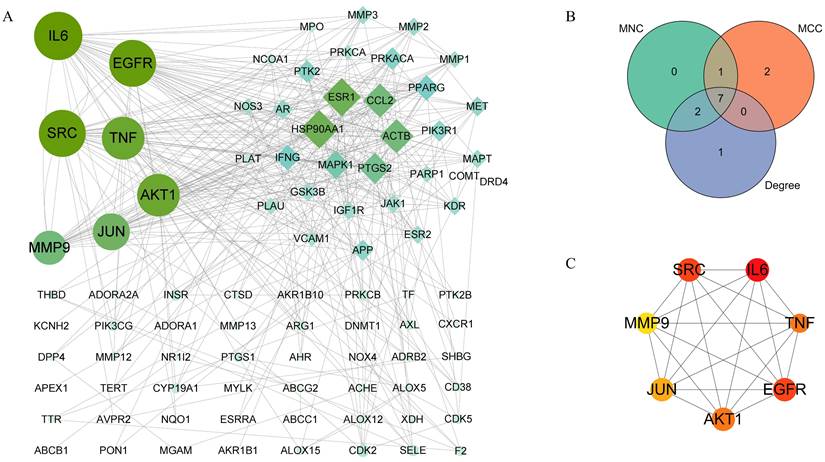
To explore core targets involved in QCT-related DW treatment, the PPI network was constructed and was visualized by using the Cytoscape software. As shown in Figure 4A, the larger nodes with deeper color indicated they had more influential. To ensure the accuracy, three prediction methods, the maximal clique centrality (MCC), maximum neighborhood component (MNC), and degree method, was used in this study. Then, we overlapped the prediction results and 7 targets in common were obtained (Figure 4B). As shown in Figure 4C, these targets were interleukin 6 (IL-6), epidermal growth factor receptor (EGFR), proto-oncogene tyrosine-protein kinase Src (SRC), tumor necrosis factor (TNF), RAC-alpha serine/threonine-protein kinase (AKT1), transcription factor jun (JUN) and matrix metalloproteinase-9 (MMP9). The degree of predicted core targets was listed in Table 2 and were chosen for further analysis.
Selection for hub genes.
| Rank | MNC | MCC | Degree |
|---|---|---|---|
| 1 | ACTB | ACTB | AKT1 |
| 2 | AKT1 | AKT1 | CCL2 |
| 3 | EGFR | EGFR | EGFR |
| 4 | ESR1 | IL6 | ESR1 |
| 5 | HSP90AA1 | JUN | HSP90AA1 |
| 6 | IL6 | MMP9 | IL6 |
| 7 | JUN | PPARG | JUN |
| 8 | MMP9 | PTGS2 | MMP9 |
| 9 | SRC | SRC | SRC |
| 10 | TNF | TNF | TNF |
Functional enrichment analysis
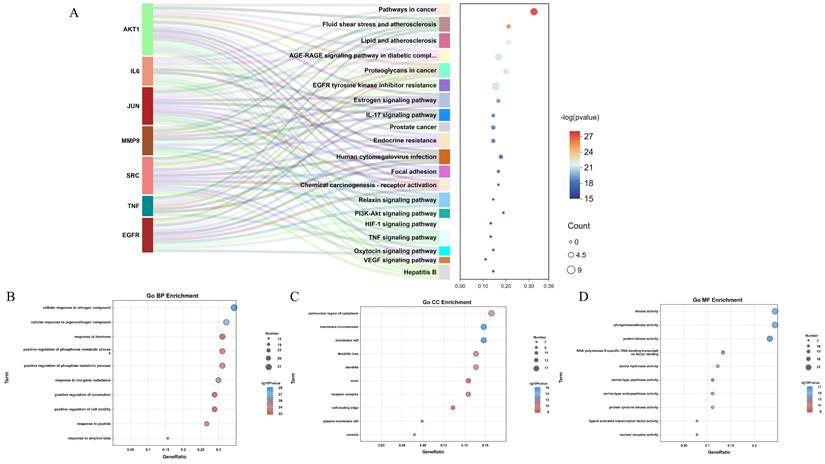
To better investigate the functional mechanism and pathways of QCT for treating DW, the predicted targets were uploaded to the Metascape database for enrichment analysis. In the present study, a total of 1419 GO terms and 178 KEGG pathways were obtained. The Sankey diagram summarized the contribution between identified core targets and enriched KEGG pathways (Figure 5A). The enriched GO biological process (BP), cellular components (CC) and molecular function (MF) was shown in Figure 5B, 5C and 5D respectively. The top 10 GO processes were listed in Table 3 and the top 20 KEGG pathways were shown in Table 4. In view of the results, we found that QCT affected a serious of biological processes including cellular response to nitrogen compound, cellular response to organonitrogen compound, response to inorganic substance, positive regulation of phosphate metabolic process, positive regulation of phosphorus metabolic process et al. Moreover, we found that the most significantly enriched KEGG pathways were pathways in cancer, fluid shear stress and atherosclerosis, Lipid and atherosclerosis, AGE-RAGE signaling pathway in diabetic complications, proteoglycans in cancer. And the most relevant pathway involved in QCT for treating DW might be the AGE-RAGE signaling pathway in diabetic complications.
The workflow of the present study.
Enriched biological processes relating to core targets.
| Term | Biological process | Contained core genes | P value |
|---|---|---|---|
| GOBP:1901699 | cellular response to nitrogen compound | EGFR, SRC, AKT1, TNF | 1.00E-29 |
| GOBP:0071417 | cellular response to organonitrogen compound | EGFR, SRC, AKT1, TNF | 1.00E-27 |
| GOBP:0010035 | response to inorganic substance | EGFR, SRC, AKT1, IL6, JUN, MMP9 | 1.00E-25 |
| GOBP:0045937 | positive regulation of phosphate metabolic process | EGFR, SRC, AKT1, IL6, MMP9, TNF | 1.00E-23 |
| GOBP:0010562 | positive regulation of phosphorus metabolic process | EGFR, SRC, AKT1, IL6, MMP9, TNF | 1.00E-23 |
| GOBP:1904645 | response to amyloid-beta | SRC, AKT1, TP53, EGFR, MMP9, TNF | 1.00E-23 |
| GOBP:1901652 | response to peptide | AKT1, MMP9, SRC, TNF | 1.00E-23 |
| GOBP:2000147 | positive regulation of cell motility | AKT1, EGFR, IL6, MMP9, SRC, TNF | 1.00E-22 |
| GOBP:0040017 | positive regulation of locomotion | AKT1, EGFR, IL6, MMP9, SRC, TNF | 1.00E-22 |
| GOBP:0009725 | response to hormone | AKT1, EGFR, IL6, SRC, TNF | 1.00E-22 |
| GOCC:0045121 | membrane raft | EGFR, SRC, TNF | 1E-16 |
| GOCC:0098857 | membrane microdomain | EGFR, SRC, TNF | 1E-16 |
| GOCC:0048471 | perinuclear region of cytoplasm | EGFR, SRC | 1E-10 |
| GOCC:0030425 | dendrite | SRC | 3.98E-10 |
| GOCC:0097447 | dendritic tree | SRC | 3.98E-10 |
| GOCC:0043235 | receptor complex | EGFR, IL6 | 7.94E-10 |
| GOCC:0044853 | plasma membrane raft | SRC | 1.58E-09 |
| GOCC:0005901 | caveola | SRC | 5.01E-09 |
| GOCC:0031252 | cell leading edge | AKT1, EGFR, SRC | 5.01E-09 |
| GOCC:0030424 | axon | SRC | 7.94E-09 |
| GOMF:0004672 | protein kinase activity | AKT1, EGFR, SRC | 1E-17 |
| GOMF:0016773 | phosphotransferase activity | AKT1, EGFR, SRC | 1E-16 |
| GOMF:0016301 | kinase activity | AKT1, SRC | 1E-16 |
| GOMF:0004713 | protein tyrosine kinase activity | SRC | 1E-11 |
| GOMF:0017171 | serine hydrolase activity | MMP9 | 1E-11 |
| GOMF:0004252 | serine-type endopeptidase activity | MMP9 | 1.26E-10 |
| GOMF:0004879 | nuclear receptor activity | - | 2E-10 |
| GOMF:0098531 | ligand-activated transcription factor activity | - | 2E-10 |
| GOMF:0008236 | serine-type peptidase activity | MMP9 | 3.16E-10 |
| GOMF:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | JUN, SRC | 5.01E-10 |
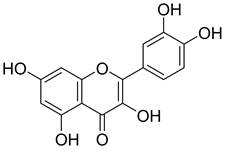
The chemical structure of QCT.
Network pharmacology predicted therapeutic targets of QCT for treating DW and PPI network construction. (A)Venn diagram of QCT and DM-related targets. The intersection of the yellow circle and red circle represented the intersecting targets. (B) Predicted therapeutic targets of QCT for treating DW. (C)The bar chart representing the top 30 targets ranked by degree.
The top 20 enriched KEGG pathways.
| Term | Pathway | Contained genes | P value |
|---|---|---|---|
| hsa05200 | Pathways in cancer | AKT1, IL6, JUN, MMP9 | 1.00E-28 |
| hsa05418 | Fluid shear stress and atherosclerosis | AKT1, SRC, JUN, TNF, MMP9 | 1.00E-26 |
| hsa05417 | Lipid and atherosclerosis | AKT1, SRC, TNF, IL6, JUN | 1.00E-22 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | AKT1, IL6, JUN, TNF | 1.00E-21 |
| hsa05205 | Proteoglycans in cancer | AKT1, EGFR, SRC, TNF, MMP9 | 1.00E-21 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | AKT1, SRC, EGFR, IL6 | 1.00E-21 |
| hsa04915 | Estrogen signaling pathway | AKT1, EGFR, JUN, SRC, MMP9 | 1.00E-19 |
| hsa04657 | IL-17 signaling pathway | TNF, MMP9, IL6, JUN | 1.00E-18 |
| hsa05215 | Prostate cancer | AKT1, MMP9, EGFR | 1.00E-18 |
| hsa01522 | Endocrine resistance | AKT1, EGFR, MMP9, SRC, JUN | 1.00E-18 |
| hsa05163 | Human cytomegalovirus infection | AKT1, EGFR, SRC, TNF, IL6 | 1.00E-17 |
| hsa04510 | Focal adhesion | AKT1, EGFR, SRC, JUN | 1.00E-16 |
| hsa05207 | Chemical carcinogenesis - receptor activation | AKT1, EGFR, JUN, SRC | 1.00E-16 |
| hsa04926 | Relaxin signaling pathway | AKT1, EGFR, JUN, SRC, MMP9 | 1.00E-16 |
| hsa04151 | PI3K-Akt signaling pathway | AKT1, IL6, EGFR | 1.00E-15 |
| hsa04066 | HIF-1 signaling pathway | AKT1, EGFR, IL6 | 1.00E-15 |
| hsa04668 | TNF signaling pathway | AKT1, IL6, JUN, MMP9, TNF | 1.00E-15 |
| hsa04921 | Oxytocin signaling pathway | SRC, EGFR, JUN | 1.00E-15 |
| hsa04370 | VEGF signaling pathway | AKT1, SRC | 1.00E-15 |
| hsa05161 | Hepatitis B | AKT1, IL6, JUN, MMP9, SRC | 1.00E-15 |
Identification of core therapeutic targets of QCT against DW. (A) Citespace analysis representing the overlapping targets. (B) Venn diagram of intersection among MNC, MCC and degree. (C) Top 7 core targets. Larger and darker represented higher degree.
Functional enrichment analysis. (A) The KEGG enrichment analysis. (B-D) The GO enrichment analysis (BP, CC and MF). The node size represented the number of targets annotated to the term. The color of nodes represented the p value.
Molecular docking analysis
In this study, molecular docking analysis were performed to evaluate the binding sites and binding energies between the QCT and macromolecules (Figure 6). The crystal structures of ligands, possible binding sites and binding energies were shown in Table 5. We found that the binding energies of QCT with core macromolecules ranged from -3.75 to -7.35. These results indicated that QCT had better affinities with these macromolecules. In view of our results, we found that QCT had the strongest affinity with MMP9 by possessing six conventional hydrogen bonds with PRO254, HIS257, PRO255, LEU243, TYR245 and GLU241; one Pi-Sigma interaction with THR251 and one Pi-Alkyl interaction with LEU243. As for TNF, QCT possessed 5 conventional hydrogen bonds with GLY24, ALA134, TRP28, ASN46, GLN25 and one Pi-Donor Hydrogen Bond with LEU26. Besides, QCT had strong affinity with IL6 by forming 6 conventional hydrogen bonds with GLU93, THR138, THR137, THR143 and ASN63; one Pi-cation interaction with GLU93; one Pi-Donor Hydrogen Bond with ASP140 and one Pi-Alkyl interaction with PRO139. As for EFGR, QCT formed 5 conventional hydrogen bonds with MET793, CYS797, ASP800; three Pi-Alkyl interactions with ALA743, VAL726 and LEU718; and one Pi-Sigma interaction with LEU844. As for AKT1, QCT formed 4 conventional hydrogen bonds with LYS17, THR87, ARG15 and LYS20; one carbon hydrogen bond with ARG15; two Pi-Alkyl interactions with LYS17 and ARG15; one Pi-Sigma interaction with LYS17. And QCT had affinity with SRC by forming 5 conventional hydrogen bonds with GLN324, Leu325, MET314, LEU317 and LYS315; one Pi-cation interaction, one Pi-Alkyl interaction and one carbon hydrogen bond with ARG318; one Pi-Sigma interaction with TRP260. As for JUN, QCT formed 4 conventional hydrogen bonds with GLN30, SER22, GLU19, GLN33; one Pi-Alkyl interaction with MET26; one Pi-Sigma interaction with PRO37. Taken together, our results showed that QCT had better affinity with predicted macromolecules and might exert therapeutic effects by forming these interactions against DW.
Molecular docking of QCT with identified core targets.
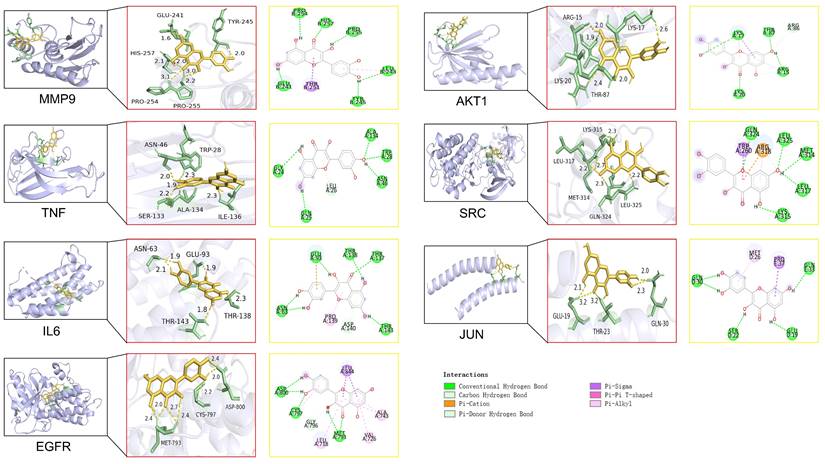
Molecular docking score.
| No | Target (PDB) | Binding sites with amino acid | Binding energy(kcal/mol) |
|---|---|---|---|
| 1 | MMP9 (4hma) | PRO254, HIS257, PRO255, LEU243, TYR245, THR251, GLU241 | -7.35 |
| 2 | TNF (1a8m) | GLY24, ALA134, TRP28, ASN46, LEU26, GLN25 | -5.48 |
| 3 | IL6 (1alu) | GLU93, THR138, THR137, THR143, ASP140, PRO139, ASN63 | -4.26 |
| 4 | EGFR (7u99) | LEU844, ALA743, VAL726, MET793, LEU718, GLY796, CYS797, ASP800 | -4.26 |
| 5 | AKT (2uzs) | LYS17, THR87, ARG86, ARG15, LYS20 | -4.16 |
| 6 | SRC (1y57) | GLN324, LEU325, TRP260, ARG318, MET314, LEU317, LYS315 | -3.85 |
| 7 | JUN (5fv8) | GLN30, MET26, PRO37, GLN33, GLU19, SER22 | -3.75 |
Treatment with QCT accelerated DW healing in vivo by reducing the activation of proinflammatory cytokines and regulating the PI3K-AKT-mTOR signaling pathway
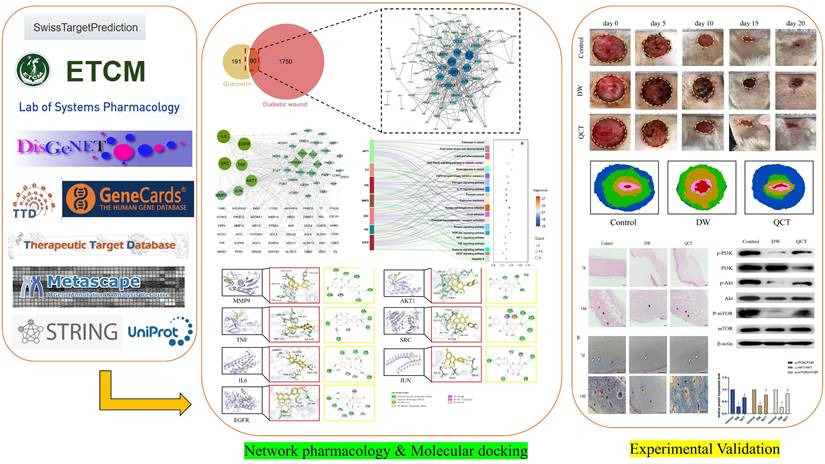
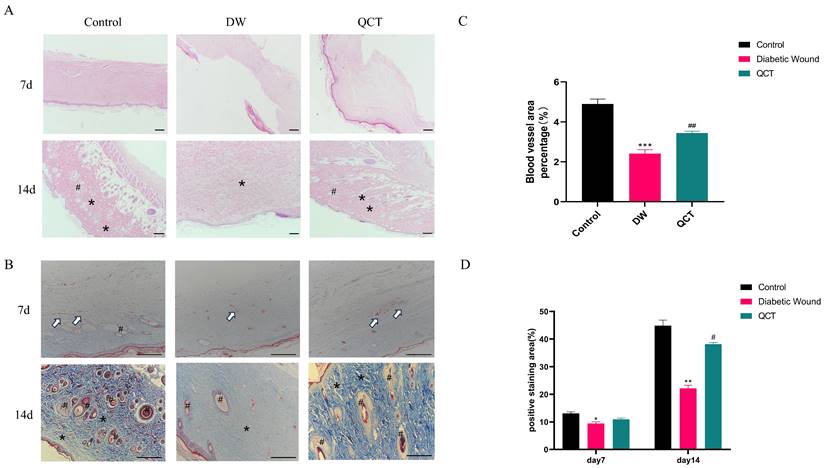
The workflow of animals was shown in Figure 7A. The back wounds were photographed on the 0th, 5th, 10th, 15th, 20th after surgery (Figure 7B). Our results indicated that the diabetic wounds without treatment had the slowest healing rate compared to those in control and QCT group. Treatment with QCT promoted wound healing at different time points (Figure 7C). As shown in Figure 7D, compared with DW group, treatment with QCT significantly improved the wound healing rate. Then, we investigated the condition of wounds at early stages. As indicated in Figure 8A, by day 7, the wounds in control group were healing well. Skin samples in control and QCT group exhibited intact epidermis. Wound tissues from DW group had poorly regenerated epidermis. As shown in Figure 8B, treatment with QCT significantly increased the density of blood vessels. By day 14, the epithelialization process has appeared in all group. Administration of QCT increased the density of skin accessory organs such as blood vessels and hair follicles. In addition, treatment with QCT significantly increased collagen fiber deposition and the presence of blood vessels and hair follicles. The quantitative analysis was shown in Figure 8C and 8D.
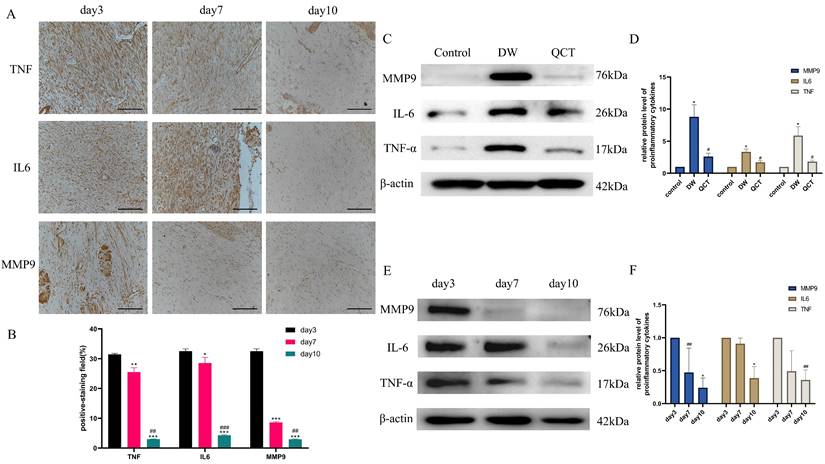
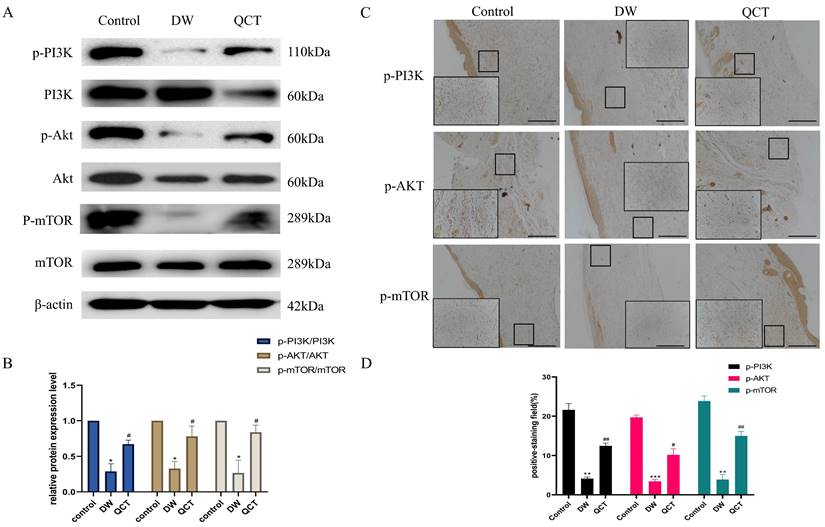
To further investigate the role of QCT for treating DW, we collected the tissues from wound margin and performed WB and IHC analysis. The IHC and WB results demonstrated that treatment with QCT significantly attenuated the production of inflammatory cytokines IL-6, TNF-α and MMP9 at days 3, 7 and 10 (Figure 9A and 9E). The quantitative analysis was shown in Figure 9B and 9F. WB analysis showed that the protein expression of proinflammatory cytokines was consistent with IHC results (Figure 9C). The quantitative analysis was shown in Figure 9D. In addition, we investigated the role of QCT in the PI3K-AKT-mTOR signaling pathway. As shown in Figure 10A, treatment with QCT significantly increased the phosphorylation of PI3k, AKT and mTOR. The IHC staining showed the expression of the above proteins was consistent with WB results (Figure 10C). The quantitative analysis was shown in Figure 10B and 10D respectively. Taken together, these results suggested that QCT might exert beneficial effects against DW by reducing the activation of proinflammatory cytokines and regulating the PI3K-AKT-mTOR signaling pathway.
QCT accelerated DW healing in diabetic rats. (A) The flow diagram of animals. (B) The images of wound healing at different time points. (C) The overlay image of wounds in different group. (D) Statistical graph of wound healing rate. The wound healing rate=(S0-Sa)/S0*100%. S0 represented the original wound area (day 0) whereas Sa represented the wound area in day a. **p<0.01 versus control group, *p<0.05 versus control group; ##p<0.01 versus DW group, #p<0.05 versus DW group.
Photomicrographs of HE and Masson staining of wounds from different groups. (A) HE staining of wounds from different groups on days 7 and 10. (B) Masson staining of wounds from different groups on days 7 and 10. (C) The corresponding quantitative analysis of blood vessel percentage at early stage in different groups. (D) The corresponding quantitative analysis of collagen fiber in different groups. Data are represented as mean ± SD(n=3). Scale bar=100μm. The white arrow indicates blood vessel; * indicates collagen; # indicates skin appendage. **p<0.01 versus control group, *p<0.05 versus control group; ##p<0.01 versus DW group, #p<0.05 versus DW group.
Effects of QCT on pro-inflammatory cytokines at early stages. (A) Immunohistochemical staining of TNF-α, IL6 and MMP9 at days 3, 7 and 10. Scale bar=50μm. (B) Quantitative analysis of TNF-α, IL6 and MMP9. (C) Representative WB images of MMP9, IL6 and TNF-α in different group. (D) Quantitative analysis of MMP9, IL6 and TNF-α in different group. (E) Representative WB images of MMP9, IL6 and TNF-α at days 3, 7 and 10. (F) Quantitative analysis of MMP9, IL6 and TNF-α at days 3, 7 and 10. ***p<0.001 versus day 3, **p<0.01 versus day 3, *p<0.05 versus day 3; ###p<0.001 versus day 7, ##p<0.01 versus day 7, #p<0.05 versus day 7.
Effects of QCT on PI3k-AKT-mTOR signaling pathway. (A) Changes of PI3k-AKT-mTOR signaling pathway after QCT treatment were detected by WB. (B) Quantitative analysis of p-PI3K/PI3K, p-AKT/AKT, p-mTOR/mTOR. (C) Changes of PI3k-AKT-mTOR signaling pathway after QCT treatment were detected by immunohistochemical staining. Scale bar=50μm. (D) Quantitative IHC analysis of p-PI3K, PI3K, p-AKT, AKT, p-mTOR, mTOR. ***p<0.001 versus control group, **p<0.01 versus control group, *p<0.05 versus control group; ##p<0.01 versus DW group, # p<0.05 versus DW group.
Discussion
Nowadays, the global incidence of DM is rapidly rising, more and more researchers have paid more attention to its associated complications. Statistics have shown that more than 15% of patients suffered from DM develop chronic non-healing wounds [4]. The common wound healing is considered as a natural physiological process, including hemostasis, inflammation, proliferation and remodeling [23]. However, the healing process in DW is usually disrupted by prolonged accumulation of inflammatory cells. Under pathological conditions, diabetic hyperglycemia triggers inflammatory cascades and lead to dysregulated angiogenesis, finally contributing to chronic non-healing wound.
Accumulating evidence suggested that naturally occurring polyphenol had remarkable health-prompting functions in a wide range of chronic diseases [24]. QCT, a naturally polyphenolic flavonoid found abundantly in various vegetables and fruits, exhibits a range of biological activities including anti-inflammatory, anti-apoptotic, anti-oxidative, anti-infective, and anti-tumor effects. Additionally, it promotes vasodilation and has therapeutic potential for the treatment of metabolic and cardiovascular diseases [25]. In recent years, the beneficial therapeutic roles of QCT in DW have been extensively discussed. A study by Latif reported that using a newly synthesized agent (QCT and its esterified complex with 4FPBA-Q) significantly accelerated wound healing in infected diabetic rats [26]. A study by Aboud et al. found that QCT combined with laser therapy attenuated the expression of pro-inflammatory cytokines and accelerated the wound healing process [27]. Besides, Asemi suggested that QCT was an effective polyphenol against DW and reviewed the effects of QCT for DW treatment. He suggested that the therapeutic effects of QCT for DW treatment might be related to its anti-inflammation and anti-oxidant properties [28]. All these findings suggested that QCT was an effective and promising polyphenol against DW. In this study, we used network pharmacological to predict and screen the possible therapeutic targets and associated signaling pathways of QCT for treating DW. Additionally, experimental methodologies were utilized to uncover the potential mechanisms of action of QCT.
Through constructing a PPI network, we identified seven pivotal hub genes essential for treating DW with QCT. IL6, a cytokine involved in inflammation and immune responses, is crucial for initiating inflammation necessary for wound healing. However, excessive IL6 activity can lead to chronic inflammation, impairing the healing process [29, 30]. MMP9, an enzyme that degrades extracellular matrix components, is involved in tissue remodeling and repair. In DWs, abnormally high MMP9 levels result in excessive matrix degradation, preventing new tissue formation and prolonging inflammation [31, 32]. AKT1, a serine/threonine kinase, promotes cell proliferation, migration, and survival, all critical for wound healing. In diabetes, impaired AKT1 signaling reduces cellular responses necessary for effective wound healing, such as keratinocyte migration and angiogenesis [33]. TNF, a cytokine regulating immune cells and systemic inflammation, is essential for initiating inflammation and mobilizing immune cells to the wound site. However, excessively high TNF levels in DWs contribute to chronic inflammation, tissue damage, and delayed healing [34, 35]. EGFR, a receptor tyrosine kinase, activates pathways promoting cell growth, survival, and proliferation. EGFR signaling is crucial for keratinocyte migration, proliferation, and differentiation, necessary for wound re-epithelialization. In diabetes, impaired EGFR signaling leads to delayed wound closure and defective tissue repair [36]. SRC, a non-receptor tyrosine kinase, regulates various cellular processes, including proliferation, differentiation, and survival. SRC influences cell migration and adhesion through responses to growth factors and integrins. Dysregulation of SRC activity in DWs impairs these processes, resulting in delayed healing and reduced tissue regeneration [37]. Finally, c-Jun, a component of the transcription factor AP-1, regulates gene expression in response to stress and cytokines. c-Jun is involved in cell proliferation, differentiation, and apoptosis, and in DWs, dysregulated c-Jun activity can impair cell proliferation and delay wound closure [38]. These hub genes play significant roles in the pathological processes affecting DW healing and are potential targets for therapeutic intervention.
Additionally, to elucidate the interactions between the target proteins and the bioactive compound, semiflexible molecular docking was conducted. The docking simulations revealed that QCT exhibited significant binding affinity towards the seven hub genes, with a particularly strong affinity for MMP9. The primary mode of interaction between QCT and the target proteins was identified as van der Waals forces.
In addition, the majority of hub genes were annotated to signaling pathways identified through GO and KEGG analyses, which are intricately associated with the wound healing process. GO enrichment analysis indicated that QCT is involved in key biological processes such as the positive regulation of cell motility and locomotion. KEGG pathway enrichment analysis demonstrated that QCT exerts therapeutic effects on DW by modulating several pathways, including the AGE-RAGE signaling pathway in diabetic complications, the IL-17 signaling pathway, the PI3K-Akt signaling pathway, and the TNF signaling pathway. The PI3K-AKT signaling pathway is integral to the wound healing process, particularly in regulating inflammation, cell proliferation, and tissue regeneration [39, 40]. Activation of PI3Ks by growth factors and cytokines leads to the production of PIP3, which recruits and activates AKT through phosphorylation [41]. AKT, in turn, influences numerous downstream targets, including the mammalian target of rapamycin (mTOR). This pathway modulates the inflammatory response by regulating the production of pro-inflammatory cytokines and growth factors, essential for the initial phase of wound healing [42]. Additionally, AKT promotes the proliferation and migration of keratinocytes and fibroblasts, crucial for re-epithelialization and tissue formation, while mTORC1 enhances protein synthesis and cell growth, aiding in tissue repair and regeneration [43]. The pathway also supports angiogenesis by upregulating vascular endothelial growth factor (VEGF), ensuring an adequate blood supply to the healing tissue. Furthermore, mTOR influences the production and remodeling of the extracellular matrix (ECM), providing structural support to new tissue [44]. Activation of this pathway is essential for promoting angiogenesis. In human umbilical vein endothelial cells (HUVECs) exposed to high glucose conditions, there is a notable decrease in the expression of p-PI3K and p-AKT. Importantly, the angiogenic capacity and functionality of HUVECs can be significantly enhanced through the upregulation of the PI3K/AKT/eNOS pathway [45, 46]. These findings suggest that targeting the PI3K-AKT pathway represents a promising therapeutic strategy for treating DWs.
In this study, QCT was chosen as the intervention for DW model rats to investigate its mechanism of action and validate the outcomes derived from network pharmacology analysis. Our data show that treatment with QCT significantly improved the wound healing rate compared with DW model group. In addition, WB results demonstrated that QCT treatment significantly reduced the protein levels of the inflammatory cytokines IL-6, TNF-α, and MMP9 at early stage. This reduction validates the therapeutic effect of QCT on DWs, as previously mentioned, where elevated levels of MMP9 and IL-6 were observed in DW tissues. Compared to the control group, the DW group exhibited a significant downregulation in the protein levels of p-AKT, p-PI3K, and mTOR. Conversely, these protein levels were significantly elevated in the QCT treatment group. Moreover, histological analysis indicated that after QCT treating, significantly increased collagen fiber deposition and the presence of blood vessels and hair follicles were observed, accompanied with decreased in the production of pro-inflammatory cytokines.
Our study suggests that QCT can treat DWs through interactions with multiple target proteins. These molecules play crucial roles in the complex process of wound healing, and their dysregulation in diabetes contributes to the chronicity and impaired healing characteristic of DWs. The therapeutic process of QCT on DW primarily involves activating the PI3K-AKT signaling pathway and reducing the inflammatory response.
Conclusion
In this study, we used network pharmacology methods combined with molecular docking to identify therapeutic targets and pathways of QCT for treating DW. In view of our results, we identified 7 potential therapeutic targets (including MMP9, TNF-α, IL-6, EGFR, AKT1, SRC and JUN) of QCT in the treatment of DW. The in silico studies showed that QCT had better affinities with these targets. Furthermore, our results proved that QCT accelerated DW healing by reducing the activation of pro-inflammatory cytokines and regulating the PI3K-AKT-mTOR signaling pathway. Our study might provide theoretical basis for further investigation of QCT in DW treatment.
Acknowledgements
Data availability
All data used to support our findings are available from the corresponding author upon request.
Ethical considerations
The animal procedures in this research were approved by the Ethics Committee of China Medical University (Ethic number: CMU20241591). All applicable institutional and governmental regulations concerning the ethical use of animals were followed.
Author contributions
All authors contributed to the revision of the study. Dongyu Li and Zhe Zhang carried out the study and contributed to data collection and analyzing and animal experiments. Lei Wang contributed to manuscript writing. Yuxi Miao contributed to data analysis. Xuan Li contributed to manuscript corrections. All authors approved the version to be published.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kallikazaros IE. Diabetes mellitus: a sweet-and-sour disease. Hellenic journal of cardiology: HJC = Hellenike kardiologike epitheorese. 2013;54:153-4
2. Ramachandran A. Know the signs and symptoms of diabetes. The Indian journal of medical research. 2014;140:579-81
3. Ceriello A, Prattichizzo F. Variability of risk factors and diabetes complications. Cardiovascular diabetology. 2021;20:101
4. Holl J, Kowalewski C, Zimek Z, Fiedor P, Kaminski A, Oldak T. et al. Chronic Diabetic Wounds and Their Treatment with Skin Substitutes. Cells. 2021;10:655
5. Huang J, Yang R, Jiao J, Li Z, Wang P, Liu Y. et al. A click chemistry-mediated all-peptide cell printing hydrogel platform for diabetic wound healing. Nature communications. 2023;14:7856
6. Louiselle AE, Niemiec SM, Zgheib C, Liechty KW. Macrophage polarization and diabetic wound healing. Translational research: the journal of laboratory and clinical medicine. 2021;236:109-16
7. Sharifiaghdam M, Shaabani E, Faridi-Majidi R, De Smedt SC, Braeckmans K, Fraire JC. Macrophages as a therapeutic target to promote diabetic wound healing. Molecular therapy: the journal of the American Society of Gene Therapy. 2022;30:2891-908
8. Shorobi FM, Nisa FY, Saha S, Chowdhury MAH, Srisuphanunt M, Hossain KH. et al. Quercetin: A Functional Food-Flavonoid Incredibly Attenuates Emerging and Re-Emerging Viral Infections through Immunomodulatory Actions. Molecules (Basel, Switzerland). 2023;28:938
9. Deepika Maurya PK. Health Benefits of Quercetin in Age-Related Diseases. Molecules (Basel, Switzerland). 2022;27:2498
10. Li Y, Yao J, Han C, Yang J, Chaudhry MT, Wang S. et al. Quercetin, Inflammation and Immunity. Nutrients. 2016;8:167
11. Nusantoro AP, Kuntaman K, Perdanakusuma DS. Management of wounds in diabetes by administering allicin and quercetin in emulsion form as wound medicine in diabetic rat models. Journal of complementary & integrative medicine. 2024
12. Badhwar R, Mangla B, Neupane YR, Khanna K, Popli H. Quercetin loaded silver nanoparticles in hydrogel matrices for diabetic wound healing. Nanotechnology. 2021 32
13. Kant V, Jangir BL, Sharma M, Kumar V, Joshi VG. Topical application of quercetin improves wound repair and regeneration in diabetic rats. Immunopharmacology and immunotoxicology. 2021;43:536-53
14. Nogales C, Mamdouh ZM, List M, Kiel C, Casas AI, Schmidt H. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends in pharmacological sciences. 2022;43:136-50
15. Ru J, Li P, Wang J, Zhou W, Li B, Huang C. et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of cheminformatics. 2014;6:13
16. Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH. et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic acids research. 2019;47:D976-d82
17. Zhou Y, Zhang Y, Zhao D, Yu X, Shen X, Zhou Y. et al. TTD: Therapeutic Target Database describing target druggability information. Nucleic acids research. 2024;52:D1465-d77
18. Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): A Knowledgebase of Human Genes and Genetic Phenotypes. Current protocols in bioinformatics. 2017;58:1.2.1-2.12
19. Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M. et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database: the journal of biological databases and curation. 2015;2015:bav028
20. Pinna C, Morazzoni P, Sala A. Proanthocyanidins from Vitis vinifera inhibit oxidative stress-induced vascular impairment in pulmonary arteries from diabetic rats. Phytomedicine: international journal of phytotherapy and phytopharmacology. 2017;25:39-44
21. Fu J, Huang J, Lin M, Xie T, You T. Quercetin Promotes Diabetic Wound Healing via Switching Macrophages From M1 to M2 Polarization. The Journal of surgical research. 2020;246:213-23
22. Li D, Ma J, Wang L, Xin S. Apigenin Prevent Abdominal Aortic Aneurysms Formation by Inhibiting the NF-κB Signaling Pathway. Journal of cardiovascular pharmacology. 2020;75:229-39
23. Zhang H, Zhang M, Wang X, Zhang M, Wang X, Li Y. et al. Electrospun multifunctional nanofibrous mats loaded with bioactive anemoside B4 for accelerated wound healing in diabetic mice. Drug delivery. 2022;29:174-85
24. Salehi B, Venditti A, Sharifi-Rad M, Kregiel D, Sharifi-Rad J, Durazzo A. et al. The Therapeutic Potential of Apigenin. International journal of molecular sciences. 2019;20:1305
25. Anand David AV, Arulmoli R, Parasuraman S. Overviews of Biological Importance of Quercetin: A Bioactive Flavonoid. Pharmacogn Rev. 2016;10:84-9
26. Abid HMU, Hanif M, Mahmood K, Aziz M, Abbas G, Latif H. Wound-Healing and Antibacterial Activity of the Quercetin-4-Formyl Phenyl Boronic Acid Complex against Bacterial Pathogens of Diabetic Foot Ulcer. ACS omega. 2022;7:24415-22
27. Ahmed OM, Mohamed T, Moustafa H, Hamdy H, Ahmed RR, Aboud E. Quercetin and low level laser therapy promote wound healing process in diabetic rats via structural reorganization and modulatory effects on inflammation and oxidative stress. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;101:58-73
28. Yan L, Vaghari-Tabari M, Malakoti F, Moein S, Qujeq D, Yousefi B. et al. Quercetin: an effective polyphenol in alleviating diabetes and diabetic complications. Critical reviews in food science and nutrition. 2023;63:9163-86
29. Hirano T. IL-6 in inflammation, autoimmunity and cancer. Int Immunol. 2021;33:127-48
30. Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2011;1813:878-88
31. Fu K, Zheng X, Chen Y, Wu L, Yang Z, Chen X. et al. Role of matrix metalloproteinases in diabetic foot ulcers: Potential therapeutic targets. Frontiers in pharmacology. 2022;13:1050630
32. Ayuk SM, Abrahamse H, Houreld NN. The Role of Matrix Metalloproteinases in Diabetic Wound Healing in relation to Photobiomodulation. Journal of Diabetes Research. 2016;2016:1-9
33. Lima MH, Caricilli AM, de Abreu LL, Araujo EP, Pelegrinelli FF, Thirone AC. et al. Topical insulin accelerates wound healing in diabetes by enhancing the AKT and ERK pathways: a double-blind placebo-controlled clinical trial. PLoS One. 2012;7:e36974
34. Nirenjen S, Narayanan J, Tamilanban T, Subramaniyan V, Chitra V, Fuloria NK. et al. Exploring the contribution of pro-inflammatory cytokines to impaired wound healing in diabetes. Front Immunol. 2023;14:1216321
35. Lin S, Wang Q, Huang X, Feng J, Wang Y, Shao T. et al. Wounds under diabetic milieu: The role of immune cellar components and signaling pathways. Biomedicine & Pharmacotherapy. 2023;157:114052
36. Hsieh M-CW, Wang W-T, Lin C-Y, Kuo Y-R, Lee S-S, Hou M-F. et al. Stem Cell-Based Therapeutic Strategies in Diabetic Wound Healing. Biomedicines. 2022;10:2085
37. Espada J, Martín-Pérez J. An Update on Src Family of Nonreceptor Tyrosine Kinases Biology. Int Rev Cell Mol Biol. 2017;331:83-122
38. Kappelmann M, Bosserhoff A, Kuphal S. AP-1/c-Jun transcription factors: Regulation and function in malignant melanoma. European Journal of Cell Biology. 2014;93:76-81
39. Hoke GD, Ramos C, Hoke NN, Crossland MC, Shawler LG, Boykin JV. Atypical Diabetic Foot Ulcer Keratinocyte Protein Signaling Correlates with Impaired Wound Healing. J Diabetes Res. 2016;2016:1586927
40. Li T, Wang G. Computer-aided targeting of the PI3K/Akt/mTOR pathway: toxicity reduction and therapeutic opportunities. International journal of molecular sciences. 2014;15:18856-91
41. He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW. et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:425
42. Roy T, Boateng ST, Uddin MB, Banang-Mbeumi S, Yadav RK, Bock CR. et al. The PI3K-Akt-mTOR and Associated Signaling Pathways as Molecular Drivers of Immune-Mediated Inflammatory Skin Diseases: Update on Therapeutic Strategy Using Natural and Synthetic Compounds. Cells. 2023;12:1671
43. Teng Y, Fan Y, Ma J, Lu W, Liu N, Chen Y. et al. The PI3K/Akt Pathway: Emerging Roles in Skin Homeostasis and a Group of Non-Malignant Skin Disorders. Cells. 2021;10:1219
44. Yuan Z, Li Y, Zhang S, Wang X, Dou H, Yu X. et al. Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments. Mol Cancer. 2023;22:48
45. Hu Y, Tao R, Chen L, Xiong Y, Xue H, Hu L. et al. Exosomes derived from pioglitazone-pretreated MSCs accelerate diabetic wound healing through enhancing angiogenesis. Journal of Nanobiotechnology. 2021;19:150
46. Chen J, Huang Y, Hu X, Bian X, Nian S. Gastrodin prevents homocysteine-induced human umbilical vein endothelial cells injury via PI3K/Akt/eNOS and Nrf2/ARE pathway. Journal of Cellular and Molecular Medicine. 2020;25:345-57
Author contact
![]() Corresponding author: Dongyu Li, Nanjingbei 155 Street, Shenyang, Liaoning, 110001, China. Email address: dyli0512edu.cn.
Corresponding author: Dongyu Li, Nanjingbei 155 Street, Shenyang, Liaoning, 110001, China. Email address: dyli0512edu.cn.

 Global reach, higher impact
Global reach, higher impact