Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(11):2201-2207. doi:10.7150/ijms.99545 This issue Cite
Research Paper
Association of long noncoding RNA GAS5 gene polymorphism with progression of diabetic kidney disease
1. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
2. Department of Family and Community Medicine, Chung Shan Medical University Hospital, Taichung, Taiwan.
3. Division of Cardiology, Department of Internal Medicine, Changhua Christian Hospital, Yunlin Branch, Yunlin, Taiwan.
4. Department of Medicine and Nursing, Hungkuang University, Taichung, Taiwan.
5. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan.
6. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
7. Division of Nephrology, Department of Internal Medicine, Chung Shan Medical University Hospital, Taichung, Taiwan.
8. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan.
9. Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan.
10. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
Received 2024-6-12; Accepted 2024-8-4; Published 2024-8-13
Abstract
Diabetic kidney disease (DKD) is a common microvascular complication of diabetes, whose complex etiology involves a genetic component. Growth arrest-specific 5 (GAS5), a long noncoding RNA (lncRNA) gene, has been recently shown to regulate renal fibrosis. Here, we aimed to explore the potential role of GAS5 gene polymorphisms in the predisposition to DKD. One single-nucleotide (rs55829688) and one insertion/deletion polymorphism (rs145204276) of GAS5 gene were surveyed in 778 DKD cases and 788 DKD-free diabetic controls. We demonstrated that diabetic subjects who are heterozygous at rs55829688 (TC; AOR, 1.737; 95% CI, 1.028-2.937; p=0.039) are more susceptible to advanced DKD but not early-staged DKD, as compared to diabetic subjects who are homozygous for the major allele of rs55829688 (TT). Carriers of at least one minor allele (C) of rs55829688 (TC and CC; AOR, 1.317; 95% CI, 1.023-1.696; p=0.033) more frequently suffer from advanced DKD than do those homozygotes for the major allele (TT). Furthermore, in comparison to those who do not carry the minor allele of rs55829688 (TT), advanced DKD patients possessing at least one minor allele of rs55829688 (TC and CC) exhibited a lower glomerular filtration rate, revealing an impact of rs55829688 on renal co-morbidities of diabetes. In conclusion, our data indicate an association of GAS5 gene polymorphisms with the progression of DKD.
Keywords: growth arrest-specific 5, gene polymorphism, diabetic kidney disease, long non-coding RNA
Introduction
Chronic kidney disease (CKD) and its progression into end-stage renal disease (ESRD) have been associated with premature mortality and recognized as a global healthcare priority [1]. As the renal tissue represents one of the key targets of microvascular damage in diabetes, diabetic kidney disease (DKD), a common complication of diabetes that develops in approximately half of cases with type 2 diabetes mellitus (T2DM) and one-third of those with type 1 diabetes mellitus (T1DM) [2], was thought to be caused by a series of metabolic, hemodynamic, and immunological dysfunctions [3]. These dysregulated reactions, involving excessive excretion of metabolites due to aberrant glucose catabolism [4], disturbance of the renin-angiotensin-aldosterone system (RAAS) [5], and activation of numerous signaling cascades that are linked to kidney fibrosis [6, 7], oxidative stress [8, 9], complement system [10], and inflammation [11, 12], collectively orchestrate the pathogenesis of DKD, leading to irreversible kidney damage. Distinct risk parameters have been recognized as contributors to DKD. In addition to several non-modifiable risks (such as age, gender, and genetic inheritance), some of these factors, including hyperglycemia, obesity, hypertension, and dyslipidemia, appear possibly modifiable through intensive diabetes care [13]. Such complexity of DKD etiology augments the heterogeneity of the disease epidemiology and treatment, thus prompting us for the discovery of novel biomarkers or manipulable pathogenic factors to improve DKD diagnosis and management.
Numerous investigations have revealed a clear genetic component to both diabetes and its co-morbidities [14]. In addition, diabetic subjects with a family history of hypertension or cardiovascular disease tend to develop DKD more frequently [15, 16], supporting the notion that genetic parameters, to some extent, confer the predisposition of DKD in T2DM patients. To date, several hundreds of genetic variants associated with T2DM and DKD have been identified from recent large-scale, multi-ancestry studies [17-21]. These genes demonstrate the genetic architecture of T2DM and provide pathogenic insights into DKD, particularly in the development of diabetes, albuminuria, and reduced kidney function in different ethnic groups [22]. Nevertheless, the spectrum of DKD susceptibility loci is highly heterogeneous and accounts for only a certain proportion of why some subjects develop CKD and some do not [23]. Therefore, identification of novel inherited factors relevant to the development and progression of DKD not only facilitates the understanding of the molecular mechanisms of DKD, but also offers reliable molecular targets for early diagnosis and effective treatment.
The growth arrest-specific 5 (GAS5) gene encodes a long noncoding RNA (lncRNA) that was originally found to regulate cell growth, differentiation, and development [24, 25]. In addition to cell growth arrest, GAS5 promotes cell apoptosis through acting as a decoy to repress activities of the glucocorticoid receptor, which is a transcription factor for inducing the expression of its target genes in diverse glucocorticoid-mediated responses, such as cell growth/survival and energy expenditure [26]. As being downregulated in a verity of malignancies, a tumor-suppressive role of GAS5 has been recognized [27, 28]. However, not only implicated in cancer development, a functional association of GAS5 with renal fibrosis, a pathological feature of CKD characterized by an excessive accumulation and deposition of extracellular matrix (ECM) components [29], was also proposed. Recently, in an animal model of diabetes, GAS5 was shown to attenuate renal interstitial fibrosis and kidney inflammation by downregulating matrix metalloproteinase-9, a key regulator of ECM remodeling [30]. Through sponging specific microRNAs, such suppressive effect of GAS5 on renal fibrosis was consistently observed [31]. These findings suggest a connection between GAS5 and renal traits in diabetic individuals. Moreover, the single-nucleotide polymorphisms (SNPs) of GAS5 were reported to be associated with risk of various cancers [32-35]. To date, the effect of GAS5 gene polymorphisms on the risk and progression of DKD remains unexplored, while a genetic association of GAS5 with two common co-morbidities of diabetes, retinopathy and coronary artery disease, has been detected [36, 37]. Here, we aimed to explore the impact of GAS5 gene variants on the development and progression of DKD.
Materials and Methods
Subject enrollment
To explore the influence of GAS5 gene polymorphisms on the risk of DKD, 778 patients with DKD were recruited in Chung Shan Medical University Hospital, Taichung, Taiwan, with the approval by the institutional review board (CSMUH No: CS2-22145). CKD was defined as either the presence of proteinuria or an estimated glomerular filtration rate (eGFR, determined by using simplified Modification of Diet in Renal Disease equation) of less than 60 mL/min/1.73 m2 in two separate visits [38]. For investigating the disease progression, DKD patients were grouped into early (n=689, CKD stage 1-3; with an eGFR ≥ 30) and advanced DKD (n=89, CKD stage 4-5; with an eGFR < 30) based on the level of renal function decline. In addition, 788 diabetic subjects with normal kidney function were enrolled for comparisons. Informed written consent was obtained from each individual participated in this study. Medical and demographic data concerning age, sex, diabetic status, hyperlipidemic condition, and kidney function were collected from each participant.
Genotyping
Two GAS5 gene variants, rs55829688 (T/C, promoter region) and rs145204276 (Ins/Del, promoter region), were surveyed based on their potential link with the risk of diverse diseases [36, 39-41]. Extraction of genomic DNA from the whole blood was carried out by using QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA, USA). Allelic discrimination of these two GAS5 variants including rs55829688 (assay ID: C_88335251_10), and rs145204276 (assay ID: C_166593916_10) was evaluated through the TaqMan assay with an ABI StepOne™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Genotyping data were then processed by SDS version 3.0 software (Applied Biosystems).
Statistical analysis
Comparisons of clinical or demographic information between DKD patients and non-DKD controls were performed by using the Mann-Whitney U test. Interactions of GAS5 genotypic frequencies with the development and progression of DKD were assessed by multiple logistic regression models after the adjustment for tentative confounding factors. Differences in GFR between groups and GAS5 expression data from the Genotype-Tissue Expression (GTEx) database [42] were determined with student t-test and one-way ANOVA, respectively. A p value of <0.05 was considered statistically significant.
Results
Subject characteristics
To explore the influence of GAS5 gene polymorphisms on the development of DKD, 778 DKD patients and 788 DKD-free diabetic controls were enrolled. Their demographic and clinical features were assessed (Table 1). The average age of DKD cases was higher than that of the controls, as no significant difference in gender was found between two groups. In addition to typical signs of renal impairment (reduced GFR, elevated levels of urinal microalbumin and serum creatinine, and increased UACR), the duration of diabetes and levels of hyperglycemia (elevation of HbA1c levels) were higher in DKD patients, as compared to the control group. Moreover, we observed that DKD group exhibited a higher systolic blood pressure and blood triglycerides level in comparison with diabetic subjects with normal kidney function.
Association of GAS5 gene variants with advanced DKD
To examine the potential interaction between GAS5 gene polymorphisms and DKD risks, one single-nucleotide (rs55829688) and one insertion/deletion polymorphism (rs145204276) of GAS5 gene were genotyped in this survey. Genotypic frequencies of each variant between DKD cases and DKD-free diabetic controls were determined. We did not detect any significant association of these two variants with the development of DKD from our study cohorts (Table 2). Subsequently, we further conducted stratification analyses based on the severity of renal impairment. We found that diabetic subjects who are heterozygous at rs55829688 (TC; AOR, 1.737; 95% CI, 1.028-2.937; p=0.039) are more likely to develop advanced DKD (Table 3), as compared to diabetic subjects who are homozygous for the major allele of rs55829688 (TT). Carriers of at least one minor allele (C) of rs55829688 (TC and CC; AOR, 1.317; 95% CI, 1.023-1.696; p=0.033) more frequently suffer from the advanced form of DKD than do those homozygotes for the major allele (TT). However, we failed to observe any association of rs55829688 with early DKD (Table 4). These results implicate a genotypic influence of GAS5 rs55829688 on promoting the progression of DKD.
Clinical and laboratory characteristics of patients with diabetic kidney disease in diabetic patients.
| Variable | No diabetic kidney disease (N=788) | Diabetic kidney disease (N=778) | p value |
|---|---|---|---|
| Age (years) | 58.80 ± 11.91 | 64.10 ± 11.66 | <0.001 |
| Male gender [n (%)] | 427 (54.2%) | 419 (53.9%) | 0.895 |
| Duration of diabetes (years) | 8.29 ± 6.65 | 12.09 ± 8.32 | <0.001 |
| HbA1c [% (mmol/mol)] | 6.96 ± 1.17 | 7.51 ± 1.51 | <0.001 |
| Body mass index [kg/m2] | 25.92 ± 4.43 | 26.26 ± 4.54 | 0.131 |
| Systolic blood pressure [mmHg] | 130.86 ± 14.52 | 137.11 ± 17.23 | <0.001 |
| Diastolic blood pressure [mm Hg] | 76.30 ± 10.62 | 76.11 ± 11.76 | 0.740 |
| Serum creatinine [mg/dL] | 0.81 ± 0.29 | 1.44 ± 1.46 | <0.001 |
| Glomerular filtration rate [ml/min] | 92.44 ± 26.78 | 63.76 ± 33.78 | <0.001 |
| Total cholesterol [mmol/L] | 161.29 ± 40.33 | 161.65 ± 48.09 | 0.873 |
| HDL cholesterol [μmol/L] | 46.79 ± 12.81 | 44.12 ± 12.85 | <0.001 |
| LDL cholesterol [μmol/L] | 87.50 ± 30.43 | 84.21 ± 31.43 | 0.037 |
| Triglycerides, [μmol/L] | 133.26 ± 185.12 | 157.54 ± 161.25 | 0.006 |
| TC/HDL ratio | 3.65 ± 1.39 | 3.94 ± 2.14 | 0.002 |
| Microalbumin (mg/dL) | 1.16 ± 1.24 | 45.86 ± 98.98 | <0.001 |
| UACR (mg/g) | 9.90 ± 7.10 | 551.87 ± 1291.52 | <0.001 |
Association between GAS5 genotypic frequencies and diabetic kidney disease.
| Variable | No diabetic kidney disease (N=788) | diabetic kidney disease (N=778) | AOR (95% CI) | p value |
|---|---|---|---|---|
| rs55829688 | ||||
| TT | 400 (50.8%) | 362 (46.5%) | 1.000 (reference) | |
| TC | 317 (40.2%) | 332 (42.7%) | 1.345 (0.841-2.153) | p=0.216 |
| CC | 71 (9.0%) | 84 (10.8%) | 1.503 (0.699-3.233) | p=0.297 |
| TC+CC | 388 (49.2%) | 416 (53.5%) | 1.172 (0.938-1.465) | p=0.163 |
| rs145204276 | ||||
| Ins/Ins | 327 (41.5%) | 324 (41.6%) | 1.000 (reference) | |
| Ins/Del | 362 (45.9%) | 371 (47.7%) | 1.020 (0.631-1.648) | p=0.936 |
| Del/Del | 99 (12.6%) | 83 (10.7%) | 0.677 (0.328-1.395) | p=0.290 |
| Ins/Del + Del/Del | 461 (58.5%) | 454 (58.4%) | 0.965 (0.769-1.210) | p=0.755 |
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models.
Effect of GAS5 rs55829688 genotypes on GFR across DKD subgroups and GAS5 expression
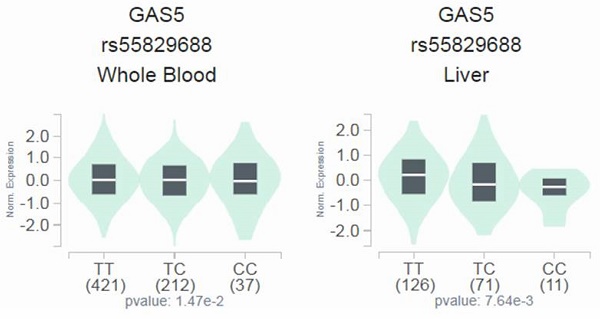
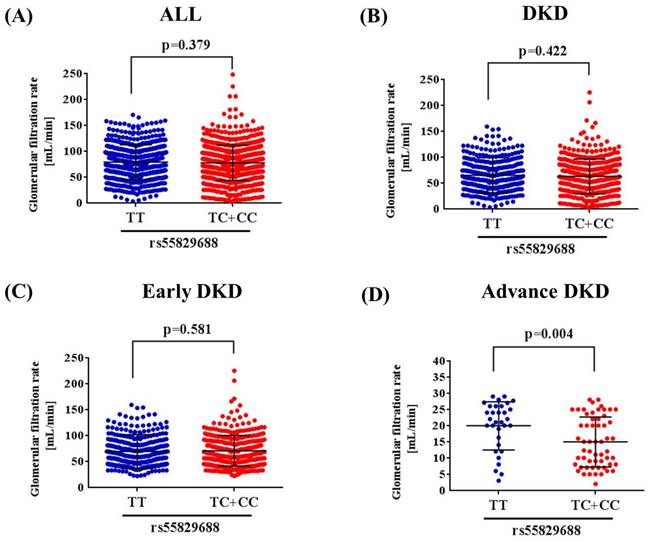
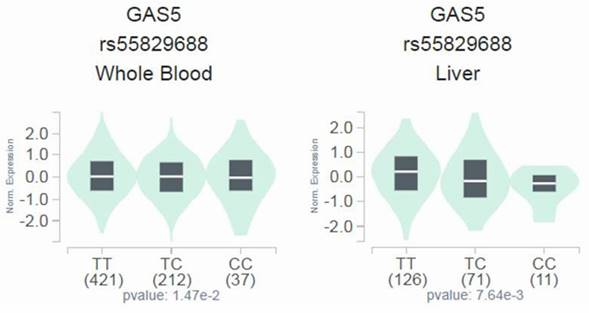
Since a genetic link between GAS5 rs55829688 and advanced DKD was noted, we next tested whether distinct rs55829688 genotypes affect kidney function of diabetic patients with different levels of renal impairment. We found that the GFR of advanced DKD patients who are homozygous for the major allele of rs55829688 (TT) was significantly higher than that of those who possess at least one minor allele (C) of rs55829688 (TC and CC) (Figure 1). Yet, no difference in GFR was seen between two genotypic groups of early DKD, all DKD, or all diabetic patients (DKD cases and DKD-free diabetic controls), suggesting an impact of rs55829688 on loss of kidney function in patients with severe renal failure. In addition, to have a preliminary assessment of the functional relevance for rs55829688, a publicly available dataset was used to evaluate the relationship between rs55829688 genotypes and GAS5 expression. We found alterations of GAS5 expression in the whole blood cells and liver tissues and among individuals who carry different rs55829688 genotypes in the Genotype-Tissue Expression (GTEx) database (Figure 2). There data suggest that changes in GAS5 expression due to genetic polymorphisms may affect the disease progression of DKD.
Discussion
Tremendous amounts of studies have indicated that the risk of DKD is modulated by the combination of inherited and acquired etiologic factors. In this investigation, by employing a candidate gene strategy, we exhibited a correlation between genotypes of GAS5 rs55829688 and the risk of developing advanced DKD. Moreover, in patients with severe renal impairment, carriers of at least one minor allele of rs55829688 (TC and CC) showed a lower glomerular filtration rate than those homozygotes for the major allele (TT), unveiling an effect of rs55829688 on renal co-morbidities of diabetes. These findings demonstrate a connection of GAS5 gene variations with the progression of DKD.
Association of advanced diabetic kidney disease with GAS5 genotypic frequencies.
| Variable | No diabetic kidney disease (N=788) | Advanced diabetic kidney disease (N=89) | AOR (95% CI) | p value |
|---|---|---|---|---|
| rs55829688 | ||||
| TT | 400 (50.8%) | 32 (36.0%) | 1.000 (reference) | |
| TC | 317 (40.2%) | 47 (52.8%) | 1.737 (1.028-2.937) | p=0.039 |
| CC | 71 (9.0%) | 10 (11.2%) | 1.727 (0.717-4.159) | p=0.223 |
| TC+CC | 388 (49.2%) | 57 (64.0%) | 1.317 (1.023-1.696) | p=0.033 |
| rs145204276 | ||||
| Ins/Ins | 327 (41.5%) | 40 (44.9%) | 1.000 (reference) | |
| Ins/Del | 362 (45.9%) | 42 (47.2%) | 0.871 (0.519-1.462) | p=0.602 |
| Del/Del | 99 (12.6%) | 7 (7.9%) | 0.558 (0.224-1.389) | p=0.210 |
| Ins/Del + Del/Del | 461 (58.5%) | 49 (55.1%) | 0.898 (0.700-1.152) | p=0.397 |
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models.
Effect of rs55829688 genotypes on glomerular filtration rate (GFR) across DKD groups. Comparisons of GFR between two rs55829688 genotypic groups of all diabetic patients (DKD cases and DKD-free diabetic controls) (A), DKD patients (B), early DKD patients (C), advanced DKD patients (D).
Effect of rs55829688 genotypes on GAS5 expression. Comparisons of GAS5 expression among rs55829688 genotypic groups in representative normal tissues based on data from the GTEx portal.
Association of early diabetic kidney disease with GAS5 genotypic frequencies.
| Variable | No diabetic kidney disease (N=788) | Early diabetic kidney disease (N=689) | AOR (95% CI) | p value |
|---|---|---|---|---|
| rs55829688 | ||||
| TT | 400 (50.8%) | 330 (47.9%) | 1.000 (reference) | |
| TC | 317 (40.2%) | 285 (41.4%) | 1.347 (0.842-2.156) | p=0.215 |
| CC | 71 (9.0%) | 74 (10.7%) | 1.505 (0.700-3.237) | p=0.295 |
| TC+CC | 388 (49.2%) | 359 (52.1%) | 1.173 (0.938-1.466) | p=0.161 |
| rs145204276 | ||||
| Ins/Ins | 327 (41.5%) | 284 (41.2%) | 1.000 (reference) | |
| Ins/Del | 362 (45.9%) | 329 (47.8%) | 1.021 (0.632-1.649) | p=0.933 |
| Del/Del | 99 (12.6%) | 76 (11.0%) | 0.677 (0.328-1.395) | p=0.290 |
| Ins/Del + Del/Del | 461 (58.5%) | 405 (58.8%) | 0.965 (0.769-1.210) | p=0.757 |
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models.
Recently, dysregulation of lncRNAs has been attracting increasing attentions on the development of DKD [43]. These DKD-associated lncRNAs regulate inflammation, programmed cell death, and epithelial-mesenchymal transition in key resident cells of the kidney, such as mesangial cells, renal endothelial cells, podocytes, and tubular epithelial cells, serving as potential therapeutic targets of DKD [44]. One of such lncRNA genes, GAS5, encompassing 12 exons and generating two mature RNA transcripts [24, 45], has been proposed as a tumor-suppressor gene [27] and a key regulator of bone diseases [46]. Currently, aberrant expression and function of GAS5 were extensively studied in the field of renal physiology and pathology. In renal tubular cells, GAS5 hampered the inflammation, oxidative stress, and pyroptosis induced by the treatment of high glucose [47]. In addition to the nephroprotective role in renal inflammation and cell death, GAS5 interfered with the expression of ECM proteins, collagen type I and fibronectin, to alleviate TGFβ-induced renal fibrosis [48]. Through diverse molecular mechanisms (e.g. sponging specific microRNAs, suppressing ECM enzymes, and regulating fibrogenic gene transcription), this inhibitory effect of GAS5 on renal fibrosis was consistently detected in various studies [30, 31, 49-51]. Moreover, knockdown of GAS5 affected the remodeling of renal arteries via altered communications between endothelial cells and vascular smooth muscle cells [52]. Collectively, these findings underline a functional relevance of GAS5 in the pathogenesis of DKD through an epigenetic regulation of nutrient metabolism, renal inflammation, and angiogenic responses.
In this case-control study, we identified a significant correlation of advanced DKD with a single-nucleotide polymorphism (SNP) of GAS5 gene, rs55829688. This SNP, located at the promoter region of the GAS5 gene, has been shown to affect the prognosis of acute myeloid leukemia (AML) [53], as specific haplotypes containing rs55829688 were linked to a higher risk of developing AML [54]. In addition to the risk and treatment outcome of AML, rs55829688 variation has been demonstrated to confer the susceptibility to colorectal carcinoma [41]. Recently, a genetic effect of rs55829688 on the treatment responses of patients with coronary artery disease, another common co-morbidities of diabetes associated with microvascular dysfunction, has also been detected [55]. Functional investigations of rs55829688 reveal that genotypes of this variant were able to regulate the expression levels of GAS5 in peripheral blood and colon cancer cells through altered binding affinities of GAS5 promoter with the transcription factor p63 [53] and Yin Yang-1 [41], respectively. Such changes in GAS5 expression simultaneously manipulated the transcriptional process of its target genes via acting as a sponge for numerous microRNAs and as a scaffold for the formation of multiple transcription factor complexes, eventually resulting in various human disorders [28, 46]. Our findings, together with the results from others, suggest that alterations of GAS5 levels owing to polymorphic alleles of rs55829688 may influence the progression of DKD.
Here, we demonstrated a connection of GAS5 gene variations with the progression of DKD. Yet, additional efforts are required to deal with several study limitations. One concern is that the highly heterogeneous complications of diabetes (e.g. diabetic retinopathy, diabetic neuropathy, diabetic cardiomyopathy, and diabetic myopathy) and their overlaying genetic architectures may lead to different discoveries regarding the association of GAS5 gene polymorphisms with advanced DKD. Nevertheless, disease-associated variants within the promoter region were commonly reported as expression quantitative trait loci [56] but we did not examine whether polymorphic alleles of rs55829688 contribute to altered GAS5 expression in relevant cell types, such as tubular epithelial cells, podocytes, mesangial cells, and renal endothelial cells. In addition, the genetic effect identified in our study might be restricted to specific cohorts if not replicated in other ethnic groups.
Taken together, our data revealed a correlation of GAS5 rs55829688 with the severe form of DKD. This genetic association links fluctuations of GAS5 expression owing to gene variations to the exacerbation of renal failure in diabetic individuals.
Acknowledgements
We are grateful to the Human Biobank of Chung Shan Medical University Hospital, Taichung, Taiwan for sample preparation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Stenvinkel P. Chronic kidney disease: a public health priority and harbinger of premature cardiovascular disease. J Intern Med. 2010;268:456-467
2. Reutens AT. Epidemiology of diabetic kidney disease. Med Clin North Am. 2013;97:1-18
3. Sinha SK, Nicholas SB. Pathomechanisms of Diabetic Kidney Disease. J Clin Med. 2023;12:7349
4. Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest. 2014;124:2333-2340
5. Roscioni SS, Heerspink HJ, de Zeeuw D. The effect of RAAS blockade on the progression of diabetic nephropathy. Nat Rev Nephrol. 2014;10:77-87
6. Wang L, Wang HL, Liu TT, Lan HY. TGF-Beta as a Master Regulator of Diabetic Nephropathy. Int J Mol Sci. 2021;22:7881
7. Zhang Y, Jin D, Kang X, Zhou R, Sun Y, Lian F. et al. Signaling Pathways Involved in Diabetic Renal Fibrosis. Front Cell Dev Biol. 2021;9:696542
8. Schaffer SW, Jong CJ, Mozaffari M. Role of oxidative stress in diabetes-mediated vascular dysfunction: unifying hypothesis of diabetes revisited. Vascul Pharmacol. 2012;57:139-149
9. Susztak K, Raff AC, Schiffer M, Bottinger EP. Glucose-induced reactive oxygen species cause apoptosis of podocytes and podocyte depletion at the onset of diabetic nephropathy. Diabetes. 2006;55:225-233
10. Flyvbjerg A. The role of the complement system in diabetic nephropathy. Nat Rev Nephrol. 2017;13:311-318
11. Matoba K, Takeda Y, Nagai Y, Kawanami D, Utsunomiya K, Nishimura R. Unraveling the Role of Inflammation in the Pathogenesis of Diabetic Kidney Disease. Int J Mol Sci. 2019;20:3393
12. Navarro-Gonzalez JF, Mora-Fernandez C, Muros de Fuentes M, Garcia-Perez J. Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat Rev Nephrol. 2011;7:327-340
13. Alicic RZ, Rooney MT, Tuttle KR. Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clin J Am Soc Nephrol. 2017;12:2032-2045
14. Cole JB, Florez JC. Genetics of diabetes mellitus and diabetes complications. Nat Rev Nephrol. 2020;16:377-390
15. Tedla FM, Brar A, Browne R, Brown C. Hypertension in chronic kidney disease: navigating the evidence. Int J Hypertens. 2011;2011:132405
16. Yeung EH, Pankow JS, Astor BC, Powe NR, Saudek CD, Kao WH. Increased risk of type 2 diabetes from a family history of coronary heart disease and type 2 diabetes. Diabetes Care. 2007;30:154-156
17. Mahajan A, Spracklen CN, Zhang W, Ng MCY, Petty LE, Kitajima H. et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat Genet. 2022;54:560-572
18. Xue A, Wu Y, Zhu Z, Zhang F, Kemper KE, Zheng Z. et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018;9:2941
19. Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ. et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41-47
20. Kwak SH, Srinivasan S, Chen L, Todd J, Mercader JM, Jensen ET. et al. Genetic architecture and biology of youth-onset type 2 diabetes. Nat Metab. 2024;6:226-237
21. van Zuydam NR, Ahlqvist E, Sandholm N, Deshmukh H, Rayner NW, Abdalla M. et al. A Genome-Wide Association Study of Diabetic Kidney Disease in Subjects With Type 2 Diabetes. Diabetes. 2018;67:1414-1427
22. Sandholm N, Dahlstrom EH, Groop PH. Genetic and epigenetic background of diabetic kidney disease. Front Endocrinol (Lausanne). 2023;14:1163001
23. Thomas MC, Brownlee M, Susztak K, Sharma K, Jandeleit-Dahm KA, Zoungas S. et al. Diabetic kidney disease. Nat Rev Dis Primers. 2015;1:15018
24. Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787-793
25. Coccia EM, Cicala C, Charlesworth A, Ciccarelli C, Rossi GB, Philipson L. et al. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol. 1992;12:3514-3521
26. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8
27. Lin G, Wu T, Gao X, He Z, Nong W. Research Progress of Long Non-Coding RNA GAS5 in Malignant Tumors. Front Oncol. 2022;12:846497
28. Yang X, Xie Z, Lei X, Gan R. Long non-coding RNA GAS5 in human cancer. Oncol Lett. 2020;20:2587-2594
29. Liu Y. Renal fibrosis: new insights into the pathogenesis and therapeutics. Kidney Int. 2006;69:213-217
30. Zhang L, Zhao S, Zhu Y. Long noncoding RNA growth arrest-specific transcript 5 alleviates renal fibrosis in diabetic nephropathy by downregulating matrix metalloproteinase 9 through recruitment of enhancer of zeste homolog 2. FASEB J. 2020;34:2703-2714
31. Ge X, Xu B, Xu W, Xia L, Xu Z, Shen L. et al. Long noncoding RNA GAS5 inhibits cell proliferation and fibrosis in diabetic nephropathy by sponging miR-221 and modulating SIRT1 expression. Aging (Albany NY). 2019;11:8745-8759
32. Hsieh MH, Lu HJ, Lin CW, Lee CY, Yang SJ, Wu PH. et al. Genetic Variants of lncRNA GAS5 Are Associated with the Clinicopathologic Development of Oral Cancer. J Pers Med. 2021;11:348
33. Lin CY, Wang SS, Yang CK, Li JR, Chen CS, Hung SC. et al. Impact of GAS5 genetic polymorphism on prostate cancer susceptibility and clinicopathologic characteristics. Int J Med Sci. 2019;16:1424-1429
34. Weng SL, Ng SC, Lee YC, Hsiao YH, Hsu CF, Yang SF. et al. The relationships of genetic polymorphisms of the long noncoding RNA growth arrest-specific transcript 5 with uterine cervical cancer. Int J Med Sci. 2020;17:1187-1195
35. Weng WC, Chen CJ, Chen PN, Wang SS, Hsieh MJ, Yang SF. Impact of Gene Polymorphisms in GAS5 on Urothelial Cell Carcinoma Development and Clinical Characteristics. Diagnostics (Basel). 2020;10:260
36. Lee CM, Yang YS, Kornelius E, Huang CN, Hsu MY, Lee CY. et al. Association of Long Non-Coding RNA Growth Arrest-Specific 5 Genetic Variants with Diabetic Retinopathy. Genes (Basel). 2022;13:584
37. Li H, Liu Y, Huang J, Liu Y, Zhu Y. Association of genetic variants in lncRNA GAS5/miR-21/mTOR axis with risk and prognosis of coronary artery disease among a Chinese population. J Clin Lab Anal. 2020;34:e23430
38. National Kidney Foundation. K/DOQI Clinical practice guidelines for Chronic Kidney Disease: Evaluation, classification and stratification. Am J Kidney Dis. 2002;39:S1-S266
39. Tang Y, Wang Y, Wang X, Liu Y, Zheng K. A Genetic Variant of rs145204276 in the Promoter Region of Long Noncoding RNA GAS5 Is Associated With a Reduced Risk of Breast Cancer. Clin Breast Cancer. 2019;19:e415-e421
40. Zheng Z, Liu S, Wang C, Han X. A Functional Polymorphism rs145204276 in the Promoter of Long Noncoding RNA GAS5 Is Associated with an Increased Risk of Ischemic Stroke. J Stroke Cerebrovasc Dis. 2018;27:3535-3541
41. Wang Y, Wu S, Yang X, Li X, Chen R. Association between polymorphism in the promoter region of lncRNA GAS5 and the risk of colorectal cancer. Biosci Rep. 2019;39:BSR20190091
42. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580-585
43. Kato M. Noncoding RNAs as therapeutic targets in early stage diabetic kidney disease. Kidney Res Clin Pract. 2018;37:197-209
44. Hu M, Ma Q, Liu B, Wang Q, Zhang T, Huang T. et al. Long Non-Coding RNAs in the Pathogenesis of Diabetic Kidney Disease. Front Cell Dev Biol. 2022;10:845371
45. Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897-6909
46. Zhou Z, Chen J, Huang Y, Liu D, Chen S, Qin S. Long Noncoding RNA GAS5: A New Factor Involved in Bone Diseases. Front Cell Dev Biol. 2021;9:807419
47. Xie C, Wu W, Tang A, Luo N, Tan Y. lncRNA GAS5/miR-452-5p Reduces Oxidative Stress and Pyroptosis of High-Glucose-Stimulated Renal Tubular Cells. Diabetes Metab Syndr Obes. 2019;12:2609-2617
48. Zhang YY, Tan RZ, Yu Y, Niu YY, Yu C. LncRNA GAS5 protects against TGF-beta-induced renal fibrosis via the Smad3/miRNA-142-5p axis. Am J Physiol Renal Physiol. 2021;321:F517-F526
49. Yu Y, Jiang H, Niu Y, Huang J, Zhang X, Liu X. et al. Long noncoding RNA-GAS5 retards renal fibrosis through repressing miR-21 activity. Exp Mol Pathol. 2020;116:104518
50. Guo Y, Li G, Gao L, Cheng X, Wang L, Qin Y. et al. Exaggerated renal fibrosis in lncRNA Gas5-deficient mice after unilateral ureteric obstruction. Life Sci. 2021;264:118656
51. Zhang X, Hu S, Xiang X, Li Z, Chen Z, Xia C. et al. Bulk and single-cell transcriptome profiling identify potential cellular targets of the long noncoding RNA Gas5 in renal fibrosis. Biochim Biophys Acta Mol Basis Dis. 2024;1870:167206
52. Wang YN, Shan K, Yao MD, Yao J, Wang JJ, Li X. et al. Long Noncoding RNA-GAS5: A Novel Regulator of Hypertension-Induced Vascular Remodeling. Hypertension. 2016;68:736-748
53. Yan H, Zhang DY, Li X, Yuan XQ, Yang YL, Zhu KW. et al. Long non-coding RNA GAS5 polymorphism predicts a poor prognosis of acute myeloid leukemia in Chinese patients via affecting hematopoietic reconstitution. Leuk Lymphoma. 2017;58:1948-1957
54. Ketab FNG, Gharesouran J, Ghafouri-Fard S, Dastar S, Mazraeh SA, Hosseinzadeh H. et al. Dual biomarkers long non-coding RNA GAS5 and its target, NR3C1, contribute to acute myeloid leukemia. Exp Mol Pathol. 2020;114:104399
55. Liu YL, Hu XL, Song PY, Li H, Li MP, Du YX. et al. Influence of GAS5/MicroRNA-223-3p/P2Y12 Axis on Clopidogrel Response in Coronary Artery Disease. J Am Heart Assoc. 2021;10:e021129
56. Mostafavi H, Spence JP, Naqvi S, Pritchard JK. Systematic differences in discovery of genetic effects on gene expression and complex traits. Nat Genet. 2023;55:1866-1875
Author contact
![]() Corresponding authors: Shun-Fa Yang, Ph.D. or Shih-Chi Su, Ph.D. Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Tel: +886-4-24739595 ext. 34253; Fax: +886-4-24723229; E-mail: ysfedu.tw (Shun-Fa Yang); E-mail: ssu1org.tw (Shih-Chi Su).
Corresponding authors: Shun-Fa Yang, Ph.D. or Shih-Chi Su, Ph.D. Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Tel: +886-4-24739595 ext. 34253; Fax: +886-4-24723229; E-mail: ysfedu.tw (Shun-Fa Yang); E-mail: ssu1org.tw (Shih-Chi Su).

 Global reach, higher impact
Global reach, higher impact