Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(14):2725-2744. doi:10.7150/ijms.98778 This issue Cite
Review
The Biomarkers in Extreme Longevity: Insights Gained from Metabolomics and Proteomics
1. Zhanjiang Key Laboratory of Human Microecology and Clinical Translation Research, the Marine Biomedical Research Institute, College of Basic Medicine, Guangdong Medical University, Zhanjiang, Guangdong, 524023, China.
2. Guangdong Key Laboratory of Age-Related Cardiac and Cerebral Diseases, Affiliated Hospital of Guangdong Medical University, Zhanjiang, 524000, China.
3. CNTTI of the Institute of Life Sciences & Anesthesia Department of the Second Affiliated Hospital, Chongqing Medical University, Chongqing, 400016, China.
4. Basic Medicine Research and Innovation Center for Novel Target and Therapeutic Intervention, Chongqing Medical University, Chongqing, 400016, China.
* These authors contributed equally to this work.
Received 2024-5-22; Accepted 2024-9-10; Published 2024-10-21
Abstract
The pursuit of extreme longevity is a popular topic. Advanced technologies such as metabolomics and proteomics have played a crucial role in unraveling complex molecular interactions and identifying novel longevity-related biomarkers in long-lived individuals. This review summarizes key longevity-related biomarkers identified through metabolomics, including high levels of omega-3 polyunsaturated fatty acids (PUFAs), short-chain fatty acids (SCFAs) and sphingolipids, as well as low levels of tryptophan. Proteomics analyses have highlighted longevity-related proteins such as apolipoprotein E (APOE) and pleiotrophin (PTN), along with lower S-nitrosylated and higher glycosylated proteins found from post-translational modification proteomics as potential biomarkers. We discuss the molecular mechanisms that could support the above biomarkers' potential for healthy longevity, including metabolic regulation, immune homeostasis maintenance, and resistance to cellular oxidative stress. Moreover, multi-omics studies of various long-lived cohorts are encompassed, focusing on how the integration of various omics technologies has contributed to the understanding of longevity. This comprehensive review aims to provide new biological insights and pave the way for promoting health span.
Keywords: Biomarkers, Longevity, Metabolomics, Proteomics, Multi-omics
1. Introduction
The ongoing trend of global aging indicates that human life expectancy will continue to rise [1]. The average life expectancy in 2019 was 72.6 years old and is expected to rise to 77.1 years old in 2050 according to the World Population Prospects (United Nations, 2019). However, the elderly may be affected by age-related diseases, so the central problem of aging is to explain why the organism cannot adapt to these endogenous stimuli, such as oxidative stress and molecular hyperfunction [2]. Instead, we believe that a central question of longevity is to explain why organisms are able to delay or avoid adverse stimuli. Hence, extending the human lifespan while achieving healthy aging to longevity simultaneously are the major goals of global aging and anti-aging research. The healthy elderly over 90 years old, who are the representatives of extreme longevity, have reached the limit of human longevity while largely avoiding and postponing major age-related diseases, thereby making them the most successful examples of healthy aging [3, 4]. As individuals who are deemed most likely to achieve extreme longevity, the offspring of healthy longevity have the unique interests of researchers [5, 6]. Therefore, the identification of potential longevity-related biomarkers in centenarians and their offspring and the exploration of the complex associations among biomarkers are critical for extending lifespan.
During the journey to seeking longevity biomarkers, metabolites and proteins are considered the ultimate effectors for longevity phenotype, besides genes and transcription factors associated with genetic factors [7, 8]. Metabolites, such as fatty acids, amino acids and vitamins, perform diverse functions including energy metabolism and signaling. Proteins, as the products of gene transcription and translation, affect physiological functions by different protein forms, such as cell membrane structure and enzymes. Moreover, post-translational modifications of proteins also have a significant impact on molecular functions. Hence, metabolites and proteins uncovered through investigations of centenarians and their offspring can be regarded as potential longevity-related biomarkers.
Metabolomics and proteomics, as advanced high-throughput omics techniques, are considered the most promising tools for identifying biomarkers [9-12]. Metabolomics typically includes untargeted and targeted metabolomics, which is widely utilized to identify metabolites produced in response to all kinds of challenges [13]. Proteomics mainly includes both general and post-translational modification (PTM) proteomics, which is applicable to identify proteins regulated by genetic and external stimuli [14]. Furthermore, the multi-omics integration of metabolomics and proteomics with genomics, transcriptomics, or microbiomics will provide more precise longevity-related biomarkers in different biomolecular levels. Here, we briefly reviewed longevity-related biomarkers, focusing on metabolomics, proteomics and multi-omics in long-live populations. Additionally, we also discuss the molecular mechanisms that underlying these biomarkers and their role in promoting longevity. We hope to deepen our understanding of the mechanisms behind biomarkers influencing individual aging, providing a theoretical basis to achieve longevity.
2. Metabolomics studies associated with longevity
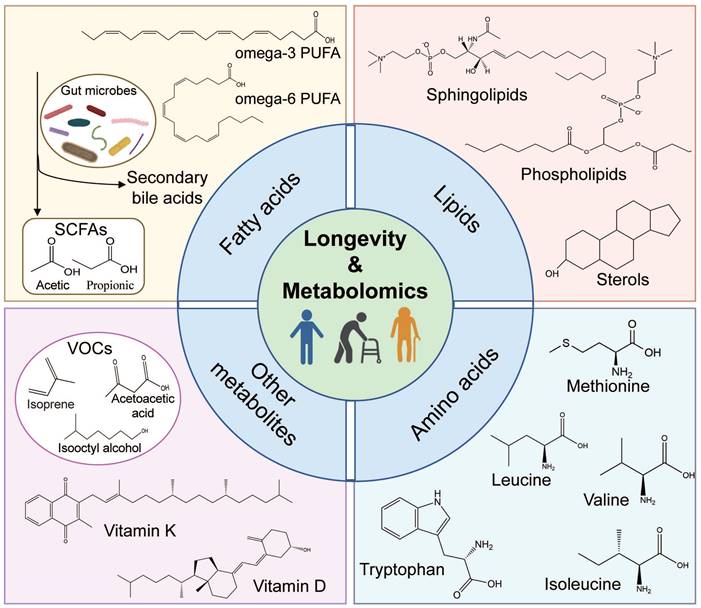
Metabolomics comprises untargeted metabolomics, involving the universal detection of small molecules, and targeted metabolomics, which selectively detects specific metabolites like lipids or amino acids [15, 16]. In this section, we briefly describe the longevity-related biomarkers discovered by metabolomics, including fatty acids, lipids, amino acids, and other metabolites (Figure 1).
2.1 Fatty acids
Fatty acids are classified as unsaturated fatty acids and saturated fatty acids. Unsaturated fatty acids, particularly monounsaturated fatty acids (MUFAs) and polyunsaturated fatty acids (PUFAs), have gradually become a focus of longevity research due to their susceptibility to lipid peroxidation compared to saturated fatty acids. Furthermore, gut-bacteria-derived fatty acids, such as bile acids and short-chain fatty acids, also frequently have been drawn focus in longevity studies. In this section, we will focus on the impact of PUFAs and gut-bacteria-derived fatty acids in long-lived populations.
2.1.1 PUFA
PUFAs can be designated as omega-3 PUFAs and omega-6 PUFAs. Eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) in omega-3 PUFA family, and arachidonic acid (AA) in omega-6 PUFA family played critical roles in maintaining the integrity of cell structures, producing signaling molecules, and regulating a variety of biological pathways [17].
In the omega-3 PUFA family, EPA and DHA frequently appeared in the study of healthy aging and longevity, which are famous for their potential benefits in reducing triglyceride levels and exerting anti-inflammatory effects [18]. EPA and DHA have been reported to be beneficial in preventing cardiovascular disease, cancer, and diabetes in the elderly [19]. Xyda et al. [20] found that plasma levels of diacylglycerols, phospholipids, and triacylglycerols (TG) decreased dramatically in the elderly taking EPA and DHA supplements. The possible effects of EPA and DHA supplementation were the reduction in total TG, representing a decrease in very low-density lipoprotein (VLDL) particles, and a modest increase in high-density lipoprotein (HDL) cholesterol.
As a member of omega-6 PUFA, AA appeared a lot of attention in longevity studies. Among the metabolites of AA, the serum levels of 8(9)-epoxyeicosatrienoic acid [8(9)-EET] was reported to elevate considerably in centenarians, as a possible result of activation of cell detoxification processes. What's more, the serum levels of 11,12-dihydroxyeicosatrienoic acid (11,12-DHET), linoleic acid, 9-hydroxyoctadecadienoic acid (9-HODE), and 9-oxo-hydroxyoctadecadienoic acid (9-oxo-HODE) were lower in centenarians, which might be a phenomenon of good antioxidant responses [21]. Moreover, the EPA/AA ratio seems to have important physiological implications for healthy aging rather than a single EPA level. In the Longevity Sciences-Longitudinal Study of Aging, greater EPA/AA ratios were inversely linked with all-cause mortality, whereas higher serum EPA levels alone were not substantially negatively correlated with mortality [22]. The EPA/AA ratio might be a protective factor for longevity through influences on membrane properties, cell signaling, and reduced cardiac events [23]. Furthermore, the EPA/AA ratio was highest in healthy centenarians, compared to healthy controls and acute myocardial infarction controls [24]. Hence, the EPA/AA ratio could be a potential predictor against major age-related disorders.
Schematic representation of longevity-related biomarkers. The schematic diagram shows the following four main longevity-related biomarkers: fatty acids, lipids, amino acids, and other metabolites. (1) fatty acids: omega-3/6 polyunsaturated fatty acids (PUFAs), secondary bile acids, and short-chain fatty acids (SCFAs, which are derived by gut bacteria utilizing vegetables and fruits); (2) lipids: phospholipids, sphingolipids, and sterols; (3) amino acids: essential and non-essential amino acids, including the branched-chain amino acids (BCAAs); (4) other metabolites: volatile organic compounds (VOCs) and vitamin (such as vitamin D/K).
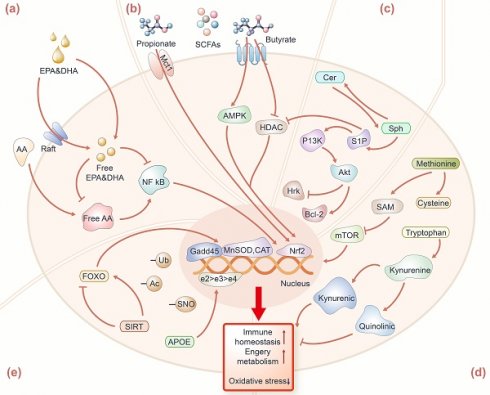
As to why EPA and DHA can affect human longevity, scientists have the following conjecture that it is generally believed that they have the regulation of the immune function on cells. The impact of EPA, DHA, AA, and their metabolites in omega-3 or 6 fatty acids are the regulation of the immune function on cells. EPA and DHA, acting as activators for anti-inflammatory transcription factors [25, 26], can compete with AA to bind to substrates in the enzymatic pathways catalyzed by cyclooxygenase (COX) and lipoxygenase (LOX), thereby inhibiting the conversion of AA into pro-inflammatory molecules such as prostaglandins and leukotrienes (Figure 2a). Studies suggest that the antioxidant properties of EPA and DHA are mediated by activating nuclear factor erythroid 2-related factor 2 (Nrf2), involving the upregulation of hemoglobin oxygenase 1 (HO-1) to protect the brain from ischemic damage [27]. Therefore, effective inducers of Nrf2 activation can be considered as an effective measure to prevent inflammation-mediated diseases. Simultaneously, the increasing EPA and DHA levels in circulation facilitate the aggregation of signaling membrane proteins and lipids to form lipid rafts [28, 29], and then reduces the transmission efficiency of cellular inflammatory signals and the activation of pro-inflammatory transcription factors such as Nuclear factor kappa B (NF-κB) [26]. Additionally, omega-3 or 6 fatty acids can regulate the balance of gut microbes, enhancing fatty acids absorption, utilization, and biotransformation [30, 31]. In conclusion, changes in human PUFAs (e.g., EPA, DHA, and AA) and the ratio of EPA/AA may be potential biomarkers of longevity.
2.1.2 Gut-bacteria-derived fatty acids
Gut bacteria have been proven to involve human health and impact longevity by fatty acid metabolic pathway [32-34]. Of these, bile acids and short-chain fatty acids (SCFAs) are famous metabolites derived from gut bacteria. Bile acids include primary and secondary bile acids, and the latter are the products formed by decomposition of primary bile acids by gut bacteria. Regarded bile acid metabolites, the levels of those (such as iso-, 3-oxo-and isoallo-lithocholic acid (LCA)) generated from Odoribacteraceae in centenarians' feces were significantly increased, and relatively low in the elderly and young controls [33]. The unique biosynthetic pathway for isoallo-LCA is that 3-oxo-Δ4-LCA conversion to isoallo-LCA is mediated by 5AR (3-oxo-5-alpha-steroid 4-dehydrogenase) and 3β-HSDH (short-chain dehydrogenase). Subsequently, Odoribacteraceae St21 was administered to mice infected with Clostridioides difficile (Gram-positive pathogens), resulting in reduced intestinal C. difficile numbers and enhanced production of isoalloLCA to defend against infection, a mechanism that could potentially delay aging. Furthermore, LCA also were enriched in nonagenarians and centenarians' plasma, which can be detoxified to form lithocholic acid sulfate via gut bacteria such as Aeromonas veronii and Butyrivibrio crossotus [35]. Overall, gut microbe alterations and bile acid metabolic differences will affect gut homeostasis.
Mechanisms of longevity-related metabolites and proteins for potentially fostering healthy longevity. (a) EPA, DHA, and AA are resistant to oxidative stress and inflammation mainly by maintaining the fluidity of the cell membrane, promoting lipid raft assembly, and further inhibiting the activation of NF-κB, and Nrf2 pathways in the cell nucleus. (b) Propionate binds to FFAR3; butyrate inhibits HDAC and promotes the expression of Mct1, all of which further mediate the Nrf2 pathway to maintain immune homeostasis. (c) S1P can reduce the gene expression of pro-apoptotic Hrk protein through the PI3K-Akt pathway and upregulate the expression of the Bcl-2 family to counteract the pro-apoptotic effect of Cer. (d) Methionine enters the methionine metabolism by being converted into SAM, and further inhibiting the mTOR signaling activation. Tryptophan produces kynurenic acid with neuroprotection through the kynurenine metabolism pathway. (e) APOE, FOXO, and SIRT regulate autophagy, DNA repair, energy metabolism, and immune homeostasis by affecting the gene expression of Gadd45, MnSOD, and CAT. Protein modifications include S-nitrosation (SNO), acetylation (Ac) and ubiquitination (Ub), which play an important role in the modification of longevity-related proteins.
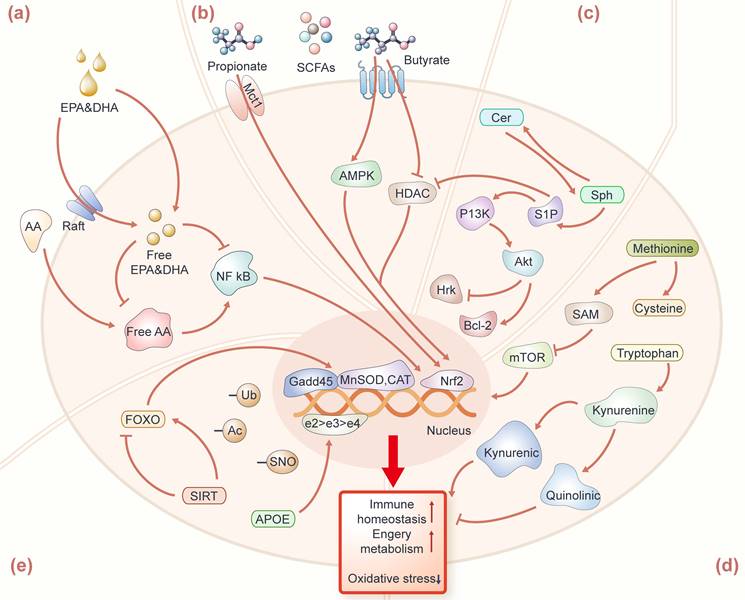
SCFAs are the major products of dietary fiber fermentation by gut bacteria. The prevalent SCFAs in humans (76-95 yrs) are mainly acetic acid, propionic acid, and butyric acid [36]. The levels of these prevalent SCFAs in fecal were decreased in the elderly (>80 yrs), compared with young adults (50-80 yrs) [37]. In nonagenarians and centenarians, these prevalent SCFAs mentioned above were over-represented with respect to their children (50-79 yrs), which were produced by Parabacteroides and further affected gut homeostasis [35]. In addition, branched SCFAs such as isobutyric and isovaleric were elevated in centenarians [33]. Existing studies have elucidated the molecular mechanisms through which SCFAs sustain human health, predominantly by modulating the integrity of cellular barriers, such as the intestinal barrier and the blood-brain barrier (Figure 2b). Among these, propionate and butyrate positively influence longevity by promoting the repair and regeneration of intestinal cells, maintaining intestinal mucosa integrity, and regulating the activity of the immune system [38]. Propionate can bind to the free fatty acid receptor 3 (FFAR 3) in the human brain endothelial cells and then protect the blood-brain barrier from oxidative stress through the Nrf2 pathway [39]. Butyrate can mediate protection against endothelial dysfunction in mice by inhibiting protein deacetyase (HDAC) -mediated activation of Nrf2 [40]. The newest article found that butyrate can also alleviate the imbalance of muscle satellite cell homeostasis caused by antibiotic-induced intestinal microflora disorders, and promote the expression of monocarboxylate transporter 1 (Mct1) to regulate muscle satellite cell homeostasis [41]. Thus, propionate and butyrate are potential biomarkers for centenarians to achieve health and longevity, and exploring more other SCFAs derived from salutary microorganisms could be beneficial to the discovery of novel longevity-related biomarkers.
Apart from the fatty acids discussed above, some intermediate products in fatty acid oxidation also play important roles in health and longevity, such as β-hydroxybutyrate [42]. Studies have shown that mice fed a high-fat diet and supplemented with the ketone body β-hydroxybutyrate displayed improvements in metabolic health and memory. These results suggest that dietary supplementation with β-hydroxybutyrate may prove to be an effective intervention for the future treatment of age-related dysfunction and further promote healthy longevity [43]. Therefore, the downstream products of fatty acids are also worthy of attention in promoting healthy longevity.
2.2 Lipids
Lipids are a broad group of organic compounds which include glycerol, fatty acids, phospholipids, sterols, and others. They exhibit diverse structures, often comprising one or more fatty acid molecules bound to other compounds. Other lipids besides fatty acids have also played an important role in longevity. This part primarily discusses lipids other than fatty acids. Lipidomics is one of the targeted metabolomics approaches, which enables the extraction and optimization of lipid molecule separation based on their lipophilic properties. Compared to untargeted metabolomics, lipidomics mainly provides a more precise detection method for some specific lipid molecules (such as phospholipids, sphingolipids, sterols, etc.) (Table 1).
Phospholipids, sphingolipids, and sterols are common types of lipids. (1) Phospholipids include phosphatidylcholine (PC), phosphatidylethanolamine (PE), ether phosphatidylcholine (PC-O), lysophosphatidylcholine (LPC), phosphatidylcholine (LPE), which maintain membrane fluidity. (2) Sphingolipids, such as sphingomyelins (SMs) and ceramides (Cers), are ubiquitous building blocks of cell membranes, which are involved in cell biological processes [44]. (3) Sterols play crucial roles in both cell proliferation as essential components and as significant signaling molecules, including cholesterol, steroids, estrogen and cholesterol ester (CEs) [45]. The species and molecular composition of lipids influence cellular distribution, metabolism and subsequently impact on cellular aging.
The abundance of lipids species in centenarians is determined by the number of carbon atoms and double bonds of lipids species. Pradas et al. analyzed plasma ether lipid profiles of centenarians, elderly and young in Spain, and identified lipids phenotype with exceptional human longevity [46]. The results revealed that fifteen LPC lipid species, such as LPC(O-24:0) and LPC(O-24:1) with lower carbon atomic numbers and double bonds in centenarians were significantly increased and predominated the presence in alkyl form. Jové's team observed an increase in the levels of phospholipids—including PS (40:3), PC (40:5), PE (33:3 34:2), LPC (18:1), LPE (24:1), and CEs—among centenarians and adults compared to other groups [47]. Similarly, the concentration of most unsaturated diacyl PC and LPC species rose significantly among centenarians and adults [21]. Collino et al. found that the concentrations of tiny PC, LPC, and PC-O remained unchanged until age 70 and underwent significant changes with longer carbon chains and double bond numbers in centenarians compared to the elderly and the young [48]. It is unclear whether these compositional differences are benefits or drawbacks for longevity.
Summary of lipid species in longevity associated study.
| Lipids | Populations | Group | Sample | Analyticaltechnique | Main findings | Year | Reference | |
|---|---|---|---|---|---|---|---|---|
| Phospholipids | Centenarians | 143 centenarians; aged; young | Serum | LC-MS/MS | PC, PC-O ↑ in centenarians and adults, compared to aged | 2013 | [21] | |
| PC, LPC, PC-O ↓ in centenarians, unchanging until 70 aged | ||||||||
| 98 centenarians; 196 aged | Serum | 1H-NMR | PC, polyunsaturated PC-O, LPC, PE, PI ↑ in centenarians, compared to aged | 2014 | [48] | |||
| saturated PC-O ↓ in centenarians, compared to elderly | ||||||||
| 27 centenarians; 31 aged, and 31 adults | Plasma | LC-MS/MS | PC, LPC, PE, LPE, PG, LPG, PA, PS ↑ in centenarians and adults compared to aged | 2017 | [47] | |||
| LPE, LPI, PA, PG, PS ↓ in centenarians and adults, compared to aged | ||||||||
| 25centenarians; 22 aged, and 21adult. | Plasma | LC-MS/MS | total ether lipids, alkyl-PE, alkenyl-lipids/PE ↓ with age | 2019 | [46] | |||
| alkyl-PC/LPC, alkenyl-PC/LPC ↑ in centenarians, compared to adults and aged | ||||||||
| Elderly | 143 centenarians; aged; young | Serum | LC-MS/MS | PC ↑ with age, LPC ↓ with age | 2013 | [21] | ||
| offspring of nonagenarians; controls | Plasma | UPLC-MS | PC-O ↑ in offspring of nonagenarians, compared to controls | 2013 | [29] | |||
| polyunsaturated PE ↓ in offspring of nonagenarians, compared to controls | ||||||||
| 150 subjects :30-49, 50-59, 60-79, and 90-99 years old | Plasma | LC-MS/MS | PC, PS ↓ with age | 2016 | [47] | |||
| 374 males (61.2±6.9); 838 females (60.7±6.6) | Plasma | LC-MS/MS | 5 PC, 1 LPC, 3 PE ↑ with age; 3 PC, 1 LPC, 1 PE ↓ with age | 2019 | [55] | |||
| Sphingolipids | Centenarians | 143 centenarians; aged; young | Serum | LC-MS/MS | SM, SM-OH ↓ in centenarians, compared to adults and aged | 2013 | [21] | |
| 98 centenarians; 196 aged | Serum | 1H-NMR | SM, Cer ↑ in centenarians, compared to aged | 2014 | [48] | |||
| 27 centenarians; 31 aged, and 31 adults | Plasma | LC-MS/MS | Cer ↑ in centenarians and adults, compared to aged | 2017 | [47] | |||
| Cer, GM3, GlcCer ↓ in centenarians and adults, compared to aged | ||||||||
| Sphingolipids | Centenarians | 25 centenarians; 22 aged, and 21adults | Plasma | LC-ESI-QQQ MS/MS | HexCer, Hex3Cer, GM3↑in centenarians, compared to aged and adults | 2022 | [49] | |
| Sulfatides, dhCer, SM (d18:0/22:0) ↓in centenarians, compared to aged and adults | ||||||||
| 15 centenarians; 15 aged, and 15 adults | Serum | Glyco/Sphingolipid LC-MS/MS | HexCer, Cer, GM3, SM ↑ in centenarians, compared to aged and adults | 2022 | [50] | |||
| SM, S1P↓in centenarians, compared to aged | ||||||||
| dhSM, dhCer, SM ↓ in centenarians, compared to aged and adults | ||||||||
| Elderly | offspring of nonagenarians; controls | Plasma | UPLC-MS | SM↑in offspring of nonagenarians, compared to controls | 2013 | [29] | ||
| 374 males (61.2±6.9); 838 females (60.7±6.6) | Plasma | LC-MS/MS | 24 SM ↑ with age, 1 SM ↓ with age | 2019 | [55] | |||
| Sterols | Centenarians | 27 centenarians; 31 aged, 31 adults | Plasma | LC-MS/MS | CE ↑ in centenarians and adults, compared to aged | 2017 | [47] | |
| Elderly | 150 subjects: 30-49, 50-59, 60-79, and 90-99 years old | Plasma | LC-MS/MS | 15-keto-prostaglandin F2α ↓ with age | 2017 | [47] | ||
| 374 male (61.2±6.9); 838 female (60.7±6.6) | Plasma | LC-MS/MS | 2 Steroids ↑ with age, 29 Steroids ↓ with age | 2019 | [55] | |||
↓decreased levels,↑increased levelsPC Phosphatidylcholine, PE phosphatidylethanolamine, SM sphingomyelin, LPC lysophosphatidylcholine, LPE lysophosphatidylethanolamine, LPG lysophosphatidylglycerol, PA phosphatidic acid, PG phosphatidylglycerol, PS phosphatidylserine, LPI lysophosphatidylinositol, PC-O ether phosphatidylcholine, CE cholesteryl ester. Specific lipids have been named and classified according to Lipid Maps (http://www.lipidmaps.org)
Nevertheless, it can be inferred that the lipid species composition in centenarians has undergone remodeling to enhance resistance against lipid peroxidation. For example, serum PC-O species are positively correlated to the HDL circulating level, supporting their role as antioxidants preventing lipoprotein oxidation [21, 46].
Some sphingolipids in long-lived individuals have also attracted the attention of scientists. Sphingolipids are categorized as structural or signal sphingolipids based on location and function. Structural sphingolipids are found primarily in cell membranes, including the myelin sheath of neurons, where they provide support and protection to the cells. Signal sphingolipids involve functions in the process of cell signal transmission. A recent study based on targeted sphingolipidomic in plasma found that centenarians have a significant increase in sphingolipid species, such as monohexosylceramides (HexCer), trihexosylceramides, and gangliosides (GM), which have structural roles [49]. Barbacini et al. also found similar levels of sphingolipids in the serum of centenarians (increased in HexCer and GM) [50]. Sphingosines, sphingosine-1-phosphate (S1P), ceramide-1-phosphate, and Cers with signaling roles remained unaltered among cohorts. Additionally, very long-chain ceramides like C24:0 and the ratio to the long-chain ceramide C16:0 (i.e., C24:0/C16:0) have been found to be associated with better cardiovascular health, lower cardiovascular event rates as well as lower all-cause mortality [51]. Therefore, structural glycosphingolipids with signaling roles may be key factors in attaining longevity. Some studies have found that the increase of S1P and the decrease of Cers are conducive to neuronal stress resistance (Figure 2c). S1P can counteract the pro-apoptotic effects of ceramides by reducing oxidative stress, decreasing the gene expression of pro-apoptotic Harakiri (Hrk) protein, and upregulating the expression of the B-cell lymphoma 2 (Bcl-2) family of pro-apoptotic proteins [52], which appear to influence cell metabolism, stress response, and immune status through the PI3K/Akt pathway [53, 54]. Enzymes involved in sphingolipid metabolism in centenarians such as sphingomyelin phosphodiesterase 3 (SMPD3) have been verified to be over-expressed at PBMC mRNA and protein levels. This study revealed that the synthesis of Cers in centenarians is obtained by SM degradation. Overall, targeted sphingolipidomic profiling of centenarians enhances our understanding of longevity mechanisms that warrant further investigation.
Since sterols are related to hormones, the study of the large cohorts has proven that sterols are affected by sex [55]. Sterols play an important role in the gender differences in longevity among centenarians. Studies have shown that estrogen can influence the immune and antioxidant systems, as well as help maintain telomere length [56-58]. Baseline data from the China Hainan Centenarian Cohort Study indicated that compared to elderly women in the perimenopausal and postmenopausal stages, female centenarians have a non-protective association between estradiol and progesterone levels above a certain threshold and all-cause mortality [59]. Therefore, in lipidomics studies of longevity cohorts, it is necessary to further evaluate hormone levels related to gender.
2.3 Amino acid metabolites
Amino acids are important signaling molecules to regulate the physiological processes in humans. Essential amino acids such as methionine and tryptophan, along with non-essential amino acids like branched-chain amino acids (BCAAs), have garnered attention in the study of longevity.
Methionine is an essential amino acid that the body cannot generate, and is one of the four common sulfur-containing amino acids (methionine, cysteine, homocysteine, and taurine). Methionine metabolism pathway has been proven to upregulate in long-lived individuals [60, 61], which involved various biochemical reactions, such as transmethylation and transsulfuration [62]. Methionine enters the methionine metabolism by being converted into S-adenosylmethionine (SAM), a universal donor for methyl transfer reactions, thus participating in the methylation process of a variety of biological molecules including DNA, RNA and proteins [63], hence SAM plays a crucial role in maintaining vital biological processes in the elderly (Figure 2d). SAM is converted to homocysteine under the action of glycine N-methyltransferase (Gnmt) and S-adenosylhomocysteine hydrolase (AHCY), and then homocysteine is converted back to methionine under the action of methionine synthase, completing the cycle of methionine. Here, we observed a unique pattern of methionine metabolism in the blood circulation of centenarians compared to other controls. Plasma methionine in centenarians did not differ between elderly and young, while methionine metabolites (such as cystathionine, cysteine and taurine) were significantly higher than the other two groups, while the homocysteine levels of centenarians were significantly lower than the other two groups, which may be beneficial to maintaining healthy lifespan and reducing the risk of chronic diseases in centenarians [61]. Understanding the unique patterns of methionine metabolism observed in centenarians prompts further investigation into its implications across different species, particularly for mice as the model animal. Methionine restriction in mice was found to reduce serum IGF-I, insulin, glucose, thyroid hormone and inflammation levels, and increase MIF (macrophage migration inhibition factor) levels in hepatocytes, leading to resistance to multiple diseases and slower aging [64]. Homocysteine can increase the level of oxidative stress and activate the TOR signaling pathway, thereby accelerating the aging process [65, 66]. Thus, methionine restriction and lowering homocysteine levels contribute to maintaining the best physiological state in centenarians. Finally, methionine intake may be decreased by vegetarian diets, which possibly promoted health benefits [67], and the dietary habits of centenarians are also worth further investigating.
Tryptophan, an essential amino acid, as the precursor of the neurotransmitters-serotonin and melatonin, plays a crucial role in human health [68]. In a plasma metabolomics study, examining centenarians, nonagenarians, elderly in longevity region and controls in non-longevity region, the results indicated that the tryptophan biosynthesis pathways were significantly enriched in centenarians [69]. Mota-martorell et al. discovered decreased levels of tryptophan in centenarians' plasma [61], and this result is consistent with previous studies of declining tryptophan levels in centenarians [21]. Additionally, tryptophan was also decreased in centenarians' urine compared to the elderly [70]. As previously reported, a low level of tryptophan is considered to be related to the rise of chronic low-grade inflammatory disorders and immune activation in centenarians [71, 72]. Tryptophan can be metabolized to kynurenine, which further produces several metabolites, including kynurenic acid and quinolinic acid. It has been reported that frail elderly have lower kynurenic acid compared to quinolinic acid [73]. Studies have shown that kynurenic acid increases energy utilization by activating G protein-coupled receptor Gpr35 [74] (Figure 2d). The elevation of kynurenic acid and Gpr35 enhance cellular respiration, and increase the levels of Rgs14 in adipocytes, which leads to enhanced energy homeostasis. Quinolinic acid has been linked to neurodegenerative diseases and has been shown to impair mitophagy, thereby promoting microglial senescence, thereby contributing to the aging process of the brain [75]. Thus, the tryptophan and its metabolism play a crucial role in maintaining health status in centenarians.
The catabolism of branched-chain amino acids (such as valine, leucine, and isoleucine) is reported to be a conserved regulator of physiological aging, which can promote aging though activating the target of rapamycin/ribosomal protein S6 kinase (TOR/S6K) signaling and the target of rapamycin complex 1 (TORC1) pathway [76, 77]. In a study based on a longevity cohort in Guangxi, China [78], the oldest-old group always had relatively low branched-chain amino acid levels. In centenarians, lower valine levels were observed with respect to adults and aged individuals [61]. Studies have found that high levels of BCAAs in the blood are associated with an increased risk of cardiovascular diseases and neurodegenerative diseases. A Mendelian randomization study using data from the UK Biobank and other databases found that individuals genetically predisposed to higher circulating levels of BCAAs had a greater risk of peripheral arterial disease and stroke [79, 80]. Moreover, there is evidence from a bidirectional Mendelian randomization study that points to Alzheimer's disease being associated with decreased BCAAs levels [81], while leucine has been demonstrated to promote AD via a mTOR-dependent mechanism [82]. Furthermore, the restriction of isoleucine has been shown to effectively reduce obesity and improve glucose tolerance, reversing the impact of age on several molecular markers of aging such as phosphatidylglycerol, promoting healthy lifespan and extending longevity in young or adult mice [83]. In other report, isoleucine restriction also promotes upregulation of fatty acid metabolism and downregulation of immune pathways in male mice, suggesting potential effects of isoleucine restriction on metabolism and inflammation in healthy aging [84]. In conclusion, lower levels of these amino acids may contribute to longevity, and the underlying mechanisms should be further studied.
2.4 Other metabolites in longevity associated study
As far as other metabolites, numerous noteworthy molecules are concerned, including vitamins and volatile organic compounds (VOCs). Vitamin D is a secosteroid (pro)-hormone that is well recognized to regulate bone metabolism, and vitamin D deficiency is frequently presented in elderly [85]. High vitamin D level is associated with better cognitive function in Ashkenazi Jewish centenarians [86]. The study of the Chinese centenarians' cohort has observed that vitamin D deficiency is a predictor of functional dependence [87]. Thus, centenarians have higher blood Vitamin D levels, which may explain their excellent health and exceptional lifespan. Vitamin K is also known as the clotting vitamin, which exists in the form of phylloquinone and menaquinones. Recently, a study described that menaquinones-4 was the predominant vitamin in brain samples from 48 decedents (aged 98-107) enrolled in the Georgia Centenarian Study [88]. Furthermore, circulating phylloquinone were significantly higher in nondemented centenarians than in demented, which might reflect intake of vitamin K-rich foods.
VOCs have been discovered by metabolomics based on GC-MS [89]. VOCs are low-weight carbon-based molecules that reflect the metabolic conditions of the individual, affected by age, gender, diet, physiological and habits. Therefore, VOCs could be considered as “odor-fingerprint” of individuals [90] and non-invasive diagnostic biomarkers [91-93]. Conte M et al. investigated the profile of VOCs in both urine and feces samples from 73 volunteers, including centenarians [94]. Surprisingly, the profile of VOCs can separate for the couples “centenarians-offspring” or the trios “centenarians-offspring-spouse”. Octanoic acid, ethanol, and phenol 4methyl can explain the majority of similarity between the couple centenarian-offspring, while cyclopentanol, benzaldeide, and acetic acid butyl ester can explain the majority of similarity between family trios (centenarian, offspring and spouse). Overall, the above metabolites are worth exploring to provide new insight to understand the mechanism of longevity.
3. Proteomics studies associated with longevity
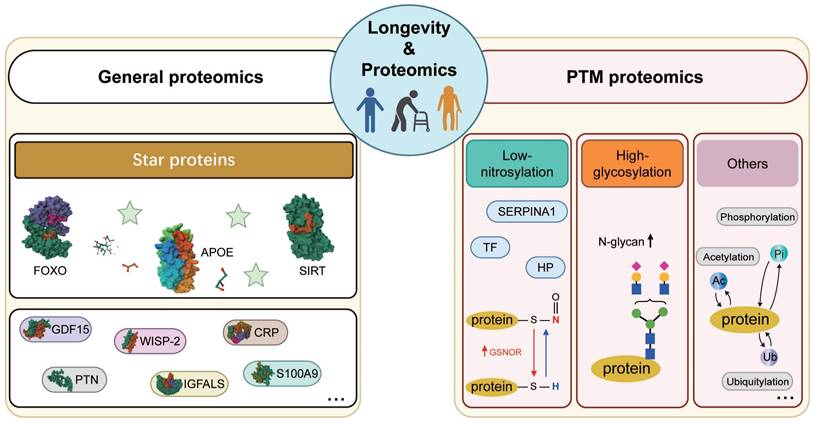
Proteomics encompasses both general proteomics, which quantitatively profiles various proteins, and post-translational modification (PTM) proteomics, which identifies chemical modifications and specific sites on polypeptide chains, such as nitrosylation and glycosylation [95, 96]. In this section, we will briefly describe longevity-related biomarkers discovered by general proteomics, including Apolipoprotein E (APOE), Forkhead box O (FOXO), and sirtuin (SIRT), which have been noticed by scientists since the 1990s [97, 98], and other vital proteins (Table 2). Moreover, PTM proteomics revealed post-translational modifications of longevity-related proteins, such as lower nitrosylation and higher N-glycan, which can further provide us with a deeper insight into longevity (Figure 3).
3.1 Longevity-related biomarkers based on general proteomics
3.1.1 "Star proteins"
In recent decades, scientists have successively discovered several longevity-related proteins through experiments involving nematode worms, mouse models, and population surveys, elucidating the mechanisms governing lifespan [99-101]. These proteins, which we refer to here as "star proteins", include APOE [102-104], FOXO [8, 105-108], and SIRT [100, 109, 110].
APOE is a transport protein found in plasma and interstitial fluid and the APOE gene has three common alleles: e2, e3, and e4, which play a crucial role in lipid metabolism, cognitive function, and immune regulation (Figure 2e). APOE is indispensable in lipid metabolism and can assist in the distribution of triglyceride-rich lipoproteins to various tissues and cells [111]. Gene APOE in different alleles may lead to different changes in cholesterol metabolism. The association between APOE gene loci and lifespan has been confirmed by various studies based on genome-wide association studies [112-116] and centenarians have a higher frequency of the APOE e2 allele. The e2 allele have a neuroprotective effect [102, 117, 118], while the e4 allele is recognized as a risk factor for Alzheimer's disease and impaired cognitive function [119-121]. Sebastiani et al. utilized the expression levels of e2, e3, and e4 alleles to correlate with serum proteins in serum proteomics targeting centenarians and their offspring [122]. The study indicated that the prevalence of e2 allele was higher in centenarians and offspring compared to controls. Studies have shown that centenarians with the APOE e2 allele have lower total cholesterol and LDL levels, and higher HDL and TG levels in plasma [123]. Building upon this foundation, Sebastiani's team conducted a meta-analysis by integrating data from five longevity cohorts on the population prevalence of APOE alleles e2 to e4 and metabolomics data. They revealed that the e2 allele might be related to changes in levels of lipid-bound polyunsaturated fatty acids, as well as with the prevalence of beneficial gut bacteria such as Akkermansia and Lachnoclostridium, thereby affecting lipid metabolism conducive to healthy longevity [124]. In conclusion, proteins associated with the APOE e2 allele appear to have a positive impact on healthy longevity, while proteins associated with the e4 allele might exert negative effects. They revealed that the e2 allele might be as well as with the prevalence of beneficial gut bacteria such as Akkermansia and Lachnoclostridium.
FOXO and SIRT are considered prominent proteins associated with longevity. FOXO can bind to specific DNA sequences, playing a crucial role in regulating gene expression related to cell growth, differentiation, metabolism and autophagy [99]. The polymorphism of the FOXO gene has been found to be closely linked to longevity in different populations from various regions, such as the Japanese (FOXO3A rs2764264, rs13217795 and rs2802292) [105], the Chinese (FOXO3 rs10499051, rs7762395, rs4946933 and rs3800230) [125], and the American (FOXO3 rs6911407 and rs2253310) [108]. Studies have shown that the copy number of the rs2802292 G allele at the FOXO3 locus is associated with a reduced incidence of age-related diseases in centenarians, and its importance in promoting reactive oxygen species (ROS) detoxification, redox balance and DNA repair was found in the human HAP1 homologous cell line (G/T) [126]. Similarly, long-lived individuals carrying the rs2802292 G allele had lower plasma TNF-α than non-carriers, suggesting that FOXO3 may be involved in oxidative stress [127]. Studies have found that FOXO3 may be essential for the lifespan-extending effects of dietary restriction (DR), as WT-DR mice live longer compared to foxo3+/- or foxo3-/--DR mice [128]. In addition, FOXO3 also contributes to mitigating intestinal neurodegeneration associated with inflammation [129]. It maintains cellular homeostasis by promoting the transcription of antioxidant genes such as manganese superoxide dismutase (MnSOD) and catalase [105, 130, 131], while also regulate growth arrest DNA damage-inducible 45 (GADD45) and DNA damage response genes to resist stress [132] (Figure 2e).
Some prominent proteins related with longevity study.
| Group | Sample | Analyticaltechnique | Description | Proteins | Year | Reference | |
|---|---|---|---|---|---|---|---|
| Centenarians | centenarians (n=77, 105.7±3.6), centenarians' offspring (n=82, 71.2±9.3), controls (n=65, 70.6±7.8) | Serum | SOMAscan© technology | Proteins related to the APOE e2 allele | BIRC2, CEP57, VPS29, PSME1, TBCA, | 2019 | [122] |
| UBA2, KMT2C, KIN, CKAP2 | |||||||
| Proteins related to the APOE e4 allele | S100A13, LRRN1, APOE, C5orf38, CTF1, APOB, CRYZL1 | ||||||
| Healthy centenarians (n=9, 100-103 yrs), controls (n=9, 67-81 yrs) | Plasma | TMT LC-MS/MS | ↑in centenarians, compared to controls | CLEC3B, CRISP3, IGFALS, TAS1R3, TGFBI | 2020 | [157] | |
| ↓in centenarians, compared to controls | AOPEP, C1S, CD14, CDKL1, CRTAC1 | ||||||
| centenarians (n=77, 105.7±3.6), centenarians' offspring (n=82, 71.2±9.3), controls (n=65, 70.6±7.8) | Serum | SOMAscan© technology | ↑in centenarians, compared to offspring and young | SFRP1, PTN, CHRDL1, QAGLN, GDF15, IGFBP2, COL28A1, SVEP, B2M FSTL3, NBL1, RSPO4, RNASE1, WFDC2, TNFRSF1B, IGFBP2, SMOC1, WISP2, IGFBP6, SPON1, DKK2, AKT2, HSP90AB1, STAT1, STAT3, SOST, DKK4, IGFBP7 | 2021 | [147] | |
| ↓in centenarians, compared to offspring and young | IGFALS, SERPINF2, ATP1B1, GDF11, CRP, CST3, GHR, IGFR, GHRL, IDE, SMAD3, FLT3, NUDT9, KLKB1, CKM | ||||||
| Semi supercentenarians (n=10, 106-109 yrs), centenarians (n=10, 100 yrs), healthy volunteers (n=10, 20-39 yrs) | Plasma | MALDI-TOF/MS | ↑in Semi centenarians, compared to young | HP-β, A2 M, CLU | 2010 | [95] | |
| centenarians (105-114 yrs), aged (76-83 yrs), young (32-44 yrs) | Serum | Nitro-DIGE | S-nitrosation levels ↑in centenarians, compared to other groups | GSNOR | 2020 | [163] | |
| S-nitrosation levels ↓in centenarians, compared to other groups | SERPINA1, SERPINA3, CP, CERCAM, HP, TTR, | ||||||
| VDBP, IDLC1, TF, TRXR1 | |||||||
| Centenarians | Semi supercentenarians (mean 106.7 yrs), aged controls (mean 71.6 yrs), young controls (mean 30.2 yrs) | Plasma | LC-MS/MS; DSA-FACE; MALDI-TOF-MS | Glycosylation levels ↑ in Semi centenarians, compared to other groups | N-glycans (dHex1 Hex6 HexNAc5 NeuNAc3) ↑ in Semi centenarians | 2015 | [164] |
| Multi-branched and highly sialylated N-glycans, as well as agalacto- and/or bisecting N-glycans | |||||||
| Glycosylation levels ↓ in Semi centenarians, compared to other groups | Biantennary N-glycans | ||||||
| Semi supercentenarians (106-109 yrs), aged controls (70-88 yrs), young controls (20-38 yrs) | Plasma | LC-MS/MS | Glycosylation levels ↑ in Semi centenarians, compared to other groups | Tri-antennary and sialylated N-glycans of haptoglobin at Asn207 and Asn211 sites were characterized in Semi centenarians | 2018 | [175] | |
| Elderly | TwinsUK cohort (n=202, 65.30±6.92), replication Cohort (n=677, 76.96±7.06) | Plasma | SOMAscan© technology | ↑with age | CHRDL1, CCDC80, PTN, FSTL3, TIMP1, MMP12, | 2015 | [149] |
| CST3, IGFBP6, ROR1, THBS4, HA VCR2 | |||||||
| ↓with age | NA | ||||||
| men and women (n=240, 22-93 yrs) | Plasma | SOMAscan© technology | ↑with age | GDF15, PTN, ADAMTS5, CGA FSHB, SOST, | 2018 | [150] | |
| CHRDL1, NPPB, EFEMP1, MMP12 | |||||||
| ↓with age | CTSV | ||||||
| offspring of parents with exceptional longevity (OPEL) (n=506, 74.5±6.1), offspring of parents with usual survival (OPUS) (n=519, 77.1±7.1) | Plasma | SOMAscan© technology | ↑in OPEL, compared to OPUS | PTN, WISP2, CRDL1, TAGLN, RSPO1, GDF15, FBLN1, SMOC1, HE4, CST3, FSTL3, RNase1, sTREM-1, URB, NPPB, SREC-II | 2020 | [151] | |
| ↓in OPEL, compared to OPUS | ERBB1/EGFR, SERPINF2 | ||||||
| Long-lived men (≥90% expected survival for 12 years) (n=554, 78.5±3.1), Not long-lived men who died earlier (n=642, 76.4±2.9) | Serum | LC-IMS-MS | ↓in long-lived men compared to non-long-lived men | C9, C7, CFD, CD5L, S100A9, MCAM, LGALS3BP, CSF1R, ALCAM, CRP, FCGBP, IGHG3, IGHM, FCGR3A, CD163, NRP1, GPLD1, B2 M, A2 M, MMP2, CST3, PTGDS, VWF, HPR, F2 | 2020 | [162] |
Abbreviations: A2 M, α2-macroglobulin; ADAMTS5, A disintegrin and metalloproteinase with thrombospondin motifs 5; ADH5/GSNOR, alcohol dehydrogenase 5/S-nitrosoglutathion; AKT2, RAC-beta serine/threonine-protein kinase; ALCAM, CD166 antigen; AOPEP, Aminopeptidase O; APOB, Apolipoprotein B; APOE, Apolipoprotein E; ATP1B1, Sodium/potassium-transporting ATPase subunit beta-1; B2 M, β2 microglobulin; BIRC2, Baculoviral IAP repeat containing 2; C1S, Complement C1s subcomponent; C5orf38, Chromosome 5 open reading frame 38; C7, Complement component C7; C9, Complement component C9; CCDC80, Coiled-coil domain- containing protein 80; CD14, Monocyte differentiation antigen CD14; CD163, Scavenger receptor cysteine-rich type 1 protein M130; CD5L, CD5 antigen-like; CDKL1, Cyclin-dependent kinase-like 1; CEP57, Centrosomal protein 57; CERCAM, Inactive glycosyltransferase 25 family member 3; CFD, Complement factor D; FSHB, Follitropin subunit beta; CHRDL1, Chordin-like protein 1; CKAP2, Cytoskeleton associated protein 2; CKM, Creatine kinase M-type; CLEC3B, Tetranectin; CLU, Clusterin precursor; COL28A1, Collagen alpha 1(XXVIII) chain; COL6A3, Collagen alpha 3(VI) chain; CP, Ceruloplasmin; CRISP3, Cysteine-rich secretory protein 3; CRP, C-reactive protein; CRTAC1, Cartilage acidic protein 1; CRYZL1, Crystallin zeta like 1; CSF1R, Macrophage colony-stimulating factor 1 receptor; CST3, Cystatin-C; CTF1, Cardiotrophin 1; CTSV, Cathepsin V; DKK2, Dickkopf-related protein 2; DKK4, Dickkopf-related protein 4; EFEMP1, Fibulin 3; ERBB1/EGFR, Epidermal growth factor receptor; F2, Prothrombin; FBLN1, EGF-containing fibulin-like extracellular matrix protein 1; FCGBP, IgG Fc-binding protein; FCGR3A, Low affinity immunoglobulin gamma Fc region receptor III A; FSTL3, Follistatin-related protein 3; GDF11, Growth/differentiation factor 11; GDF15, Growth/differentiation factor 15; GHR, Growth hormone receptor; GHRL, Appetite-regulating hormone; GPLD1, Phosphatidylinositol-glycan-specific phospholipase D; HA VCR2, Hepatitis A virus cellular receptor 2; HE4, WAP four-disulfide core domain protein 2; HP, Haptoglobin; HPR, Haptoglobin-related protein; HP-β, Haptoglobin β chain; HSP90AB1, Heat shock protein HSP 90-beta; IDE, Insulin-degrading enzyme; IGFALS, Insulin-like growth factor-binding protein complex acid labile subunit; IGFBP2, Insulin-like growth factor-binding protein 2; IGFBP6, Insulin-like growth factor-binding protein 6; IGFBP7, Insulin-like growth factor-binding protein 7; IGFR, IGF-like family receptor; IGHG3, Immunoglobulin heavy constant gamma 3; IGHM, Immunoglobulin heavy constant mu; IGLC1, immunoglobulin light chain 1; KIN, Kin17 DNA and RNA binding protein; KLKB1, Plasma kallikrein; KMT2C, Lysine methyltransferase 2C; LGALS3BP, Galectin-3-binding protein; LRRN1, Leucine rich repeat neuronal 1; MCAM, Cell surface glycoprotein; MMP12, matrix metallopeptidase 12; MMP2, matrix metallopeptidase 2; MSR1, Macrophage scavenger receptor types I and II;NBL1, Neuroblastoma suppressor of tumorigenicity 1; NPPB, N-terminal pro-BNP; NRP1, Neuropilin-1; NUDT9, ADP-ribose pyrophosphatase, mitochondrial; PCSK1, Neuroendocrine convertase 1; PGD2 synthase, Prostaglandin-H2 D-isomerase; PON1, Paraoxonase/arylesterase 1; PSME1, Proteasome activator subunit 1; PTGDS, Prostaglandin-H2 D-isomerase; PTN, Pleiotrophin; RNase 1, Ribonuclease pancreatic; ROR1, Tyrosine-protein kinase transmembrane receptor ROR1; RSPO1, R-spondin-1; RSPO4, R-spondin-4; S100A13, S100 calcium binding protein A13; S100A9, Protein S100-A9; SERPINA1, α1-antitripsin; SERPINA3, α1-antichimotripsin; SERPINF2, α2-antiplasmin; SFRP1, Secreted frizzled-related protein 1; SMAD3, Mothers against decapentaplegic homolog 3; SMOC1, SPARC-related modular calcium-binding protein 1; SOST, Sclerostin; SPON1, Spondin-1; SREC-II, Scavenger receptor class F member 2; STAT1, Signal transducer and activator of transcription 1-alpha/beta; STAT3, Signal transducer and activator of transcription 3; sTREM-1, Triggering receptor expressed on myeloid cells 1; SVEP, Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1; TAGLN, Transgelin; TAS1R3, Taste receptor type 1 member 3; TBCA, Tubulin folding cofactor A; TF, serotransferrin; TGFBI, Transforming growth factor-beta-induced protein ig-h3; THBS4, Thrombospondin-4; TIMP1, Metalloproteinase inhibitor 1; TNFRSF1B, Tumor necrosis factor receptor superfamily member 1B; TRXR1, thioredoxin reductase 1; TTR, transthyretin; UBA2, Ubiquitin like modifier activating enzyme 2; URB, Unhealthy ribosome biogenesis protein homolog; VDBP, vitamin D-binding protein; VPS29, Vacuolar protein sorting-associated protein 29; VWF, von Willebrand factor; WFDC2, WAP four-disulfide core domain protein 2; WISP2, WNT1-inducible signaling pathway protein 2.
Schematic representation of longevity-related biomarkers. Schematic depictions of major longevity-related biomarkers identified from both general and post-translational modification (PTM) proteomics. (1) Longevity-related biomarkers identified from general proteomics: apolipoprotein E (APOE), forkhead box gene, group O proteins (FOXO), sirtuin proteins (SIRT). Besides, there are other longevity-related protein biomarkers repeatedly identified from general proteomics in different studies, such as GDF15, PTN, SFRP1, WISP-2, IGFALS, and SERPINF2. (2) Longevity-related biomarkers identified from PTM proteomics: low-nitrosylated related proteins (such as TF, HP, and SERPINA1) and high-denitrosylated related proteins (GSNOR), as well as high N-glycan related proteins. These longevity-related proteins mentioned above are identified by nitrosoproteomics and glycoproteomics. In addition, other longevity-related biomarkers are expected to be obtained through the following PTM proteomics, such as phospho-proteomics, acetyl-proteomics, and ubiquitin-proteomics. Abbreviations: PTN: Pleiotrophin; protein 2; IGFALS: Insulin-like growth factor-binding protein complex acid labile subunit; SERPINF2: Alpha-2-antiplasmin; TF: Serotransferrin, HP: Haptoglobin, SERPINA1: α1-antitrypsin; GSNOR: nitrosoglutathione reductase. The structure of N-glycan is cited in the paper (Miura, Yuri et al., 2015). N-glycan: blue square, N-acetylglucosamine; yellow circle, galactose; green circle, mannose; purple diamond, N-acetylneuraminic acid.
SIRT is a family of nicotinamide adenosine dinucleotide (NAD) dependent histone deacetylases and adenosine diphosphate (ADP) ribose transferases [133]. SIRT can interact with the FOXO family to better respond to cellular oxidative stress [134-136]. Interestingly, genetic polymorphisms in the SIRT3 and SIRT6 genes have been found to be closely associated with human longevity [137-139], in which SIRT6 has been proven to be related to enhance neuroplasticity and improve cognitive function [140]. Studies have shown that brain-specific Sirt1-overexpressing (BRASTO) transgenic mice exhibit significant lifespan extension in both males and females [141]. Aged BRASTO mice demonstrate phenotypes consistent with delayed aging: they exhibit significant enhancements in physical activity, body temperature, and quality of sleep, compared to age-matched control mice. Additionally, mitochondria morphology and function of skeletal muscle appear younger. This is attributed to Sirt1 enhancing Ox2r promoter activity through Nkx2-1 deacetylation to regulate bodily functions. Moreover, FOXO and SIRT regulate autophagy genes expression, which can promote the degradation of harmful proteins and alleviate oxidative stress [142, 143]. Knockdown of SIRT4 gene accelerated cell senescence and led to an increase in the senescence-associated secretory phenotype (SASP), a finding that suggests SIRT4 as a potential target for extending lifespan [144]. On the other hand, they regulate insulin signaling pathways and metabolic enzymes expression to prevent age-related metabolic diseases [145]. FOXO and SIRT can also reduce inflammation by inhibiting NF-κB pathway [146]. To sum up the aforementioned findings, FOXO and SIRT are potential targets in the regulation of longevity, although their roles in the proteomics of long-lived populations have not been reported thus far.
3.1.2 Other vital proteins
Nowadays, general proteomics is utilized to identify other proteins associated with extreme longevity in large-scale longevity research cohorts, such as Pleiotrophin (PTN), WNT1-Inducible Signaling Pathway Protein 2 (WISP-2), and Growth/Differentiation Factor 15 (GDF15), Insulin-like Growth Factor-Binding Protein Complex Acid Labile Subunit (IGFALS).
PTN, WISP-2, and GDF15 have been identified as potential longevity-related biomarkers. Sebastiani et al. analyzed plasma protein profiles of the New England Centenarian Study cohort by SomaLogic technology, in which PTN and WISP-2 were found to increase with age and were highly elevated in centenarians compared to control groups [147]. Elevated levels of PTN have also been positively correlated with healthy aging in the previous cohort studies [148-150]. In long-lived offspring, WISP-2 levels increase with age, especially in females [151]. WISP-2 inhibits cardiac hypertrophy and fibrosis [152] and promotes the survival of vascular smooth muscle cells [153-155]. Thus, upregulated WISP-2 could be speculated to foster cell growth and proliferation, further exerting a positive influence on longevity. Additionally, Sebastiani et al. found lower levels of GDF15 expression in individuals with an expected lifespan exceeding 11 years, compared to those with an expected lifespan of less than 10 years, which implied that lower GDF15 levels had a favorable link to an extended lifespan [147]. High levels of GDF15 are negatively correlated with muscle strength in the elderly in another research [156]. In short, lower levels of GDF15 might be considered characteristic proteins for achieving longevity.
IGFALS was found to significantly elevate in the plasma of centenarians, which was also discovered to be linked to some biological processes like vascular generation and B-cell mediated immune response [157]. Another study indicated the connection between IGFALS and longevity though its interaction with Insulin-like Growth Factor Binding Protein 3 (IGFBP3); and IGFBP3 variant (rs11977526) was related to the longevity [158]. Additionally, IGFBP-3 is associated with the induction of cellular aging [159, 160], which shows lower levels in the plasma of long-lived individuals (>95 yrs), suggesting a potential mechanism for alleviating cellular aging [161]. The significance of IGFALS and IGFBP3 in the context of longevity need further validation by more studies of model animals and cohorts.
Moreover, inflammation-related proteins were found to be down-regulated through proteomics in the long-lived population of the MrOS cohort and the LonGenity cohort, such as TNFR (TNF Receptor), complement C9, C7, S100A9 (S100 calcium binding protein A9), and CRP (C-reactive protein), which are mostly involved in inflammatory or complement activation pathways [151, 162].
3.2 Longevity-Related Biomarkers Based on Post-translational modification (PTM) Proteomics
Protein modifications commonly include nitrosylation, glycosylation, phosphorylation, acetylation, and ubiquitination. Lower levels of nitrosylation and higher levels of N-glycosylation are reported to be closely associated with longevity in centenarians [163, 164]. Exploring the functions of proteins that undergo various modifications by PTM proteomics enables a deeper comprehension of the molecular mechanisms in proteins.
Nitrosoproteomics investigates the impact of protein nitrosylation on structure and function, aiming to understand the function of nitrosylated protein in cells and organisms [165, 166]. During cell senescence, cells were influenced by oxidative stress caused by the excessive accumulation of ROS and nitric oxide (NO), which in turn affects the nitrosylation levels of certain proteins [167, 168]. Capitanio et al. employed S-nitrosoproteomics to analyze plasma proteins in centenarians and adults [163], revealing reduced S-nitrosylation levels in certain plasma proteins, including transferrin (TF), haptoglobin (HP) and alpha-1-antitrypsin (SERPINA1), among centenarians compared to controls. In other word, lower S-nitrosylation levels in centenarians may indicate a beneficial state of maintaining lower levels of ROS and NO. Furthermore, denitrosylation is the reverse process of S-nitrosylation, which is mediated by S-nitroso glutathione reductase (GSNOR) [169]. Due to limitations in techniques, there is currently no corresponding proteomics study reported for denitrosylation. To further investigate the dynamic changes in S-nitrosylation and denitrosylation, Rizza et al. found that mRNA levels of GSNOR significantly rised in centenarians and adults compared to the elderly [170]. And they further displayed that the absence of GSNOR protein will increase S-nitrosylation levels through animal experiments. In summary, lower S-nitrosylation and higher denitrosylation levels may enhance the ability to cope oxidative stress from cellular aging.
Glycoproteomics enables the assessment of glycoprotein structures, glycosylation sites, and the heterogeneity of glycosylation sites [171]. Many researchers discovered glycosylation-related factors associated with aging by glycoproteomics, like IgG N-glycosylation [172, 173], and advanced glycation end products [174]. Miura was the first to discover a link between plasma protein N-glycan levels and extreme longevity in long-lived populations [164]. Semi-supercentenarians (SSC, >105 yrs) exhibited heightened anti-inflammatory responses characterized by increased multi-branched and highly sialylated N-glycan in plasma proteins, as well as agalacto- and/or bisecting N-glycans, distinguishing from both the elderly and young populations. Subsequently, Miura conducted further analysis of characteristic glycopeptide linked to longevity, uncovering a noteworthy augmentation in tri-antennary and sialylated N-glycans at Asn207 and Asn211 sites in haptoglobin among SSC. This extension further prolonged the half-life of haptoglobin, allowing it to bind to free hemoglobin and act as an antioxidant [175]. Therefore, N-glycan with high glycosylated levels serves as a characteristic biomarker of centenarians.
Additionally, there are relatively few research reports in longevity cohorts using proteomics such as phosphorylation, acetylation, and ubiquitination. These methods are primarily employed to study the modifications of proteins associated with diseases. Phosphorylation and acetylation proteomics have been highlighted in Alzheimer's disease [176, 177]. Furthermore, the phosphorylation of AMP-activated protein kinase (AMPK) [178], acetylation of FOXO proteins [179], and ubiquitination of ubiquitin ligase CHIP [180] are linked to longevity, impacting protein structural, energy metabolism, and stress response. With technological advancements, these PTM proteomics can be employed to explore biomarkers in longevity cohorts.
4. Multi-omics studies associated with longevity
Advancement in omics technology reveals that single omics cannot fully address the objectives of scientific study. Multi-omics, integrating genomics, transcriptomics, proteomics, metabolomics, and microbiomics data, is a comprehensive strategy to investigate the causes of healthy aging and lifespan extension in humans. Recently, several recent studies using multi-omics have made significant progress in elucidating the intricate factors contributing to longevity.
Multi-omics integrated analysis using metabolomics and proteomics has gained considerable traction in various fields, such as cognitive impairment [181], dementia [182], and Alzheimer's disease [183]. In a study conducted by Ahadi, longitudinal and deep multi-omics profiling in transcripts, proteins, metabolites, cytokines, and microbes was performed on 106 healthy individuals aged 29 to 75 [184]. And this study revealed distinct age-associated trends and levels of association for 184 molecules across multi-omics, including Clostridium cluster IV Fetuin-B and PROS1. In addition, different types of aging patterns are defined according to the molecules level of changes over 2-3 years, called “ageotypes”. Similarly, Tebani et al. also conducted a longitudinal multi-omics study over 2-3 years with a cohort of 100 people, aiming at interpret differences between individuals [185]. The findings indicate that the alpha subunit of glycoprotein hormones (CGA) exhibits higher expression in women compared to men. Furthermore, γ-tocopherol has been identified as one of the most stable metabolites in this cohorts. Overall, each population or individual exhibits a distinct molecular profile; thus, centenarians, representing a notably long-lived demographic, merit detailed examination of their unique molecular biomarkers.
Centenarians have younger epigenetic characteristics. At the protein level, centenarians may show more favorable histone modification patterns, such as histone acetylation and methylation, which are often closely related to gene expression regulation [186, 187]. At the nucleic acid level, centenarians tend to have lower global DNA methylation levels, which is associated with slower biological aging and longer lifespan [188, 189]. For example, both centenarians and their offspring resulted significantly epigenetically younger than the control though epigenetic age measures by DNA methylation values of CpG sites, with mean epigenetic age discrepancy equal to -6.45 and -1.65 years, respectively [188]. Additionally, they may also show more regulatory transcriptome expression patterns, and these transcription RNAs can affect gene transcription and translation [190, 191]. In addition, telomere length and activity also show longer length and higher activity in healthy centenarians [192]. Here, we take a simple example. In long-lived individuals, correlation analysis based on transcriptomics translation into proteomics has been reported. By developing a genome-wide precision metabolic modeling method with serum metabolomics and proteomics derived from transcriptomics, Li et al. concluded that elevated long- chain fatty acid beta-oxidation (FAO) is the most significant metabolic feature in centenarians [193]. And they hypothesized that the elevated FAO would induce the consumption of long-chain fatty acids, and serum long-chain fatty acids appear at low levels in centenarians. Further serum metabolomics studies showed that 83 down-regulated metabolites (include 67 fatty acid-like metabolites) in centenarians. Thus, the unique metabolic characteristic of centenarians is the elevated FAO.
Multi-omics approaches also include the integration of metabolomics or proteomics with other omics, and the most typical composition is to integrate metabolomics with microbial diversity. Wilmanski et al. adopted three independent cohorts comprising over 9,000 individuals and discovered that the gut microbiome uniqueness measures in healthy individuals (>80 yrs) increased with age, and the plasma metabolome was characterized by raising level of tryptophan metabolites (3-indoxyl sulfate, 6-hydroxyindole sulfate, indoleacetate and indolepropionate), and these tryptophan metabolites were thought to have a positive impact on healthy aging [194]. Moreover, the metabolites derived from tryptophan metabolism are essential for the gut bacteria to adapt to aging in human hosts. Studies have shown that tryptophan metabolites have been shown to be beneficial to health and extend survival in a number of animal models [195]. In addition to the previously mentioned secondary bile acids [33], SCFAs [35] and tryptophan metabolites [194]; exopolysaccharides [196] and polyamines [197] among other microbial-related metabolites, play crucial roles in health and longevity. Overall, beyond a certain age, long-lived individuals exhibit increasing gut microbiome uniqueness, which may result in a broader range of microbial-related metabolites entering the bloodstream to promote healthy aging.
In the past three years, a number of studies have examined the interaction between gut microbiome and metabolisms of long-lived individuals, such as the combination of fecal microbiota diversity and fecal metabolomics. Sato et al. found that centenarians have a distinct gut microbiome using fecal microbiome and bile acid metabolomics, and these gut bacteria were capable of generating unique secondary bile acids, such as isoallo-lithocholic acid (LCA) [33]. This study reported for the first time that Odoribacteraceae relied on 5AR and 3βHSDH to produce isoalloLCA, and isoalloLCA could be used to against infections. Altogether, this study illustrates that gut bacteria from centenarians participated in a unique bile acid metabolism.
The effect of gut microbiome on host metabolism was analyzed by integrating fecal microbial diversity and blood metabolomics. Xu et al. compared the gut microbiome and blood metabolome of long-lived individuals (94-105 yrs) to that of offspring (50-79 yrs) in 116 Han Chinese families and found extensive metagenomic and metabolomic remodeling [35]. The 16sRNA sequencing and metagenomic displayed nonagenarians and centenarians have greater microbial diversity than their descendants and have identified "longevous gut microbiota signature", including Bifidobacterium and Blautia. The targeted metabolomics showed that acetic acid, butyric acid, and propionic acid derived from Bacteroides over-expressed in centenarians. In general, nonagenarians and centenarians exhibited gut-related metabolic alterations. Our current research for centenarians analyzes serum metabolites and gut bacteria in fecal samples by the multi-omics integrating metabolomics, 16S rRNA, and metagenomics (unpublished data). Preliminary findings suggest that tryptophan metabolites such as 5-methoxyindole-3-acetic acid and indole-3-pyruvic acid were significantly elevated in healthy centenarians compared to frail centenarians. Furthermore, a significant positive correlation between elevated metabolite like oxindole and gut bacteria like Christensenella in centenarians have been observed. To sum up, several other omics studies in long-lived individuals are also worthwhile exploring, including those in the fields of genomics [198, 199] and microbiome [200].
5. Conclusion
In this review, we integrate longevity-related biomarkers discovered by metabolomics and proteomics and further categorize them based on different classes. The metabolites and proteins mentioned above are considered crucial signaling molecules for prolonging lifespan and alleviating age-related diseases. The mechanisms of longevity-related metabolites have been elucidated, especially for specific fatty acids like EPA, DHA, and SCFAs, which effect lifespan by reducing inflammation and activating the Nrf2 pathway. The mechanisms underlying the health benefits of the changes in certain metabolites are still largely unknown. For example, the mechanism of some isomers of secondary bile acids affects the body's immunity remains to be further studied. Additionally, the metabolic pathways and products of metabolites should also be considered. Some intermediates (such as kynurenic acid) have neuroprotective effects, which were produced from tryptophan. Regarding star proteins, APOE, FOXO, and SIRT are essential signaling proteins for cell survival, which can regulate cell proliferation, metabolism, inflammation, and stress responses by influencing multiple signaling pathways, including PI3K/Akt, NF-κB, etc. Moreover, post-translational modifications such as nitrosylation and glycosylation have important effects on the function and communication of proteins. The interaction between various modifications and star proteins creates a complex network that modulates cell survival to extend lifespan. Therefore, integrating candidate longevity-related biomarkers to conduct a "biomarker library of health and longevity" can further grasp the profile of centenarians or extreme longevity in humans and provide a theoretical foundation for anti-aging.
Metabolomics, proteomics, and multi-omics provide researchers with scientific methods to integrate information at different molecular layers, and further understand the origin and fates of small molecule substances. However, there are certain challenges and limitations in longevity cohort studies using metabolomics and proteomics, and the following interfering factors need to be carefully considered in study design and data interpretation.
Longevity cohort studies typically require large-scale population samples to obtain reliable results. Currently, several countries are conducting various longevity cohorts, such as the European Union Longevity Genetics Consortium, the New England Centenarian Study (NECS), and the Chinese Longitudinal Healthy Longevity Survey (CLHLS). Establishing longevity cohorts should consider the diversity of the population, including variations in age span, gender ratio, geographical location, and lifestyle, to ensure the accuracy and broad applicability of the results. For instance, in Chinese centenarians, females are disproportionately represented, necessitating the maintenance of gender balance to eliminate potential differences caused by gender in longevity studies. Due to the hereditary nature of certain longevity traits, such as the APOE e2 gene, understanding the participants' kinship is crucial for identifying genetic factors.
Appropriate analytical methods are crucial for different research objects based on the research question and sample characteristics. In metabolomics, untargeted and targeted metabolomics both have different advantages and disadvantages. It is worth noting that a certain degree of lipid metabolism dysfunction and neural functional damage happens during the aging process. Therefore, targeted metabolomics focusing on specific metabolites such as short-chain fatty acids, bile acids, and neurotransmitters can better reflect the physiological status of the elderly. In proteomics, general proteomics can provide a more comprehensive protein map for the longevity population. With an increase in age, protein homeostasis gradually declines and results in wrong translation modification such as nitrosylation and glycosylation. Consequently, post-translational modifications of proteins are considered crucial indicators affecting the function of proteins. The study of longevity cohorts based on untargeted metabolomics and general proteomics has been extensively reported. We regard that targeted metabolomics and PTM proteomics focusing on specific biomolecules will attract more attention in aging studies to discover more valuable longevity-related biomarkers, whether metabolites or proteins. Moreover, blood and fecal samples are commonly preferred for biomarker discovery due to their relatively easy access. If tissue-specific characteristics are exhibited in the liver or muscle tissues of centenarians, it potentially leads to obvious alterations in circulating blood metabolites. However, there are significant challenges in obtaining tissue samples such as liver and muscle from living individuals. We wish that better technology may have appeared in the future to offer the possibility to analyze the tissue or organ specificity of centenarians.
In terms of data analysis, standardization and uniformity are crucial steps to ensure experiment reproducibility and result comparability. Due to objective or unavoidable factors, such as gender differences, statistical or machine learning methods can be employed to establish an effective and reasonable model to eliminate the impact of non-research factors. Subsequently, validation through animal experiments is a crucial prerequisite for translating potential longevity biomarkers into clinical diagnosis and treatment. Some extreme longevity-related biomarkers should be replicated and verified in large cohorts or model animals such as nematodes, zebrafish, mice, and naked mole rats with an extreme lifespan.
Finally, combining and integrating different omics techniques has played an increasingly important role in scientific research. By effectively integrating multiple layers of biological information, we will be able to deepen our understanding of the biological mechanisms that regulate aging and prolong life. For example, metabolomics and proteomics are used in combination with other omics such as genomics, transcriptomics, and microbiomics to analyze the characteristics of biomarkers (metabolites, proteins, genes, transcription factors, microorganisms), and the connections of molecules mentioned above in functional annotations and signaling pathways in different clinical samples from long-lived populations. We look forward to integrating different omics and conducting association analyses in the future to discover biomarkers related to extreme longevity.
Acknowledgements
We thank the Key Lab of Zhanjiang for R&D Marine Microbial Resources in the Beibu Gulf Rim and the Suixi County People's Hospital, Zhangjiang, Guangdong.
Funding
This work was supported by the National Natural Science Foundation of China (82301752), Guangdong Province Science, the Doctoral Initial Funding of Guangdong Medical University (4SG23183G), the Science and Technology Program of Guangdong Province (2019B090905011), Guangdong University Youth Innovation Talent Project (2020KQNCX023), Special Science and Technology Innovation Project of Guangdong Province, China (2019A01005), Special Funds for Economic Development of Marine Economy of Guangdong Province, China (GDNRC[2022]38), the Technology Innovation Strategy Special Foundation of Zhanjiang (2023A103-1), the Fund of Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang) (ZJW-2019-007), and the Public Service Platform of South China Sea for R&D Marine Biomedicine Resources (2017C8A), Discipline Construction Project of Guangdong Medical University (2043K2022005).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Robine JM, Cubaynes S. Worldwide demography of centenarians. Mech Ageing Dev. 2017;165:59-67
2. Lissek T. Aging as a Consequence of the Adaptation-Maladaptation Dilemma. Adv Biol (Weinh). 2024;8:e2300654
3. Caruso C, Aiello A, Accardi G, Ciaglia E, Cattaneo M, Puca A. Genetic Signatures of Centenarians: Implications for Achieving Successful Aging. Curr Pharm Des. 2019;25:4133-8
4. Borras C, Ingles M, Mas-Bargues C, Dromant M, Sanz-Ros J, Román-Domínguez A. et al. Centenarians: An excellent example of resilience for successful ageing. Mech Ageing Dev. 2020;186:111199
5. Murabito JM, Beiser AS, Decarli C, Seshadri S, Wolf PA, Au R. Parental longevity is associated with cognition and brain ageing in middle-aged offspring. Age Ageing. 2014;43:358-63
6. Shadyab AH, Manson JE, Li W, Gass M, Brunner RL, Naughton MJ. et al. Parental longevity predicts healthy ageing among women. Age Ageing. 2018;47:853-60
7. Srivastava S. Emerging Insights into the Metabolic Alterations in Aging Using Metabolomics. Metabolites. 2019;9:301
8. Moaddel R, Ubaida-Mohien C, Tanaka T, Lyashkov A, Basisty N, Schilling B. et al. Proteomics in aging research: A roadmap to clinical, translational research. Aging Cell. 2021;20:e13325
9. Tu C, Li J, Sheng Q, Zhang M, Qu J. Systematic assessment of survey scan and MS2-based abundance strategies for label-free quantitative proteomics using high-resolution MS data. J Proteome Res. 2014;13:2069-79
10. Meleady P. Two-Dimensional Gel Electrophoresis and 2D-DIGE. Methods Mol Biol. 2018;1664:3-14
11. Hoffman JM, Lyu Y, Pletcher SD, Promislow DEL. Proteomics and metabolomics in ageing research: from biomarkers to systems biology. Essays Biochem. 2017;61:379-88
12. Sobsey CA, Ibrahim S, Richard VR, Gaspar V, Mitsa G, Lacasse V. et al. Targeted and Untargeted Proteomics Approaches in Biomarker Development. Proteomics. 2020;20:e1900029
13. Wishart DS. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol Rev. 2019;99:1819-75
14. Ke M, Shen H, Wang L, Luo S, Lin L, Yang J. et al. Identification, Quantification, and Site Localization of Protein Posttranslational Modifications via Mass Spectrometry-Based Proteomics. Adv Exp Med Biol. 2016;919:345-82
15. Di Minno A, Gelzo M, Stornaiuolo M, Ruoppolo M, Castaldo G. The evolving landscape of untargeted metabolomics. Nutr Metab Cardiovasc Dis. 2021;31:1645-52
16. Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J Am Soc Mass Spectrom. 2016;27:1897-905
17. Das UN. "Cell Membrane Theory of Senescence" and the Role of Bioactive Lipids in Aging, and Aging Associated Diseases and Their Therapeutic Implications. Biomolecules. 2021;11:241
18. Janssen CI, Kiliaan AJ. Long-chain polyunsaturated fatty acids (LCPUFA) from genesis to senescence: the influence of LCPUFA on neural development, aging, and neurodegeneration. Prog Lipid Res. 2014;53:1-17
19. Troesch B, Eggersdorfer M, Laviano A, Rolland Y, Smith AD, Warnke I. et al. Expert Opinion on Benefits of Long-Chain Omega-3 Fatty Acids (DHA and EPA) in Aging and Clinical Nutrition. Nutrients. 2020;12:2555
20. Xyda SE, Vuckovic I, Petterson XM, Dasari S, Lalia AZ, Parvizi M. et al. Distinct Influence of Omega-3 Fatty Acids on the Plasma Metabolome of Healthy Older Adults. J Gerontol A Biol Sci Med Sci. 2020;75:875-84
21. Collino S, Montoliu I, Martin FP, Scherer M, Mari D, Salvioli S. et al. Metabolic signatures of extreme longevity in northern Italian centenarians reveal a complex remodeling of lipids, amino acids, and gut microbiota metabolism. PLoS One. 2013;8:e56564
22. Otsuka R, Tange C, Nishita Y, Tomida M, Kato Y, Imai T. et al. Fish and Meat Intake, Serum Eicosapentaenoic Acid and Docosahexaenoic Acid Levels, and Mortality in Community-Dwelling Japanese Older Persons. Int J Environ Res Public Health. 2019;16:1806
23. de Carvalho C, Caramujo MJ. The Various Roles of Fatty Acids. Molecules. 2018;23:2583
24. Pareja-Galeano H, Sanchis-Gomar F, Santos-Lozano A, Garatachea N, Fiuza-Luces C, Lucia A. et al. Serum eicosapentaenoic acid to arachidonic acid ratio is associated with cardio-healthy exceptional longevity. Int J Cardiol. 2015;184:655-6
25. Mildenberger J, Johansson I, Sergin I, Kjøbli E, Damås JK, Razani B. et al. N-3 PUFAs induce inflammatory tolerance by formation of KEAP1-containing SQSTM1/p62-bodies and activation of NFE2L2. Autophagy. 2017;13:1664-78
26. Sung J, Jeon H, Kim IH, Jeong HS, Lee J. Anti-Inflammatory Effects of Stearidonic Acid Mediated by Suppression of NF-κB and MAP-Kinase Pathways in Macrophages. Lipids. 2017;52:781-7
27. Zhang M, Wang S, Mao L, Leak RK, Shi Y, Zhang W. et al. Omega-3 fatty acids protect the brain against ischemic injury by activating Nrf2 and upregulating heme oxygenase 1. J Neurosci. 2014;34:1903-15
28. Stulnig TM, Huber J, Leitinger N, Imre EM, Angelisova P, Nowotny P. et al. Polyunsaturated eicosapentaenoic acid displaces proteins from membrane rafts by altering raft lipid composition. J Biol Chem. 2001;276:37335-40
29. Ye S, Tan L, Ma J, Shi Q, Li J. Polyunsaturated docosahexaenoic acid suppresses oxidative stress induced endothelial cell calcium influx by altering lipid composition in membrane caveolar rafts. Prostaglandins Leukot Essent Fatty Acids. 2010;83:37-43
30. Shama S, Liu W. Omega-3 Fatty Acids and Gut Microbiota: A Reciprocal Interaction in Nonalcoholic Fatty Liver Disease. Dig Dis Sci. 2020;65:906-10
31. Fu Y, Wang Y, Gao H, Li D, Jiang R, Ge L. et al. Associations among Dietary Omega-3 Polyunsaturated Fatty Acids, the Gut Microbiota, and Intestinal Immunity. Mediators Inflamm. 2021;2021:8879227
32. Santoro A, Ostan R, Candela M, Biagi E, Brigidi P, Capri M. et al. Gut microbiota changes in the extreme decades of human life: a focus on centenarians. Cell Mol Life Sci. 2018;75:129-48
33. Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM. et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature. 2021;599:458-64
34. Ragonnaud E, Biragyn A. Gut microbiota as the key controllers of "healthy" aging of elderly people. Immun Ageing. 2021;18:2
35. Xu Q, Wu C, Zhu Q, Gao R, Lu J, Valles-Colomer M. et al. Metagenomic and metabolomic remodeling in nonagenarians and centenarians and its association with genetic and socioeconomic factors. Nature Aging. 2022;2:438-52
36. Cuervo A, Salazar N, Ruas-Madiedo P, Gueimonde M, González S. Fiber from a regular diet is directly associated with fecal short-chain fatty acid concentrations in the elderly. Nutr Res. 2013;33:811-6
37. Salazar N, Arboleya S, Fernández-Navarro T, de Los Reyes-Gavilán CG, Gonzalez S, Gueimonde M. Age-Associated Changes in Gut Microbiota and Dietary Components Related with the Immune System in Adulthood and Old Age: A Cross-Sectional Study. Nutrients. 2019;11:1765
38. González-Bosch C, Boorman E, Zunszain PA, Mann GE. Short-chain fatty acids as modulators of redox signaling in health and disease. Redox Biol. 2021;47:102165
39. Hoyles L, Snelling T, Umlai UK, Nicholson JK, Carding SR, Glen RC. et al. Microbiome-host systems interactions: protective effects of propionate upon the blood-brain barrier. Microbiome. 2018;6:55
40. Wu J, Jiang Z, Zhang H, Liang W, Huang W, Zhang H. et al. Sodium butyrate attenuates diabetes-induced aortic endothelial dysfunction via P300-mediated transcriptional activation of Nrf2. Free Radic Biol Med. 2018;124:454-65
41. Shujie C, Liujing H, Bingdong L, Huimin D, Ze L, Yifan L. et al. Dynamic Changes in Butyrate Levels Regulate Satellite Cell Homeostasis by Preventing Spontaneous Activation During Aging. SCI CHINA LIFE SCI. 2024;67:745-64
42. Cooper ID, Kyriakidou Y, Petagine L, Edwards K, Elliott BT. Bio-Hacking Better Health-Leveraging Metabolic Biochemistry to Maximise Healthspan. Antioxidants (Basel). 2023;12:1749
43. Fan SZ, Lin CS, Wei YW, Yeh SR, Tsai YH, Lee AC. et al. Dietary citrate supplementation enhances longevity, metabolic health, and memory performance through promoting ketogenesis. Aging Cell. 2021;20:e13510
44. Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58-67
45. Suárez Y, Fernández C, Ledo B, Martín M, Gómez-Coronado D, Lasunción MA. Sterol stringency of proliferation and cell cycle progression in human cells. Biochim Biophys Acta. 2005;1734:203-13
46. Pradas I, Jové M, Huynh K, Puig J, Ingles M, Borras C. et al. Exceptional human longevity is associated with a specific plasma phenotype of ether lipids. Redox Biol. 2019;21:101127
47. Jové M, Naudí A, Gambini J, Borras C, Cabré R, Portero-Otín M. et al. A Stress-Resistant Lipidomic Signature Confers Extreme Longevity to Humans. J Gerontol A Biol Sci Med Sci. 2017;72:30-7
48. Montoliu I, Scherer M, Beguelin F, DaSilva L, Mari D, Salvioli S. et al. Serum profiling of healthy aging identifies phospho- and sphingolipid species as markers of human longevity. Aging (Albany NY). 2014;6:9-25
49. Pradas I, Jové M, Huynh K, Ingles M, Borras C, Mota-Martorell N. et al. Long-lived Humans Have a Unique Plasma Sphingolipidome. J Gerontol A Biol Sci Med Sci. 2022;77:728-35
50. Barbacini P, Torretta E, Arosio B, Ferri E, Capitanio D, Moriggi M. et al. Novel Insight into the Serum Sphingolipid Fingerprint Characterizing Longevity. Int J Mol Sci. 2022;23:2428
51. Zatloukal J, Zylla S, Markus MRP, Ewert R, Gläser S, Völzke H. et al. The Association Between C24:0/C16:0 Ceramide Ratio and Cardiorespiratory Fitness is Robust to Effect Modifications by Age and Sex. Adv Biol (Weinh). 2024;8:e2300633
52. Czubowicz K, Strosznajder R. Ceramide in the molecular mechanisms of neuronal cell death. The role of sphingosine-1-phosphate. Mol Neurobiol. 2014;50:26-37
53. Osawa Y, Banno Y, Nagaki M, Brenner DA, Naiki T, Nozawa Y. et al. TNF-alpha-induced sphingosine 1-phosphate inhibits apoptosis through a phosphatidylinositol 3-kinase/Akt pathway in human hepatocytes. J Immunol. 2001;167:173-80
54. Selman C, Partridge L, Withers DJ. Replication of extended lifespan phenotype in mice with deletion of insulin receptor substrate 1. PLoS One. 2011;6:e16144
55. Darst BF, Koscik RL, Hogan KJ, Johnson SC, Engelman CD. Longitudinal plasma metabolomics of aging and sex. Aging (Albany NY). 2019;11:1262-82
56. Gubbels Bupp MR. Sex, the aging immune system, and chronic disease. Cell Immunol. 2015;294:102-10
57. Oertelt-Prigione S. The influence of sex and gender on the immune response. Autoimmun Rev. 2012;11:A479-85
58. Viña J, Borrás C, Gambini J, Sastre J, Pallardó FV. Why females live longer than males? Importance of the upregulation of longevity-associated genes by oestrogenic compounds. FEBS Lett. 2005;579:2541-5
59. Zhu Q, Chen SM, Li HW, Li RR, Yang SS, Wang SS. et al. Association analysis between sex hormone levels and all-cause mortality in Hainan female centenarians. Zhonghua Liu Xing Bing Xue Za Zhi. 2023;44:1245-50
60. Johnson LC, Parker K, Aguirre BF, Nemkov TG, D'Alessandro A, Johnson SA. et al. The plasma metabolome as a predictor of biological aging in humans. Geroscience. 2019;41:895-906
61. Mota-Martorell N, Jové M, Borrás C, Berdún R, Obis È, Sol J. et al. Methionine transsulfuration pathway is upregulated in long-lived humans. Free Radic Biol Med. 2021;162:38-52
62. Brosnan JT, Brosnan ME. The sulfur-containing amino acids: an overview. J Nutr. 2006;136:1636s-40s
63. Ouyang Y, Wu Q, Li J, Sun S, Sun S. S-adenosylmethionine: A metabolite critical to the regulation of autophagy. Cell Prolif. 2020;53:e12891
64. Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119-25
65. Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A. et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524-8
66. Lee BC, Kaya A, Gladyshev VN. Methionine restriction and life-span control. Ann N Y Acad Sci. 2016;1363:116-24
67. Norman K, Klaus S. Veganism, aging and longevity: new insight into old concepts. Curr Opin Clin Nutr Metab Care. 2020;23:145-50
68. Paeslack N, Mimmler M, Becker S, Gao Z, Khuu MP, Mann A. et al. Microbiota-derived tryptophan metabolites in vascular inflammation and cardiovascular disease. Amino Acids. 2022;54:1339-56
69. Li H, Ren M, Li Q. 1H NMR-Based Metabolomics Reveals the Intrinsic Interaction of Age, Plasma Signature Metabolites, and Nutrient Intake in the Longevity Population in Guangxi, China. Nutrients. 2022;14:2539
70. Cai D, Zhao Z, Zhao L, Dong Y, Wang L, Zhao S. et al. The Age-Accompanied and Diet-Associated Remodeling of the Phospholipid, Amino Acid, and SCFA Metabolism of Healthy Centenarians from a Chinese Longevous Region: A Window into Exceptional Longevity. Nutrients. 2022;14:4420
71. Gupta NK, Thaker AI, Kanuri N, Riehl TE, Rowley CW, Stenson WF. et al. Serum analysis of tryptophan catabolism pathway: correlation with Crohn's disease activity. Inflamm Bowel Dis. 2012;18:1214-20
72. Castro-Portuguez R, Sutphin GL. Kynurenine pathway, NAD(+) synthesis, and mitochondrial function: Targeting tryptophan metabolism to promote longevity and healthspan. Exp Gerontol. 2020;132:110841
73. Solvang SH, Hodge A, Watne LO, Cabral-Marques O, Nordrehaug JE, Giles GG. et al. Kynurenine Pathway Metabolites in the Blood and Cerebrospinal Fluid Are Associated with Human Aging. Oxid Med Cell Longev. 2022;2022:5019752
74. Agudelo LZ, Ferreira DMS, Cervenka I, Bryzgalova G, Dadvar S, Jannig PR. et al. Kynurenic Acid and Gpr35 Regulate Adipose Tissue Energy Homeostasis and Inflammation. Cell Metab. 2018;27:378-92.e5
75. Dongol A, Chen X, Zheng P, Seyhan ZB, Huang XF. Quinolinic acid impairs mitophagy promoting microglia senescence and poor healthspan in C. elegans: a mechanism of impaired aging process. Biol Direct. 2023;18:86
76. Mirisola MG, Taormina G, Fabrizio P, Wei M, Hu J, Longo VD. Serine- and threonine/valine-dependent activation of PDK and Tor orthologs converge on Sch9 to promote aging. PLoS Genet. 2014;10:e1004113
77. Mansfeld J, Urban N, Priebe S, Groth M, Frahm C, Hartmann N. et al. Branched-chain amino acid catabolism is a conserved regulator of physiological ageing. Nat Commun. 2015;6:10043
78. Zhang Y, Zhou Q, Yang R, Hu C, Huang Z, Zheng C. et al. Serum branched-chain amino acids are associated with leukocyte telomere length and frailty based on residents from Guangxi longevity county. Sci Rep. 2020;10:10252
79. Xu H, Wang X, Geng G, Xu X, Liu L, Zhang Y. et al. Association of Circulating Branched-Chain Amino Acids with Cardiovascular Diseases: A Mendelian Randomization Study. Nutrients. 2023;15:1580
80. Jiang W, Lu K, Zhuang Z, Wang X, Tang X, Huang T. et al. Mendelian Randomization Analysis Provides Insights into the Pathogenesis of Serum Levels of Branched-Chain Amino Acids in Cardiovascular Disease. Metabolites. 2023;13:403
81. Qian XH, Liu XL, Zhang B, Lin Y, Xu JH, Ding GY. et al. Investigating the causal association between branched-chain amino acids and Alzheimer's disease: A bidirectional Mendelian randomized study. Front Nutr. 2023;10:1103303
82. Li H, Ye D, Xie W, Hua F, Yang Y, Wu J. et al. Defect of branched-chain amino acid metabolism promotes the development of Alzheimer's disease by targeting the mTOR signaling. Biosci Rep. 2018;38:BSR20180127
83. Yeh CY, Chini LCS, Davidson JW, Garcia GG, Gallagher MS, Freichels IT. et al. Late-life isoleucine restriction promotes physiological and molecular signatures of healthy aging. bioRxiv. 2024
84. Green CL, Trautman ME, Chaiyakul K, Jain R, Alam YH, Babygirija R. et al. Dietary restriction of isoleucine increases healthspan and lifespan of genetically heterogeneous mice. Cell Metab. 2023;35:1976-95.e6
85. Ferri E, Casati M, Cesari M, Vitale G, Arosio B. Vitamin D in physiological and pathological aging: Lesson from centenarians. Rev Endocr Metab Disord. 2019;20:273-82
86. Milman S, Schulder-Katz M, Deluty J, Zimmerman ME, Crandall JP, Barzilai N. et al. Individuals with exceptional longevity manifest a delayed association between vitamin D insufficiency and cognitive impairment. J Am Geriatr Soc. 2014;62:153-8
87. Yao Y, Fu S, Shi Q, Zhang H, Zhu Q, Zhang F. et al. Prevalence of functional dependence in Chinese centenarians and its relationship with serum vitamin D status. Clin Interv Aging. 2018;13:2045-53
88. Tanprasertsuk J, Ferland G, Johnson MA, Poon LW, Scott TM, Barbey AK. et al. Concentrations of Circulating Phylloquinone, but Not Cerebral Menaquinone-4, Are Positively Correlated with a Wide Range of Cognitive Measures: Exploratory Findings in Centenarians. J Nutr. 2020;150:82-90
89. Lubes G, Goodarzi M. Analysis of Volatile Compounds by Advanced Analytical Techniques and Multivariate Chemometrics. Chem Rev. 2017;117:6399-422
90. Shirasu M, Touhara K. The scent of disease: volatile organic compounds of the human body related to disease and disorder. J Biochem. 2011;150:257-66
91. Ahmed I, Greenwood R, Costello B, Ratcliffe N, Probert CS. Investigation of faecal volatile organic metabolites as novel diagnostic biomarkers in inflammatory bowel disease. Aliment Pharmacol Ther. 2016;43:596-611
92. Liu D, Zhao N, Wang M, Pi X, Feng Y, Wang Y. et al. Urine volatile organic compounds as biomarkers for minimal change type nephrotic syndrome. Biochem Biophys Res Commun. 2018;496:58-63
93. Bond A, Greenwood R, Lewis S, Corfe B, Sarkar S, O'Toole P. et al. Volatile organic compounds emitted from faeces as a biomarker for colorectal cancer. Aliment Pharmacol Ther. 2019;49:1005-12
94. Conte M, Conte G, Martucci M, Monti D, Casarosa L, Serra A. et al. The smell of longevity: a combination of Volatile Organic Compounds (VOCs) can discriminate centenarians and their offspring from age-matched subjects and young controls. Geroscience. 2020;42:201-16
95. Miura Y, Sato Y, Arai Y, Abe Y, Takayama M, Toda T. et al. Proteomic analysis of plasma proteins in Japanese semisuper centenarians. Exp Gerontol. 2011;46:81-5
96. Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF. et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev. 1996;13:19-50
97. Schächter F, Faure-Delanef L, Guénot F, Rouger H, Froguel P, Lesueur-Ginot L. et al. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6:29-32
98. Giannakou ME, Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 2004;14:408-12
99. Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196-207
100. Lee SH, Lee JH, Lee HY, Min KJ. Sirtuin signaling in cellular senescence and aging. BMB Rep. 2019;52:24-34
101. Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63:287-303
102. Verghese PB, Castellano JM, Holtzman DM. Apolipoprotein E in Alzheimer's disease and other neurological disorders. Lancet Neurol. 2011;10:241-52
103. Brooks-Wilson AR. Genetics of healthy aging and longevity. Hum Genet. 2013;132:1323-38
104. Garatachea N, Marín PJ, Santos-Lozano A, Sanchis-Gomar F, Emanuele E, Lucia A. The ApoE gene is related with exceptional longevity: a systematic review and meta-analysis. Rejuvenation Res. 2015;18:3-13
105. Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K. et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987-92
106. Flachsbart F, Caliebe A, Kleindorp R, Blanché H, von Eller-Eberstein H, Nikolaus S. et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700-5
107. Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY. et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897-904
108. Bae H, Gurinovich A, Malovini A, Atzmon G, Andersen SL, Villa F. et al. Effects of FOXO3 Polymorphisms on Survival to Extreme Longevity in Four Centenarian Studies. J Gerontol A Biol Sci Med Sci. 2018;73:1439-47
109. Robert L, Fulop T. Longevity and its regulation: centenarians and beyond. Interdiscip Top Gerontol. 2014;39:198-211
110. Longo VD, Antebi A, Bartke A, Barzilai N, Brown-Borg HM, Caruso C. et al. Interventions to Slow Aging in Humans: Are We Ready? Aging Cell. 2015;14:497-510
111. Mahley RW, Rall SC Jr. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507-37
112. Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L. et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686-98
113. Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanché H, Junge O. et al. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech Ageing Dev. 2011;132:324-30
114. Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW. et al. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848
115. Deelen J, Beekman M, Uh HW, Broer L, Ayers KL, Tan Q. et al. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum Mol Genet. 2014;23:4420-32
116. Zeng Y, Nie C, Min J, Liu X, Li M, Chen H. et al. Novel loci and pathways significantly associated with longevity. Sci Rep. 2016;6:21243
117. Garatachea N, Emanuele E, Calero M, Fuku N, Arai Y, Abe Y. et al. ApoE gene and exceptional longevity: Insights from three independent cohorts. Exp Gerontol. 2014;53:16-23
118. Lindahl-Jacobsen R, Tan Q, Mengel-From J, Christensen K, Nebel A, Christiansen L. Effects of the APOE ε2 allele on mortality and cognitive function in the oldest old. J Gerontol A Biol Sci Med Sci. 2013;68:389-94
119. Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106-18
120. Huq AJ, Fransquet P, Laws SM, Ryan J, Sebra R, Masters CL. et al. Genetic resilience to Alzheimer's disease in APOE ε4 homozygotes: A systematic review. Alzheimers Dement. 2019;15:1612-23
121. R AA. Risk factors for Alzheimer's disease. Folia Neuropathol. 2019;57:87-105
122. Sebastiani P, Monti S, Morris M, Gurinovich A, Toshiko T, Andersen SL. et al. A serum protein signature of APOE genotypes in centenarians. Aging Cell. 2019;18:e13023
123. Wolters FJ, Yang Q, Biggs ML, Jakobsdottir J, Li S, Evans DS. et al. The impact of APOE genotype on survival: Results of 38,537 participants from six population-based cohorts (E2-CHARGE). PLoS One. 2019;14:e0219668
124. Sebastiani P, Song Z, Ellis D, Tian Q, Schwaiger-Haber M, Stancliffe E. et al. A metabolomic signature of the APOE2 allele. Geroscience. 2023;45:415-26
125. Lin R, Zhang Y, Yan D, Liao X, Wang X, Fu Y. et al. Genetic Association Analysis of Common Variants in FOXO3 Related to Longevity in a Chinese Population. PLoS One. 2016;11:e0167918
126. Grossi V, Forte G, Sanese P, Peserico A, Tezil T, Lepore Signorile M. et al. The longevity SNP rs2802292 uncovered: HSF1 activates stress-dependent expression of FOXO3 through an intronic enhancer. Nucleic Acids Res. 2018;46:5587-600
127. Willcox BJ, Morris BJ, Tranah GJ, Chen R, Masaki KH, He Q. et al. Longevity-Associated FOXO3 Genotype and its Impact on Coronary Artery Disease Mortality in Japanese, Whites, and Blacks: A Prospective Study of Three American Populations. J Gerontol A Biol Sci Med Sci. 2017;72:724-8
128. Shimokawa I, Komatsu T, Hayashi N, Kim SE, Kawata T, Park S. et al. The life-extending effect of dietary restriction requires Foxo3 in mice. Aging Cell. 2015;14:707-9
129. Becker L, Nguyen L, Gill J, Kulkarni S, Pasricha PJ, Habtezion A. Age-dependent shift in macrophage polarisation causes inflammation-mediated degeneration of enteric nervous system. Gut. 2018;67:827-36
130. Chang ZS, Xia JB, Wu HY, Peng WT, Jiang FQ, Li J. et al. Forkhead box O3 protects the heart against paraquat-induced aging-associated phenotypes by upregulating the expression of antioxidant enzymes. Aging Cell. 2019;18:e12990
131. Willcox BJ, Tranah GJ, Chen R, Morris BJ, Masaki KH, He Q. et al. The FoxO3 gene and cause-specific mortality. Aging Cell. 2016;15:617-24
132. Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ Jr, DiStefano PS. et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530-4
133. Xinxin QI, Li S. Sirtuin family and its biological characteristics. Acta Medicinae Sinica. 2016;29:169-174
134. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y. et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011-5
135. Kobayashi Y, Furukawa-Hibi Y, Chen C, Horio Y, Isobe K, Ikeda K. et al. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med. 2005;16:237-43
136. Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758-71
137. Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F. et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258-63
138. Li Y, Qin J, Wei X, Liang G, Shi L, Jiang M. et al. Association of SIRT6 Gene Polymorphisms with Human Longevity. Iran J Public Health. 2016;45:1420-6
139. Hirvonen K, Laivuori H, Lahti J, Strandberg T, Eriksson JG, Hackman P. SIRT6 polymorphism rs117385980 is associated with longevity and healthy aging in Finnish men. BMC Med Genet. 2017;18:41
140. Kastberger B, Winter S, Brandstätter H, Biller J, Wagner W, Plesnila N. Treatment with Cerebrolysin Prolongs Lifespan in a Mouse Model of Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy. Adv Biol (Weinh). 2024;8:e2300439
141. Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED. et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416-30
142. Akasaki Y, Alvarez-Garcia O, Saito M, Caramés B, Iwamoto Y, Lotz MK. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014;66:3349-58
143. D'Adamo S, Cetrullo S, Guidotti S, Borzì RM, Flamigni F. Hydroxytyrosol modulates the levels of microRNA-9 and its target sirtuin-1 thereby counteracting oxidative stress-induced chondrocyte death. Osteoarthritis Cartilage. 2017;25:600-10
144. Sun X, Li Q, Tang Y, Hu W, Chen G, An H. et al. Epigenetic activation of secretory phenotypes in senescence by the FOXQ1-SIRT4-GDH signaling. Cell Death Dis. 2023;14:481
145. Jin JH, Wen DT, Chen YL, Hou WQ. Muscle FOXO-Specific Overexpression and Endurance Exercise Protect Skeletal Muscle and Heart from Defects Caused by a High-Fat Diet in Young Drosophila. Front Biosci (Landmark Ed). 2023;28:16
146. Kim S, Byun J, Jung S, Kim B, Lee K, Jeon H. et al. Sirtuin 7 Inhibitor Attenuates Colonic Mucosal Immune Activation in Mice-Potential Therapeutic Target in Inflammatory Bowel Disease. Biomedicines. 2022;10:2693
147. Sebastiani P, Federico A, Morris M, Gurinovich A, Tanaka T, Chandler KB. et al. Protein signatures of centenarians and their offspring suggest centenarians age slower than other humans. Aging Cell. 2021;20:e13290
148. Türker Duyuler P, Duyuler S, Gök M, Kundi H, Topçuoğlu C, Güray Ü. Pleiotrophin levels are associated with improved coronary collateral circulation. Coron Artery Dis. 2018;29:68-73
149. Menni C, Kiddle SJ, Mangino M, Viñuela A, Psatha M, Steves C. et al. Circulating Proteomic Signatures of Chronological Age. J Gerontol A Biol Sci Med Sci. 2015;70:809-16
150. Tanaka T, Biancotto A, Moaddel R, Moore AZ, Gonzalez-Freire M, Aon MA. et al. Plasma proteomic signature of age in healthy humans. Aging Cell. 2018;17:e12799
151. Sathyan S, Ayers E, Gao T, Weiss EF, Milman S, Verghese J. et al. Plasma proteomic profile of age, health span, and all-cause mortality in older adults. Aging Cell. 2020;19:e13250
152. Grünberg JR, Elvin J, Paul A, Hedjazifar S, Hammarstedt A, Smith U. CCN5/WISP2 and metabolic diseases. J Cell Commun Signal. 2018;12:309-18
153. Chowdhury S, Wang X, Srikant CB, Li Q, Fu M, Gong YJ. et al. IGF-I stimulates CCN5/WISP2 gene expression in pancreatic β-cells, which promotes cell proliferation and survival against streptozotocin. Endocrinology. 2014;155:1629-42
154. Liu JL, Kaddour N, Chowdhury S, Li Q, Gao ZH. Role of CCN5 (WNT1 inducible signaling pathway protein 2) in pancreatic islets. J Diabetes. 2017;9:462-74
155. Brown BA, Connolly GM, Mill CEJ, Williams H, Angelini GD, Johnson JL. et al. Aging differentially modulates the Wnt pro-survival signalling pathways in vascular smooth muscle cells. Aging Cell. 2019;18:e12844
156. Alcazar J, Frandsen U, Prokhorova T, Kamper RS, Haddock B, Aagaard P. et al. Changes in systemic GDF15 across the adult lifespan and their impact on maximal muscle power: the Copenhagen Sarcopenia Study. J Cachexia Sarcopenia Muscle. 2021;12:1418-27
157. Santos-Lozano A, Valenzuela PL, Llavero F, Lista S, Carrera-Bastos P, Hampel H. et al. Successful aging: insights from proteome analyses of healthy centenarians. Aging (Albany NY). 2020;12:3502-15
158. He YH, Lu X, Yang LQ, Xu LY, Kong QP. Association of the insulin-like growth factor binding protein 3 (IGFBP-3) polymorphism with longevity in Chinese nonagenarians and centenarians. Aging (Albany NY). 2014;6:944-56
159. Elzi DJ, Lai Y, Song M, Hakala K, Weintraub ST, Shiio Y. Plasminogen activator inhibitor 1-insulin-like growth factor binding protein 3 cascade regulates stress-induced senescence. Proc Natl Acad Sci U S A. 2012;109:12052-7
160. Sanada F, Taniyama Y, Muratsu J, Otsu R, Iwabayashi M, Carracedo M. et al. Activated Factor X Induces Endothelial Cell Senescence Through IGFBP-5. Sci Rep. 2016;6:35580
161. Siino V, Ali A, Accardi G, Aiello A, Ligotti ME, Mosquim Junior S. et al. Plasma proteome profiling of healthy individuals across the life span in a Sicilian cohort with long-lived individuals. Aging Cell. 2022;21:e13684
162. Orwoll ES, Wiedrick J, Nielson CM, Jacobs J, Baker ES, Piehowski P. et al. Proteomic assessment of serum biomarkers of longevity in older men. Aging Cell. 2020;19:e13253
163. Capitanio D, Barbacini P, Arosio B, Guerini FR, Torretta E, Trecate F. et al. Can Serum Nitrosoproteome Predict Longevity of Aged Women? Int J Mol Sci. 2020;21:9009
164. Miura Y, Hashii N, Tsumoto H, Takakura D, Ohta Y, Abe Y. et al. Change in N-Glycosylation of Plasma Proteins in Japanese Semisupercentenarians. PLoS One. 2015;10:e0142645
165. López-Sánchez LM, Muntané J, de la Mata M, Rodríguez-Ariza A. Unraveling the S-nitrosoproteome: tools and strategies. Proteomics. 2009;9:808-18
166. López-Sánchez LM, López-Pedrera C, Rodríguez-Ariza A. Proteomic approaches to evaluate protein S-nitrosylation in disease. Mass Spectrom Rev. 2014;33:7-20
167. Wink DA, Miranda KM, Espey MG, Pluta RM, Hewett SJ, Colton C. et al. Mechanisms of the antioxidant effects of nitric oxide. Antioxid Redox Signal. 2001;3:203-13
168. Fernando V, Zheng X, Walia Y, Sharma V, Letson J, Furuta S. S-Nitrosylation: An Emerging Paradigm of Redox Signaling. Antioxidants (Basel). 2019;8:404
169. Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721-32
170. Rizza S, Cardaci S, Montagna C, Di Giacomo G, De Zio D, Bordi M. et al. S-nitrosylation drives cell senescence and aging in mammals by controlling mitochondrial dynamics and mitophagy. Proc Natl Acad Sci U S A. 2018;115:E3388-e97
171. Wada Y, Tajiri M, Yoshida S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal Chem. 2004;76:6560-5
172. Catera M, Borelli V, Malagolini N, Chiricolo M, Venturi G, Reis CA. et al. Identification of novel plasma glycosylation-associated markers of aging. Oncotarget. 2016;7:7455-68
173. Yu X, Wang Y, Kristic J, Dong J, Chu X, Ge S. et al. Profiling IgG N-glycans as potential biomarker of chronological and biological ages: A community-based study in a Han Chinese population. Medicine (Baltimore). 2016;95:e4112
174. Ebert H, Lacruz ME, Kluttig A, Simm A, Greiser KH, Tiller D. et al. Association between advanced glycation end products, their soluble receptor, and mortality in the general population: Results from the CARLA study. Exp Gerontol. 2020;131:110815
175. Miura Y, Hashii N, Ohta Y, Itakura Y, Tsumoto H, Suzuki J. et al. Characteristic glycopeptides associated with extreme human longevity identified through plasma glycoproteomics. Biochim Biophys Acta Gen Subj. 2018;1862:1462-71
176. Butterfield DA. Phosphoproteomics of Alzheimer disease brain: Insights into altered brain protein regulation of critical neuronal functions and their contributions to subsequent cognitive loss. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2031-9
177. Zhang K, Schrag M, Crofton A, Trivedi R, Vinters H, Kirsch W. Targeted proteomics for quantification of histone acetylation in Alzheimer's disease. Proteomics. 2012;12:1261-8
178. Burkewitz K, Zhang Y, Mair WB. AMPK at the nexus of energetics and aging. Cell Metab. 2014;20:10-25
179. Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG. et al. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol. 2009;5:664-72
180. Ronnebaum SM, Wu Y, McDonough H, Patterson C. The ubiquitin ligase CHIP prevents SirT6 degradation through noncanonical ubiquitination. Mol Cell Biol. 2013;33:4461-72
181. Han Y, Quan X, Chuang Y, Liang Q, Li Y, Yuan Z. et al. A multi-omics analysis for the prediction of neurocognitive disorders risk among the elderly in Macao. Clin Transl Med. 2022;12:e909
182. Semba RD, Tian Q, Carlson MC, Xue QL, Ferrucci L. Motoric cognitive risk syndrome: Integration of two early harbingers of dementia in older adults. Ageing Res Rev. 2020;58:101022
183. Navas-Carrillo D, Rivera-Caravaca JM, Sampedro-Andrada A, Orenes-Piñero E. Novel biomarkers in Alzheimer's disease using high resolution proteomics and metabolomics: miRNAS, proteins and metabolites. Crit Rev Clin Lab Sci. 2021;58:167-79
184. Ahadi S, Zhou W, Schüssler-Fiorenza Rose SM, Sailani MR, Contrepois K, Avina M. et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat Med. 2020;26:83-90
185. Tebani A, Gummesson A, Zhong W, Koistinen IS, Lakshmikanth T, Olsson LM. et al. Integration of molecular profiles in a longitudinal wellness profiling cohort. Nat Commun. 2020;11:4487
186. Assi R, Cherifi C, Cornelis FMF, Zhou Q, Storms L, Pazmino S. et al. Inhibition of KDM7A/B histone demethylases restores H3K79 methylation and protects against osteoarthritis. Ann Rheum Dis. 2023;82:963-73
187. Pedro Ferreira J, Pitt B, Zannad F. Histone deacetylase inhibitors for cardiovascular conditions and healthy longevity. Lancet Healthy Longev. 2021;2:e371-e9
188. Gensous N, Sala C, Pirazzini C, Ravaioli F, Milazzo M, Kwiatkowska KM. et al. A Targeted Epigenetic Clock for the Prediction of Biological Age. Cells. 2022;11:4044
189. Dec E, Clement J, Cheng K, Church GM, Fossel MB, Rehkopf DH. et al. Centenarian clocks: epigenetic clocks for validating claims of exceptional longevity. Geroscience. 2023;45:1817-35
190. Dong C, Miao YR, Zhao R, Yang M, Guo AY, Xue ZH. et al. Single-Cell Transcriptomics Reveals Longevity Immune Remodeling Features Shared by Centenarians and Their Offspring. Adv Sci (Weinh). 2022;9:e2204849
191. Karagiannis TT, Dowrey TW, Villacorta-Martin C, Montano M, Reed E, Belkina AC. et al. Multi-modal profiling of peripheral blood cells across the human lifespan reveals distinct immune cell signatures of aging and longevity. EBioMedicine. 2023;90:104514
192. Tedone E, Huang E, O'Hara R, Batten K, Ludlow AT, Lai TP. et al. Telomere length and telomerase activity in T cells are biomarkers of high-performing centenarians. Aging Cell. 2019;18:e12859
193. Li GH, Han F, Xiao FH, Gu KS, Shen Q, Xu W. et al. System-level metabolic modeling facilitates unveiling metabolic signature in exceptional longevity. Aging Cell. 2022;21:e13595
194. Wilmanski T, Diener C, Rappaport N, Patwardhan S, Wiedrick J, Lapidus J. et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat Metab. 2021;3:274-86
195. Dang H, Castro-Portuguez R, Espejo L, Backer G, Freitas S, Spence E. et al. On the benefits of the tryptophan metabolite 3-hydroxyanthranilic acid in Caenorhabditis elegans and mouse aging. Nat Commun. 2023;14:8338
196. Han B, Sivaramakrishnan P, Lin CJ, Neve IAA, He J, Tay LWR. et al. Microbial Genetic Composition Tunes Host Longevity. Cell. 2017;169:1249-62 e13
197. Madeo F, Hofer SJ, Pendl T, Bauer MA, Eisenberg T, Carmona-Gutierrez D. et al. Nutritional Aspects of Spermidine. Annu Rev Nutr. 2020;40:135-59
198. Nie C, Li Y, Li R, Yan Y, Zhang D, Li T. et al. Distinct biological ages of organs and systems identified from a multi-omics study. Cell Rep. 2022;38:110459
199. Horgusluoglu E, Neff R, Song WM, Wang M, Wang Q, Arnold M. et al. Integrative metabolomics-genomics approach reveals key metabolic pathways and regulators of Alzheimer's disease. Alzheimers Dement. 2022;18:1260-78
200. Li C, Luan Z, Zhao Y, Chen J, Yang Y, Wang C. et al. Deep insights into the gut microbial community of extreme longevity in south Chinese centenarians by ultra-deep metagenomics and large-scale culturomics. NPJ Biofilms Microbiomes. 2022;8:28
Author contact
![]() Corresponding authors: Xuemeng Li, Zhanjiang Key Laboratory of Human Microecology and Clinical Translation Research, The Marine Biomedical Research Institute, College of Basic Medicine, Guangdong Medical University, Dongguan, China. Emails: lixuemengedu.cn. Jun-Yan Liu, Center for Novel Target & Therapeutic Intervention (CNTTI), Institute of Life Sciences, Chongqing Medical University, Chongqing, China. Email: jyliuedu.cn. Hui Luo, Zhanjiang Key Laboratory of Human Microecology and Clinical Translation Research, The Marine Biomedical Research Institute, College of Basic Medicine, Guangdong Medical University, Dongguan, China. Emails: luohuiedu.cn.
Corresponding authors: Xuemeng Li, Zhanjiang Key Laboratory of Human Microecology and Clinical Translation Research, The Marine Biomedical Research Institute, College of Basic Medicine, Guangdong Medical University, Dongguan, China. Emails: lixuemengedu.cn. Jun-Yan Liu, Center for Novel Target & Therapeutic Intervention (CNTTI), Institute of Life Sciences, Chongqing Medical University, Chongqing, China. Email: jyliuedu.cn. Hui Luo, Zhanjiang Key Laboratory of Human Microecology and Clinical Translation Research, The Marine Biomedical Research Institute, College of Basic Medicine, Guangdong Medical University, Dongguan, China. Emails: luohuiedu.cn.

 Global reach, higher impact
Global reach, higher impact