Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(8):1575-1588. doi:10.7150/ijms.91584 This issue Cite
Review
F-box proteins and gastric cancer: an update from functional and regulatory mechanism to therapeutic clinical prospects
1. Postgraduate Training Base Alliance of Wenzhou Medical University (Zhejiang Cancer Hospital), Hangzhou, Zhejiang, 310022, China.
2. Zhejiang Cancer Hospital, Hangzhou, Zhejiang, 310005, China.
Received 2023-10-27; Accepted 2024-5-20; Published 2024-6-3
Abstract
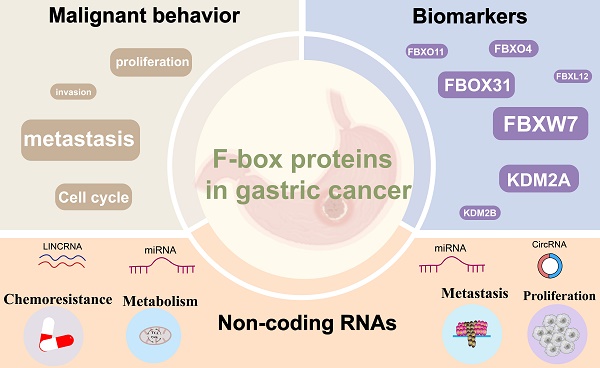
Gastric cancer (GC) is a prevalent malignancy characterized by significant morbidity and mortality, yet its underlying pathogenesis remains elusive. The etiology of GC is multifaceted, involving the activation of oncogenes and the inactivation of antioncogenes. The ubiquitin-proteasome system (UPS), responsible for protein degradation and the regulation of physiological and pathological processes, emerges as a pivotal player in GC development. Specifically, the F-box protein (FBP), an integral component of the SKP1-Cullin1-F-box protein (SCF) E3 ligase complex within the UPS, has garnered attention for its prominent role in carcinogenesis, tumor progression, and drug resistance. Dysregulation of several FBPs has recently been observed in GC, underscoring their significance in disease progression. This comprehensive review aims to elucidate the distinctive characteristics of FBPs involved in GC, encompassing their impact on cell proliferation, apoptosis, invasive metastasis, and chemoresistance. Furthermore, we delve into the emerging role of FBPs as downstream target proteins of non-coding RNAs(ncRNAs) in the regulation of gastric carcinogenesis, outlining the potential utility of FBPs as direct therapeutic targets or advanced therapies for GC.
Keywords: F-box proteins, Gastric cancer, Mechanism, Non-coding RNA, Chemoresistance
Introduction
Gastric cancer (GC) poses a significant global health challenge, imposing a substantial burden on public health. In 2020 alone, GC accounted for over 1 million new cases and caused more than 768,000 deaths, ranking it as the third leading cause of cancer-related mortality worldwide [1, 2]. Notably, the incidence and mortality rates of GC are highest in East Asian countries, particularly in China, where GC has become the second leading cause of cancer-related deaths [3-5], this trend can be attributed to dietary habits and environmental factors [6]. Surgical resection is the preferred treatment approach for early-stage GC patients (stages I to III) [7, 8]. However, due to the often-asymptomatic progression in the early stages, diagnosis is frequently delayed, resulting in the majority of GC cases being diagnosed at advanced stages (>80%). Consequently, many patients miss the opportunity for surgery, leading to poor prognosis and increased mortality [9, 10]. For patients who are ineligible for surgical resection or have advanced metastases, the combination of chemotherapy, targeted therapy, and immunotherapy represents the primary treatment option. However, treatment failure is common due to resistance and limited efficacy [11, 12]. Despite advancements in therapeutic development, the overall 5-year survival rate for advanced GC patients remains below 40% [13]. Thus, there is an urgent need for novel treatment strategies for GC. Recent studies have indicated that the ubiquitin-proteasome system-mediated degradation of oncogenes and oncoproteins plays a crucial role in the development and progression of GC.
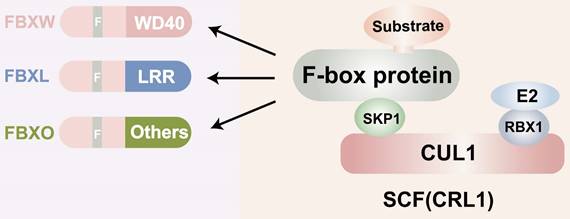
F-box proteins, though structurally simple, exhibit exceptional functional. They complexity consist of three main domains: the N-terminal F-box domain, the middle linker domain, and the C-terminal functional domain (Figure 1). The F-box domain, approximately 50 amino acids in length, is highly conserved and plays a critical role in identifying FBPs. It interacts with the Skp1 protein to form the foundation of the SCF (Skp1-Cullin1-F-box protein) complex [14]. The linker domain, generally composed of about 30 amino acids, acts as a bridge between the F-box domain and the C-terminal functional structure. The size and function of the C-terminal domain vary across different F-box proteins and can be categorized into various types, such as RING finger structures with ubiquitin ligase activity and WD40 repeat structures involved in protein degradation [15, 16]. Furthermore, the functional diversity of F-box proteins and their interactions constitute significant structural features. F-box proteins participate in numerous vital physiological and pathological processes in organisms, including cell cycle regulation, cell proliferation, gene expression and regulation, apoptosis, and signal transduction [17]. They engage in a wide array of protein-protein interactions, including interactions with phosphatases, regulation of activity through second messengers, and involvement in protein subcellular localization [18-20]. Given that each F-box protein has multiple substrates, determining whether they exert anti-tumor or tumor-promoting effects is a complex question and may depend on the cellular context. A large body of data now suggests that FBPs have oncogenic or tumor suppressor activity [21]. Some of them, such as FBXW7, are mutated or show high-frequency expression deregulation in a large number of human malignant tumors, suggesting a key role in cancer development or progression. For example, SKP2 is overexpressed in breast, prostate, colorectal, and pancreatic cancers as well as lymphoma, melanoma, and nasopharyngeal carcinoma, and is highly correlated with poor tumor prognosis [22-24]. Furthermore, Lu et al. found that FBXO44 regulates BRCA1 gene stability in breast cancer [25]. This reflects the important role of FBPs in cancer development. In this paper we will focus on the functional mechanism of FBPs in GC. Several F-box protein members, such as FBXO31, FBXL7, FBXO9, FBXO44, and FBXW11, have been found to be aberrantly expressed in human GC, suggesting their close association with gastric carcinogenesis and development. Additionally, certain F-box subfamilies, such as SKP2 and β-TrCP, are implicated in GC development and are considered prognostic factors with potential value for GC treatment.
Structure of the F-box proteins. CUL: Cullin; SCF: Skp1-Cullin-F box complexes; CRL1: cullin-RING ubiquitin ligase1; RBX1; RING-domain-containing partner.
This review paper aims to elucidate the signaling pathways, underlying mechanisms, and functional roles of FBPs in GC. Firstly, we comprehensively illustrate the stimulatory effects exerted by FBPs on proliferation, apoptosis, and invasive metastasis within the context of GC. Furthermore, we highlight their significant involvement in the development of chemoresistance. Secondly, we provide a comprehensive summary of the pivotal molecular mechanisms through which FBPs function as downstream target proteins of ncRNAs to orchestrate the process of gastric carcinogenesis. Lastly, we engage in a thorough discussion regarding the potential utility of targeting FBPs directly or employing them as advanced therapeutic strategies for the management of GC.
1. Overview of F-box proteins
1.1 The ubiquitin-proteasome system and the F-box proteins
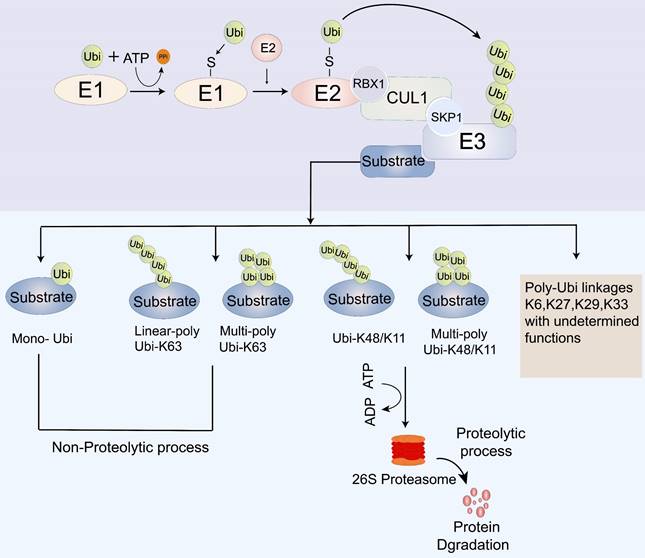
Cancer development entails the transformation of normal cells into cancer cells in response to abnormal cellular stimuli. This process is tightly regulated by the transcription, translation, post-translational modification, and degradation of key regulatory proteins, which play a vital role in maintaining cellular homeostasis. Within cells, two main systems are responsible for protein degradation: the autophagic lysosomal system and the ubiquitin-proteasome system (UPS) [26]. The lysosomal pathway facilitates the degradation of extracellular proteins introduced into the cell through endocytosis or cytokinesis, while the UPS controls the degradation of intracellular proteins [27]. Ubiquitin modification, a post-translational modification, involves a series of essential components, including ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), ubiquitin ligases (E3s), deubiquitinating enzymes (DUBs), and the 26S proteasome [28-31]. The UPS carries out its biological function through a cascade of three enzymatic reactions catalyzed by the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and the ubiquitin-protein E3 ligase [28]. Initially, E1s activate ubiquitin (Ub) and form a thioester bond between the sulfhydryl group of the active cysteine residue of the E1s and the carboxyl group of Ub, which is then delivered to the E2s. Ultimately, through the synergy between substrate binding, E3 ligase, and ubiquitin-charged E2, polyubiquitinated target proteins are produced for degradation in the 26S proteasome [31-36] (Figure 2).
E3 ligase, a crucial component of the ubiquitination cascade, determines substrate specificity for ubiquitination and subsequent degradation. Based on their structural characteristics, E3 ligases can be primarily classified into three categories: RING (really interesting new gene, including U-box E3 with similar topology), HECT (homologous to E6AP C-terminus), and RBR (RING-in-between-RING). The largest family among them is the cullin-RING E3 ligase (CRL) complex family, which comprises eight members (CRL1, 2, 3, 4A, B, 5, 7, and 9) [37, 38]. These ligases are recognized as key regulators of several cellular processes, including cell cycle progression, such as S-phase entry and G2/M-phase exit. Among them, CRL1, also known as the SCF E3 ligase complex, is the best-characterized member of the E3 ligase family [39].
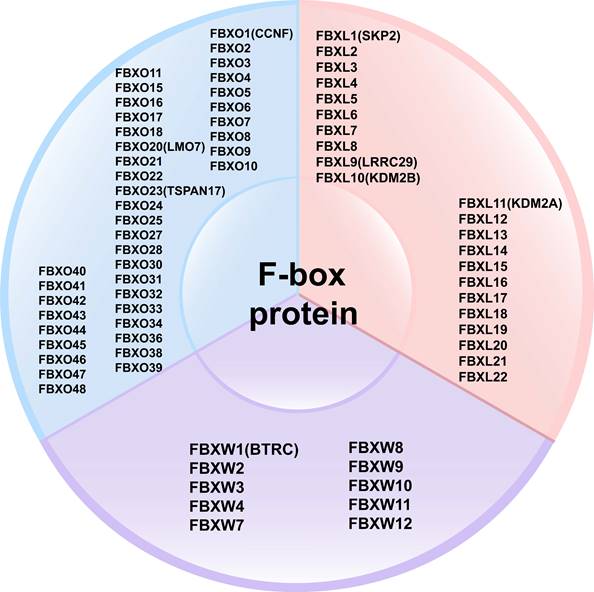
FBPs can be classified into three subclasses based on the presence of specific substrate recognition structural domains: 10 FBXWs with WD40 repeat domains, 22 FBXLs containing leucine-rich repeats, and the remaining 37 FBXOs with domains not observed in FBXWs and FBXLs subtypes [40-42] (Figure 3). FBPs can bind to other proteins to form an E3 ubiquitin ligase complex that mediates the ubiquitination and degradation of the target protein [20, 39]. Currently, several FBPs have been implicated in cell cycle progression, drug resistance, cell growth, repair, and differentiation [43-45].
The process of ubiquitination modification. E1: ubiquitin-activating enzymes; E2: ubiquitin-conjugating enzymes; E3: ubiquitin ligases; Ubi: Ubiquitin; CUL: Cullin; SCF: Skp1-Cullin-F box complexes; CRL1: cullin-RING ubiquitin ligase1; RBX1; RING-domain-containing partner.
Classification of F-box proteins: 10 FBXWs with WD40 repeat domains, 22 FBXLs containing leucine-rich repeats, and the remaining 37 FBXOs with domains not observed in FBXWs and FBXLs subtypes.
1.2 Cell cycle and the F-box proteins
FBPs are actively involved in the regulation of the UPS and can promote the ubiquitination and degradation of target proteins, thereby influencing the cell cycle. Some studies have found the destabilization of E2F1 by the SCFSKP2 ligase may be important to limit its activity in S and G2 phases of the cell cycle44. Further studies revealed that SKP2 promotes not only the G1/S transition but also the G2/M transition through targeted protein hydrolysis of p27 and p21[46, 47]. In breast cancer cells and melanoma, elevated levels of FBXO31 protein induce degradation of cell cycle protein D1, leading to cell cycle arrest in G1[42], Similarly β-TRCP1/2 proteins can regulate cell cycle progression by modulating CDK1 kinase activity [44]. FBXL2 by facilitating the ubiquitin-mediated degradation of crucial cell cycle regulators including cyclin D2, cyclin D3 and Aurora B. D-type cyclins partner with CDK4 and CDK6 to drive G1-to-S cell-cycle progression [48]. This suggests that FBPs have important regulatory roles in various processes of the tumor cell cycle. In GC, FBPs play a significant role in cell cycle regulation through two major mechanisms: Firstly, by regulating the ubiquitination of cell cycle regulatory molecules, FBPs can recognize, bind, and ubiquitinate a series of relevant proteins, thereby participating in cell cycle regulation [49, 50]. For instance, F-box proteins regulate the degradation and stabilization of proteins such as the CDK inhibitors p27 [51], p21 [52] and CDC6[53], which in turn control the cell cycle. Secondly, F-box proteins participate in signaling pathways to regulate the cell cycle. They serve as crucial regulatory molecules in various signaling pathways, such as Wnt [54], NF-κB [55] and HIF-1α pathway [56] by mediating the degradation of key molecules through ubiquitination, thereby influencing the cell cycle and other biological processes. In summary, FBPs play a pivotal role in cell cycle regulation by targeting key molecules and signaling pathways.
1.3 Therapeutic drug targets and the F-box proteins
A growing body of evidence now supports the promising development of FBPs for tumor therapy. A universal proteasome inhibitor velcade (bortezomib) for the treatment of multiple myeloma, targeting the UPS for tumor therapeutic purposes proved to be a promising approach [57]. However, non-selective inhibition of protein degradation causes undesirable side effects, limiting the use of this approach. Since the substrate specificity of UPS is achieved by E3 ligases, such proteins offer new avenues for tumor therapy. Interestingly, some studies found that FBPs can impact the metabolism and clearance rate of drugs by regulating the expression and activity of drug-metabolizing enzymes, thereby modulating the efficacy and toxicity of drugs and enhancing the effectiveness and safety of drug therapy [58, 59]. Furthermore FBPs also can influence the therapeutic effects of drugs by modulating the degradation rate of target proteins [60]. For example, Tang et al. demonstrated that β-TrCP-deficient cells are more sensitive to various anticancer drugs (e.g., adriamycin, tamoxifen, and paclitaxel) on human mammary tumor cells [61]. Several studies have demonstrated that targeting FBPs can suppress the growth and metastasis of tumor cells [28]. Yang et al. developed a chemical genetics approach to overexpress SKP2 to anti-proliferative activity by restoring p27(Kip1) in prostate cancer cells[62]. Similarly, Wu et al. screened and identified a small molecule inhibitor specific for SCF-SKP2 activity, which selectively inhibits SKP2-mediated p27 degradation in cancer cells by reducing p27 binding via key compound-receptor contacts, thereby inhibiting tumor growth [63]. However, most of the current inhibitors specific for FBPs remain in preclinical studies [60]. It's worth mentioning that, agonists targeting F-box proteins are being investigated for the treatment of neurological diseases, cardiovascular diseases, and other conditions [28, 64]. Therefore, the development of therapeutic agents targeting FBPs holds great potential for delivering excellent therapeutic outcomes in cancer clinical treatment.
2. Expression of the F-box proteins in gastric cancer
A genomic profiling revealed that FBXW7 mutations were observed in 9.2%-18.5% of GC tumors and 4.7% in IM (Intestinal Metaplasia), and suggested that FBXW7 mutations in IM are likely to functionally contribute to IM and GC development [65]. The expressions of FBPs are generally decreased in GC, including FBXW7, FBXL2, and FBXL5. However, FBXW5, KDM2A, and FBXO2 exhibit high expression levels in GC. Considering that each FBPs can target multiple substrates, their effects on tumorigenesis can be complex and dependent on the cellular context (Table 1).
Several studies have demonstrated that differential expression of FBPs is associated with poor prognosis in GC. Calcagno et al. reported that dysregulation of FBXW7 mRNA expression correlated with lymph node metastasis and advanced stages of GC, suggesting that FBXW7 may serve as an indicator of poor prognosis in GC [80]. Similarly, altered expression of FBXW7 in the presence of P53 mutations was associated with poor prognosis in GC [81]. Immunohistochemical analysis conducted by Li et al. showed that low FBXW7 expression in primary GC was associated with poorly differentiated tumor cells, shorter overall survival, and reduced response to adjuvant chemotherapy [82]. Highlighted the impact of aberrant expression of FBPs on GC progression. Specifically, FBPs have an anticancer role in GC. A study by Kogure demonstrated that low FBXO45 expression was associated with increased cancer progression and poorer prognosis in GC patients [83]. Similarly, low FBX8 expression was associated with shorter overall survival and poorer prognosis [71]. In contrast, FBPs are also pro-cancer in GC, overexpression of FBXO11 in GC was associated with larger tumor size, lymph node metastasis, advanced TNM stage, and shorter survival [77].
Expression of FBPs in GC.
| FBPs | Expression in tissue | Sample size | Expression in cancer cells | Cancer cell lines | Relative normal cell lines | Ref. |
|---|---|---|---|---|---|---|
| FBXW7 | Down | 60 | Down | AZ-521, MGC-803, BGC-823, SGC-7901 | GES-1 | [66] |
| Down | 66 | Down | AGS, HGC-27, BGC-823, MGC-803, MKN-45 | GES-1 | [67] | |
| FBXL2 | down | 15 | down | NCI-N87 | - | [68] |
| FBXL5 | down | 20 | down | SNU-5, AGS | - | [69] |
| FBXO31 | down | 77 | down | BGC-823, SGC-7901 | - | [70] |
| FBX8 | down | 136 | down | MGC-803, BGC-823, MKN45, AGS, SGC-7901 | - | [71] |
| FBXO21 | down | 21 | down | SGC-7901, BGC-823, MGC-803, MKN-45, MKN-28, AGS | GES-1 | [72] |
| FBXW5 | UP | 16 | UP | AGS, MKN-45, HGC-27, MGC-803, BGC-823, SGC-7901 | GES-1 | [73] |
| - | - | UP | CLS145, MKN1, AGS, SNU1 | - | [74] | |
| FBXL11(KDM2A) | UP | 61 | UP | AGS, BGC 803, MGC-823, SGC 7901 | GES-1 | [75] |
| FBXO2 | - | 89 | - | MGC-803, AGS, SGC-7901, MKN-28 | - | [76] |
| FBXO11 | Up | 80 | Up | SGC-7901, MGC-803, MKN-28, and BGC-823 | GES-1 | [77] |
| FBXO50 | - | 200 | - | MKN1, MKN45, MKN74, NUGC2, NUGC3, NUGC4, SC-6-JCK, AGS, KATOIII, N87 | - | [78] |
| FBXL10(KDM2B) | - | - | - | MKN-45, SGC-7901, N-87, HGC-27 | GES-1 | [79] |
High expression of FBXW5 was also correlated with poor prognosis [73]. Interestingly, FBXO50 was found to be highly expressed in GC, and patients with high FBXO50 expression had a significantly higher prevalence of recurrence after curative gastrectomy and shorter overall survival [78]. These findings suggest that differential expression of FBPs in GC not only promotes tumor progression but also inhibits it, emphasizing the dual role of FBPs in GC. Overall, FBPs play a significant role in the progression of GC.
Relationship between F-box proteins and gastric cancer
Multiple events contribute to the malignant characteristics of cells, including sustained growth, resistance to cell death, induced invasion and metastasis, and increased resistance to chemotherapy [84]. Mutations in oncogenes and tumor suppressor genes are characteristic of cancer. In recent years, FBPs have garnered attention for their crucial functions in mediating oncogenes and tumor suppressors in GC, thereby regulating various cancer-related features (Table 2).
In vitro functional characterization of FBPs in gastric cancer
| FBPs | Substrate protein | Effect on viability/ proliferation | Effect on apoptosis | Effect on invasion/ metastasis | Reference |
|---|---|---|---|---|---|
| FBXW7 | GFI1 | inhibiting | - | - | [85] |
| FBXW7 | Snail 1 / ZEB 1 | inhibiting | promote | inhibiting | [66] |
| FBXW7 | Brg1 | - | - | inhibiting | [86] |
| FBXL2 | FOXM1 | inhibiting | - | inhibiting | [68] |
| FBXL5 | Snail 1 | - | - | inhibiting | [69] |
| FBXO31 | Snail1 | - | - | inhibiting | [70] |
| FBX8 | - | inhibiting | - | inhibiting | [71] |
| FBXO21 | Nr2f2 | inhibiting | - | inhibiting | [72] |
| FBXW5 | LATS1 | promoting | - | promoting | [73] |
| FBXW5 | - | - | - | promoting | [74] |
| KDM2A | - | promoting | - | promoting | [75] |
| FBXO2 | - | - | - | promoting | [76] |
| FBXO11 | PTEN | promoting | - | promoting | [77] |
| FBXO50 | - | promoting | - | promoting | [78] |
| KDM2B | - | promoting | - | promoting | [79] |
3.1 F-box proteins are involved in the proliferation of GC
FBPs have been shown to regulate the growth and proliferation of GC cells. For example, FBXW7 mediates the degradation of GFI1, inhibiting the proliferation of GC cells [85], Similarly, overexpression of FBXL2 inhibits GC proliferation by degrading ubiquitinated fork head box M1 (FOXM1) transcription factor in GC cell lines [68]. Knockdown of FBX8 significantly promotes the proliferation and invasion of BGC823 cells [71]. In GC, Fbxo21 can inhibit proliferation, in part, by down-regulating Nr2f2 [72]. Additionally, FBPs can regulate proliferation by modulating other cell death modalities. Knockdown of KDM2B (FBXL10) immediately induces autophagy and subsequently inhibits GC cell proliferation [79]. FBPs have been found to regulate GC proliferation through multiple distinct pathways, indicating their potential as critical upstream targets within the GC proliferation cascade.
3.2 F-box proteins are involved in invasion and metastasis of gastric cancer
Metastasis is a major contributor to the poor prognosis of GC. Several studies have demonstrated that FBPs can affect GC invasion and metastasis by controlling epithelial-mesenchymal transition (EMT) (Figure 4). FBXW7 downregulates the RhoA signaling pathway, inhibiting EMT in GC [66]. FBXW7 mediates Brg1 degradation, thus inhibiting GC metastasis [86]. FBXO21 inhibits EMT, in part, through down-regulating Nr2f2 [72]. FBXL5, FBXO31, and FBXW7 negatively regulate EMT-enhancing factors such as Snail 1 or ZEB 1, inhibiting GC metastasis [66, 69, 70]. In a xenograft model of nude mice, FBX8 was found to be sufficient to inhibit metastasis [71]. Moreover, FBPs can regulate GC metastasis and invasion through the modulation of signaling pathways. For instance, FBXW5 inactivates the Hippo signaling pathway by enhancing LATS1 ubiquitination and degradation, promoting GC cell invasion and metastasis [73]. FBXW5 promotes tumorigenesis and metastasis in GC through activation of the FAK-Src signaling pathway [74]. FBXO11 acts as an oncogene by suppressing PTEN and activating the PI3K/AKT pathway, thus promoting EMT in GC [77]. Multiple FBPs have been shown to be highly correlated with metastatic invasion of GC, and different types of FBPs mediate metastatic invasion by different mechanisms, and those evidences support that FBPs are key upstream targets of EMT.
3.3 F-box proteins are involved in Chemoresistance of GC
Chemoresistance remains a major challenge in the treatment of advanced GC [87, 88]. Understanding the molecular mechanisms underlying chemoresistance is crucial. FBXW7 has been identified as a tumor suppressor gene that reduces important oncoproteins, associated oncogenic effects, and cell cycle progression. Clinical data have shown that macrophage-derived exosomal miR-223 promotes doxorubicin resistance in GC cells by inhibiting FBXW7 [89]. Exosomal miR-500a-3p has been found to promote resistance to cisplatin and enhance stemness properties of GC cells by targeting FBXW7 [90]. Increased expression of miR-363 promotes cell proliferation and chemoresistance through direct targeting of the tumor suppressor FBXW7 [91]. FBXW7 has also been identified as a direct and functional target gene of miR-223, mediating DDP resistance in human GC [92]. The miR-223/FBXW7 pathway has been shown to regulate the sensitivity of HER2-positive GC cell lines to trastuzumab [93]. Other FBPs have also been implicated in chemoresistance in GC. Wu demonstrated that depletion of FBXL5 enhances cisplatin resistance in GC cells through ERK and p38 activation [94]. Knockdown of FBXO32 enhances 5-FU cytotoxicity in GC cells that acquired prior resistance to 5-FU [95]. Downregulation of FBXL7 by Aurora Kinase A (AURKA) inhibits Survivin degradation, leading to enhanced drug resistance [96] (Table 3). This demonstrates the potent role of FBPs in the drug resistance mechanism of GC, suggesting that FBPs could represent a novel mechanism of chemotherapy resistance in this malignancy. Hence, the pursuit of targeted drugs specifically designed to counteract FBPs or their synergistic utilization holds great promise as an avenue to enhance the present treatment paradigm for GC.
4. Key molecules regulating F-box proteins in gastric cancer
Numerous distinct non-coding RNA (ncRNA) sequences are abundantly present within cells. Initially regarded as mere "junk" transcription products, ncRNAs have emerged as functional regulatory molecules that orchestrate essential cellular processes encompassing chromatin remodeling, transcriptional regulation, post-transcriptional modification, and signal transduction. By participating in intricate networks, ncRNAs possess the ability to influence multiple molecular targets, thereby eliciting specific cellular responses and determining cellular fate [98, 99].
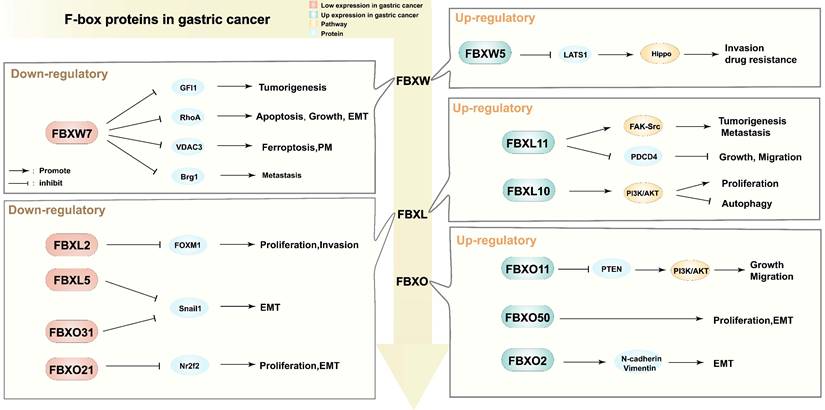
Different regulatory functions of F-box proteins in gastric cancer. PM: peritoneal metastasis; EMT: epithelial-mesenchymal transition; GFI1: Growth factor independence 1; RhoA: Ras homolog gene family member A; VDAC3: Voltage dependent anion channel 3; Brg1: Brahma related gene 1; FOXM1: Forkhead box M1; Snail1: snail family transcriptional repressor 1; Nr2f2: Nuclear receptor subfamily 2 group F member 2; LATS1: Large Tumor Suppressor Kinase 1; Hippo: Hippo signaling pathway; FAK-Src: focal adhesion kinase and c-Src signaling pathway; PDCD4: programmed cell death 4; PI3K/AKT: phosphatidylinositol 3-kinase/protein kinase B signaling pathway; PTEN: Phosphatase and tensin homolog; N-cadherin/Vimentin: the expression of interstitial markers.
The role of F-box protein in chemoresistance of gastric cancer
| F-box protein | Substrate protein | Chemotherapeutic drugs | Effects on chemosensitivity | References |
|---|---|---|---|---|
| FBXW7 | - | Cisplatin | Decreasing | [92] |
| N-cadherin, vimentin | doxorubicin | Decreasing | [89] | |
| - | trastuzumab | Decreasing | [93] | |
| c-Myc/Mcl-1/cyclin E/c-Jun | DCF | Decreasing | [91] | |
| CD133, CD44 and SOX2 | Cisplatin | Decreasing | [90] | |
| FBXL5 | RhoGDI2 | Cisplatin | Decreasing | [94] |
| FBXO32 | - | 5-Fu | Decreasing | [95] |
| FBXL1 | P27 | Acyinom | Decreasing | [97] |
| FBXL7 | survivin | doxorubicin | Decreasing | [96] |
DCF: docetaxel + cisplatin + 5-fluorouracil; 5-Fu,5-fluorouracil
Consequently, ncRNAs assume pivotal roles as regulators of physiological programs during both normal development and disease states. Notably, ncRNA genes are increasingly being recognized as valuable therapeutic targets for cancer treatment [100], opening up new avenues for tumor diagnostics. Moreover, emerging evidence highlights the involvement of ncRNAs in governing the expression of FBPs in human cancers. Specifically, microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) have been implicated in the modulation of FBPs expression in malignant tumors (Table 4). This section focuses on elucidating the mechanisms by which miRNAs target FBPs and contribute to the pathogenesis and progression of GC. Additionally, we briefly outline the regulatory roles of lncRNAs, circRNAs, and other biomarkers in relation to FBPs in GC. These collective findings underscore the potential of targeting ncRNAs as a novel approach to regulate FBPs for anti-GC therapy.
Biomarkers targeting FBPs in gastric cancer
| Biomarker | Expression in GC | Effects on FBP | FBPs | FBPs Functions | Reference |
|---|---|---|---|---|---|
| miR-20a/miR-17 | Up | Inhibiting | FBXO31 | suppressor | [101] |
| miR-25 | Up | Inhibiting | FBXW7 | suppressor | [102] |
| Up | Inhibiting | FBXW7 | suppressor | [103] | |
| miR-223 | Up | Inhibiting | FBXW7 | suppressor | [104] |
| Up | Inhibiting | FBXW7 | - | [89] | |
| Up | Inhibiting | FBXW7 | - | [93] | |
| Up | Inhibiting | FBXW7 | - | [92] | |
| miR-448 | Up | Inhibiting | KDM2B | suppressor | [105] |
| miR-29b | Down | Inhibiting | KDM2A | suppressor | [106] |
| miR-363 | Up | Inhibiting | FBXW7 | - | [91] |
| miR-500a-3p | Up | Inhibiting | FBXW7 | - | [90] |
| LncRNA MT1JP | Down | Inhibiting | FBXW7 | suppressor | [107] |
| lncRNA BDNF-AS | Up | Inhibiting | FBXW7 | suppressor | [67] |
| LINC 01436 | Up | promoting | FBOX11 | Promoter | [108] |
| LINC00511 | Up | promoting | KDM2A | Promoter | [109] |
| circSMARCA5 | Down | Inhibiting | FBXL2 | suppressor | [110] |
| circDYRK1A | Down | Inhibiting | FBXO4 | suppressor | [111] |
| STYX | Up | Inhibiting | FBXO31 | suppressor | [112] |
| STAT3 | Down | Inhibiting | FBXL1 | Promoter | [113] |
| MECP2 | Up | Inhibiting | FBXW7 | suppressor | [114] |
4.1 The regulatory roles of miRNA on F-box proteins in gastric cancer
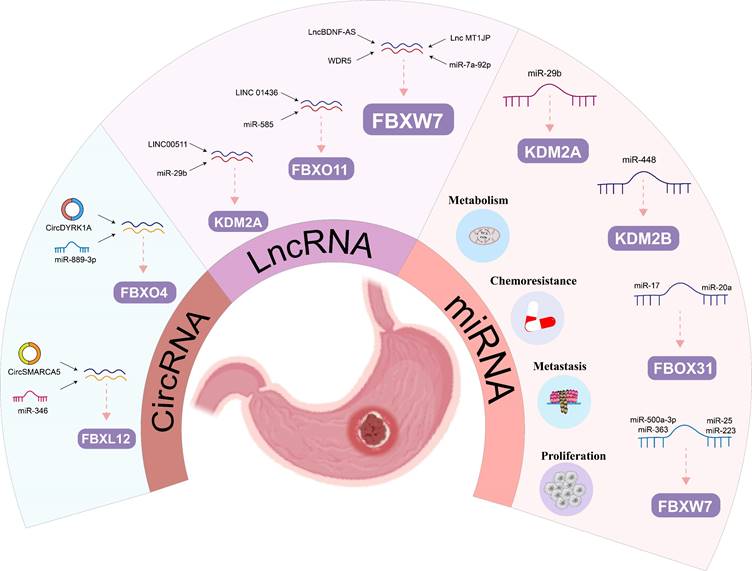
MiRNAs have been increasingly recognized in recent years as regulatory genes that can bind mRNA through sequence complementation and inhibit protein translation and/or mRNA degradation, ultimately affecting human tumor progression and patient prognosis. The regulation of GC progression by mRNAs through FBPs is now gradually being demonstrated (Figure 5). Many studies showed in vivo and in vitro experiments that MiR-92a, miR-25 and miR-223 could promote the cell proliferation, invasion and migration through FBXW7 in GC [102, 104, 115]. Meanwhile, MiR-25 could also have an antiapoptotic effect on GC by inhibiting FBXW7-promoting oncogenes, such as CCNE1 and MYC [103]. FBXW7 also plays an important function in miRNA-mediated drug resistance in GC. Zhang et al. demonstrated that increased miR-363 expression was shown to promote GC proliferation and chemoresistance by directly targeting the tumor suppressor FBXW7[91]. Two studies showed that FBXW7 is a key target of MiR-500a-3p and miR-223 in mediating DDP resistance in GC [90, 92].
In addition to FBXW7, miRNA also regulates other members of the FBPs, for example, Zhang et al. found that miR-20a and miR-17 exerted pro-cancer effects by directly binding to the 3'-UTR of FBXO31 to inhibit FBXO31 expression [101]. Furthermore, Hong et al. demonstrated that FBXL10 was the target of miR-448 that inhibited glycolysis and promoted oxidative phosphorylation [105]. Ye et al identified that RUNX3 could mediate miR-29b up-regulation to inhibit the proliferation and migration of GC cells by targeting KDM2A [106]. Thus, this growing evidence suggests that FBPs are key target protein for miRNAs regulating human GC.
4.2 The regulatory roles of lncRNAs on F-box proteins in gastric cancer
Besides the miRNAs mentioned in the appeal, other ncRNAs can also regulate FBPs (such as, lncRNAs and circRNAs) (Figure 5). LncRNAs play important roles in genomic transcription, translation, and post-translational modifications [116]. Previous studies have shown that lncRNAs are involved in a variety of biological behaviors in GC, including proliferation, invasion, and metastasis [117-119]. Huang et al. demonstrated that lncBDNF-AS can regulate FBXW7 expression by recruiting WDR5, thus affecting FBXW7 transcription, which regulates protein expression of VDAC3 through ubiquitination to protect GC cells from ferroptosis and promote the peritoneal metastasis (PM) [67]. Zhang et al. found that higher lncRNA MT1JP was significantly associated with lymph node metastasis and progression, and MT1JP regulates GC progression by competitively binding to miR-92a-3p as a competitive endogenous RNA (ceRNA) and regulating FBXW7 expression [107]. Similarly, another research team pointed out that LINC01436 promotes proliferation and metastasis of GC cells by regulating miR-585 and FBOX11 [108]. Furthermore, LINC00511/miR-29b/KDM2A axis could also be used as a diagnostic and therapeutic target for GC [109].
4.3 The regulatory roles of circRNAs on F-box proteins in gastric cancer
The expression and discovery of circRNA in tumors has become the latest research hotspot in the field of tumor RNA. Compared with traditional linear RNA, circRNA molecules have a closed-loop structure and are not affected by RNA exonucleases, and their expression is more stable and not easily degraded. Functionally, it mainly plays the role of miRNA sponge in the cell, which in turn relieves the repressive effect of miRNA on target genes and elevates the expression level of target genes [120]. This provides a theoretical basis for circRNA regulation of FBPs (Figure 5). Li et al. confirmed that overexpression of circSMARCA5 inhibited GC cell proliferation, migration and invasion, mainly because circSMARCA5 could act as a miR-346 sponge that regulates the expression of FBXL2 [110]. Similarly, circDYRK1A could act as a miR-889-3p sponge to upregulate FBXO4 expression and inhibit glutamine metabolism in GC, thereby promoting its progression [111].
4.4 The regulatory roles of others on F-box proteins in gastric cancer
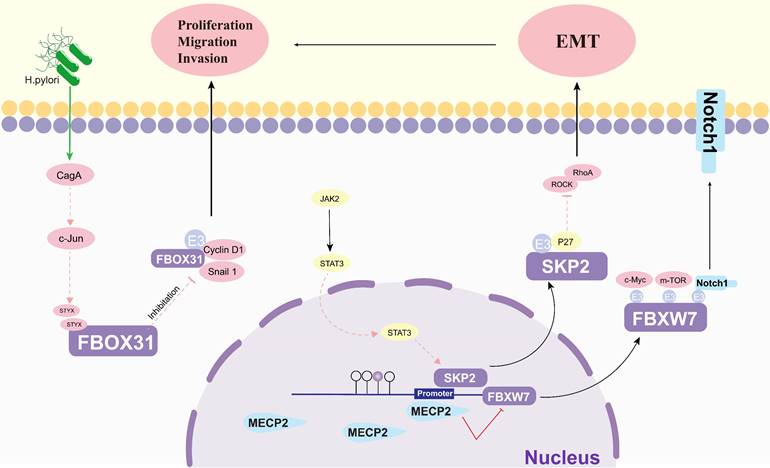
Protein cancer biomarkers have multiple clinical purposes during disease progression, both in early and late stages (Figure 6). The search for new and better biomarkers has become an integral part of contemporary cancer research [121]. Methyl-CpG-binding protein 2 (MECP2), an epigenetic regulatory factor, promotes the carcinogenesis and progression of a number of cancers. Zhao et al. found that MECP2 could regulate the Notch1/C-MYC/mTOR signaling pathway by inhibiting FBXW7 transcription, thereby promoting GC cell migration and invasion [114]. The catalytically inactive pseudophosphatase serine/threonine/tyrosine interacting protein (STYX) is a member of the protein tyrosine phosphatase family. Lui et al. suggested that STYX plays an oncogenic role in GC mainly by inhibiting FBXO31. Interestingly, both transcription factor c-Jun and Helicobacter pylori (H. pylori) infection were found to enhance the expression of STYX in GC [112]. This suggests that FBPs can be a key factor in the development of H. pylori-induced GC. Lei et al. showed that SerpinB5 was expressed at higher levels in GC tissues than in corresponding normal tissues and was associated with GC progression, and further studies revealed that KHDRBS 3 and FBXO32 are key molecules of SerpinB5 in GC carcinogenesis [122]. One study found that the interaction between STAT3 and FBXL1(SKP2)/p27/p21 pathway - plays an important role in mediating the motility, migration and invasion of GC cells [113].
The regulatory roles of ncRNA on FBPs in gastric cancer: The regulatory roles of miRNA, lncRNAs and circRNAs on FBPs in gastric cancer.
The regulatory roles of others on FBPs in gastric cancer. EMT: epithelial-mesenchymal transition; H. Pylori: helicobacter pylori; CagA: The cytotoxin-associated gene A; c-jun: AP-1 transcription Factor; STYX: Serine/threonine/tyrosine-interacting protein; Cyclin D1: A cell cycle protein; JAK2: Janus kinase 2; Snail1: snail family transcriptional repressor 1; STAT3: signal transducer and activator of transcription 3; SKP2: S-phase kinase-associated protein 2; MECP2: Methyl CpG binding protein 2; ROCK: Rho-associated coiled-coil forming protein kinase; RhoA: Ras homolog gene family member A; p27(SKIP1): The Cyclin-dependent kinase regulator; c-Myc: BHLH Transcription Factor; Notch1: Notch Homolog Protein 1.
5. Prospects of the F-box proteins in the treatment of gastric cancer
The development of new drugs with specific targets for GC has always been an important part of oncology treatment, such as human epidermal growth factor receptor 2(HER-2) targeting agent Trastuzumab [123], vascular epidermal growth factor receptor (VEGFR) targeting agent Ramucirumab [124, 125], VEGFR-2 targeting agent Apatinib [126] and so on. It has been widely used in the treatment of GC patients, bringing enough benefits to GC patients. However, the existence of inter- and intra-patient heterogeneity, as well as poor efficacy and drug resistance, bring great challenges to targeted therapy, so it is crucial to develop new targeted drugs [127].
In GC as researchers have studied FBPs in depth, targeted blockers have now been developed for some key FBPs or related pathways, providing a theoretical basis for the development of targeted drugs for GC. Ueda et al. showed that O-GlcNAcase inhibitor Thiamet G (TMG) could promote GC progression by inhibiting FBXL2-mediated FOXM1 degradation [128]. Wu et al. found that AICAR (an AMPK activator) could increase the expression of tumor suppressor genes FBXW7 and enhanced the pro-apoptotic effect of 5-FU in SGC-7901 cells [129]. Soichiro et al. found that NS398 (a COX-2 inhibitor) induced inhibition of cell proliferation through cell cycle arrest and suppressed the expression of FBXL1 in COX-2-expressing GC cells [130]. FBXL10 is normally expressed in GC, and downregulation of FBXL10 regulates the PI3K/AKT/mTOR signaling pathway to induce autophagy and subsequently inhibit proliferation, while the compound 3-methyladenine (3-MA), an inhibitor of autophagy, is able to reverse this process [79]. BK697, a chemical inhibitor of FIRΔexon2, reversed the inhibitory effect of FIRΔexon2 on FBXW7 and inhibited progression in GC via the FBXW7/BRG1/Snai1 axis [131]. Lycorine hydrochloride (LH), a derivative of lycorine, is an isoquinoline alkaloid extracted from lycoris[132-134]. Li et al. found that LH inhibits cell proliferation and induces apoptosis through promoting FBXW7-MCL1 axis in GC [135]. These studies provide a new research direction for the development of small molecule drugs targeting FBPs in GC.
6. Discussion
In conclusion, this review provides convincing evidence for the role of the FBPs in GC progression. It shows the functional diversity of the FBPs. On the one hand some oncogenic members of the FBPs, which are down-regulated in GC, block the ubiquitination degradation process of some key oncogenic proteins and thus promote tumor metastasis invasion and drug resistance. On the other hand, some oncogenic FBPs are upregulated in GC and promote metastasis and invasion by activating some key oncogenic pathways (e.g. PI3K/Akt/mTOR). Thus, FBPs and their specific protein substrates may represent promising drug targets or biomarkers for GC, however, targeting FBPs remains challenging. Most previous studies have focused on the function of protein substrates of FBPs, and little is known about the regulatory role of FBPs or CRL itself in tumorigenesis. Moreover, most FBPs have multiple protein substrates, and some FBPs promote the degradation of oncoproteins and oncoproteins in GC, thus the function of FBPs is cellular environment dependent. A better understanding of the complex regulatory network of FBPs in GC, the involvement of their protein substrates and kinases in their post-translational modifications, and the mechanisms of cross-interaction with other signaling pathways are urgently needed for future studies.
Three E3-targeting small-molecule drugs, thalidomide, lenalidomide and pomalidomide, which bind a substrate receptor of the E3 ligase cereblon (CRBN)25, have been approved by the US Food and Drug Administration (FDA) [136]. But with limited success in oncology treatment. Therefore, selective inhibitors targeting specific ubiquitin ligases and their protein substrates may be a better and more effective strategy for the treatment of GC. Growing evidence for modulation of FBPs and chemotherapy sensitivity by anticancer natural products [135]. MLN4924 is an antitumor agent, Zhang et al. highlights the potential combination of MLN4924 and P27 inhibition to improve GC therapeutic efficacy [137]. This suggests that develop natural products alone or in combination with modulators of FBPs and/or chemotherapeutic agents could show promising efficacy in human cancers, particularly drug-resistant cancers. However, further studies are necessary to identify the specific molecular targets of these natural products and to examine the efficacy and safety of these strategies in clinically relevant cancer models.
Acknowledgements
Funding
This article was supported by Zhejiang Provincial Basic Public Welfare Research Program (LGF22H160084).
Author contributions
Yanzhen Yang: Conceptualization, Writing - original draft, Writing review & editing, Visualization.
Jingli Xu & Xie Qu: Resources, Writing - original draft, Writing - review & editing.
Can Hu: Writing - review & editing.
Lei Chen: Conceptualization, Writing - review & editing.
Cong Luo & Li Yuan: Conceptualization, Writing - review & editing, Supervision.
Abbreviations
GC: gastric cancer
UPS: the ubiquitin-proteasome system
FBP: the F-box protein
SCF: the Skp1-Cullin1-F- box protein
ncRNAs: non-coding RNAs
E1s: ubiquitin-activating enzymes
E2s: ubiquitin-conjugating enzymes
E3s: ubiquitin ligases
DUBs: deubiquitinating enzymes
Ub: ubiquitin
Lys: lysine
Gly: glycine
RING: really interesting new gene
HECT: homologous to E6AP C-terminus
RBR: RING-in-between-RING
CRL: cullin-RING E3 ligase
APC/C: complex/cyclosome
SKP1: s-phase kinase-associated protein 1
ARID1A: AT-rich interactive domain 1A
EMT: epithelial-mesenchymal transition
LATS1/2: large tumor suppressor kinases 1/2
OS: overall survival
miRNA: microRNAs
lncRNA: long non-coding RNAs
circRNAs: circular RNAs
PM: peritoneal metastasis
DDP: cisplatin
DCF: docetaxel + cisplatin + 5-fluorouracil
5-Fu: 5-fluorouracil
UTRs: untranslated regions
ceRNA: competitive endogenous RNA
MECP2: methyl-CpG-binding protein 2
PM: peritoneal metastasis
LH: lycorine hydrochloride
STYX: serine/threonine/tyrosine interacting protein
H. Pylori: helicobacter pylori
Cul1: cullin1
RhoGDI2: Rho GDP dissociation inhibitor 2
AURKA: aurora Kinase A
HER-2: human epidermal growth factor receptor 2
VEGFR: vascular epidermal growth factor receptor
TMG: thiamet G
COX-2: cyclooxygenase -2
3-MA: 3-methyladenine
FDA: Food and Drug Administration
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C. et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2022;20:167-92
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
3. Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends-An Update. Cancer Epidemiol Biomarkers Prev. 2016;25:16-27
4. GBD 2017 Stomach Cancer Collaborators. The global, regional, and national burden of stomach cancer in 195 countries, 1990-2017: a systematic analysis for the Global Burden of Disease study 2017. Lancet Gastroenterol Hepatol. 2020;5:42-54
5. Chen W. Cancer statistics: updated cancer burden in China. Chin J Cancer Res. 2015;27:1
6. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-48
7. Petrelli F, Zaniboni A, Ghidini A, Ghidini M, Turati L, Pizzo C. et al. Timing of Adjuvant Chemotherapy and Survival in Colorectal, Gastric, and Pancreatic Cancer. A Systematic Review and Meta-Analysis. Cancers (Basel). 2019;11:550
8. Chan WL, Lam KO, So TH, Lee VH, Kwong LD. Third-line systemic treatment in advanced/metastatic gastric cancer: a comprehensive review. Ther Adv Med Oncol. 2019;11:1758835919859990
9. Zheng L, Wu C, Xi P, Zhu M, Zhang L, Chen S. et al. The survival and the long-term trends of patients with gastric cancer in Shanghai, China. BMC Cancer. 2014;14:300
10. Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432-9
11. Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264-79
12. Biagioni A, Skalamera I, Peri S, Schiavone N, Cianchi F, Giommoni E. et al. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev. 2019;38:537-48
13. Talebi A, Celis-Morales CA, Borumandnia N, Abbasi S, Pourhoseingholi MA, Akbari A. et al. Predicting metastasis in gastric cancer patients: machine learning-based approaches. Sci Rep. 2023;13:4163
14. Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:Reviews3002
15. Kumar A, Paietta JV. The sulfur controller-2 negative regulatory gene of Neurospora crassa encodes a protein with beta-transducin repeats. Proc Natl Acad Sci U S A. 1995;92:3343-7
16. Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW. et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263-74
17. Lee EK, Diehl JA. SCFs in the new millennium. Oncogene. 2014;33:2011-8
18. Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243-56
19. Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD. et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514-21
20. Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369-81
21. Heo J, Eki R, Abbas T. Deregulation of F-box proteins and its consequence on cancer development, progression and metastasis. Semin Cancer Biol. 2016;36:33-51
22. Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M. et al. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J Clin Invest. 2016;126:4387
23. Asmamaw MD, Liu Y, Zheng YC, Shi XJ, Liu HM. Skp2 in the ubiquitin-proteasome system: A comprehensive review. Med Res Rev. 2020;40:1920-49
24. Yokoi S, Yasui K, Mori M, Iizasa T, Fujisawa T, Inazawa J. Amplification and overexpression of SKP2 are associated with metastasis of non-small-cell lung cancers to lymph nodes. Am J Pathol. 2004;165:175-80
25. Lu Y, Li J, Cheng D, Parameswaran B, Zhang S, Jiang Z. et al. The F-box protein FBXO44 mediates BRCA1 ubiquitination and degradation. J Biol Chem. 2012;287:41014-22
26. Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB. et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859-63
27. Pla-Prats C, Thomä NH. Quality control of protein complex assembly by the ubiquitin-proteasome system. Trends Cell Biol. 2022;32:696-706
28. Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233-47
29. Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659-71
30. Chen S, Liu Y, Zhou H. Advances in the Development Ubiquitin-Specific Peptidase (USP) Inhibitors. Int J Mol Sci. 2021;22:4546
31. Li X, Elmira E, Rohondia S, Wang J, Liu J, Dou QP. A patent review of the ubiquitin ligase system: 2015-2018. Expert Opin Ther Pat. 2018;28:919-37
32. Kwon YT, Ciechanover A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem Sci. 2017;42:873-86
33. Leestemaker Y, Ovaa H. Tools to investigate the ubiquitin proteasome system. Drug Discov Today Technol. 2017;26:25-31
34. Pohl C, Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366:818-22
35. Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435-67
36. Oh E, Akopian D, Rape M. Principles of Ubiquitin-Dependent Signaling. Annu Rev Cell Dev Biol. 2018;34:137-62
37. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399-434
38. Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9-20
39. Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739-51
40. D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP. et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138-42
41. Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF. et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90-3
42. Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722-5
43. Yan L, Lin M, Pan S, Assaraf YG, Wang ZW, Zhu X. Emerging roles of F-box proteins in cancer drug resistance. Drug Resist Updat. 2020;49:100673
44. Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438-49
45. Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83-93
46. Buschbeck M, Uribesalgo I, Wibowo I, Rué P, Martin D, Gutierrez A. et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol. 2009;16:1074-9
47. Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM. et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19-30
48. Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M. et al. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood. 2012;119:3132-41
49. Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369-81
50. Zheng N, Wang Z, Wei W. Ubiquitination-mediated degradation of cell cycle-related proteins by F-box proteins. Int J Biochem Cell Biol. 2016;73:99-110
51. Li P, Li C, Zhao X, Zhang X, Nicosia SV, Bai W. p27(Kip1) stabilization and G(1) arrest by 1,25-dihydroxyvitamin D(3) in ovarian cancer cells mediated through down-regulation of cyclin E/cyclin-dependent kinase 2 and Skp1-Cullin-F-box protein/Skp2 ubiquitin ligase. J Biol Chem. 2004;279:25260-7
52. Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A. 1998;95:11324-9
53. Walter D, Hoffmann S, Komseli ES, Rappsilber J, Gorgoulis V, Sørensen CS. SCF(Cyclin F)-dependent degradation of CDC6 suppresses DNA re-replication. Nat Commun. 2016;7:10530
54. Guo CJ, Ma XK, Xing YH, Zheng CC, Xu YF, Shan L. et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell. 2020;181:621-36.e22
55. Fan W, Liu X, Zhang J, Qin L, Du J, Li X. et al. TRIM67 Suppresses TNFalpha-Triggered NF-kB Activation by Competitively Binding Beta-TrCP to IkBa. Front Immunol. 2022;13:793147
56. Cohen M, Amir S, Golan M, Ben-Neriah Y, Mabjeesh NJ. β-TrCP upregulates HIF-1 in prostate cancer cells. Prostate. 2019;79:403-13
57. Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Invest. 2004;22:304-11
58. Li Z. et al. F-box only protein 11 promotes the ubiquitination of CYP3A4 for ubiquitin-proteasome degradation in HepG2 cells. Chemico-biological interactions. 2019;312:108784
59. Yuan H. et al. F-box only protein 11 promotes the ubiquitination of andrographolide-induced CYP3A4 for ubiquitin-proteasome degradation in HepG2 cells. Journal of Ethnopharmacology. 2021;265:113382
60. Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. 2014;13:889-903
61. Tang W, Li Y, Yu D, Thomas-Tikhonenko A, Spiegelman VS, Fuchs SY. Targeting beta-transducin repeat-containing protein E3 ubiquitin ligase augments the effects of antitumor drugs on breast cancer cells. Cancer Res. 2005;65:1904-8
62. Rico-Bautista E, Yang CC, Lu L, Roth GP, Wolf DA. Chemical genetics approach to restoring p27Kip1 reveals novel compounds with antiproliferative activity in prostate cancer cells. BMC Biol. 2010;8:153
63. Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515-24
64. Jeong HS, Jung ES, Sim YJ, Kim SJ, Jang JW, Hong KS. et al. Fbxo25 controls Tbx5 and Nkx2-5 transcriptional activity to regulate cardiomyocyte development. Biochim Biophys Acta. 2015;1849:709-21
65. Huang KK, Ramnarayanan K, Zhu F, Srivastava S, Xu C, Tan ALK. et al. Genomic and Epigenomic Profiling of High-Risk Intestinal Metaplasia Reveals Molecular Determinants of Progression to Gastric Cancer. Cancer Cell. 2018;33:137-50.e5
66. Li H, Wang Z, Zhang W, Qian K, Xu W, Zhang S. Fbxw7 regulates tumor apoptosis, growth arrest and the epithelial-to-mesenchymal transition in part through the RhoA signaling pathway in gastric cancer. Cancer Lett. 2016;370:39-55
67. Huang G, Xiang Z, Wu H, He Q, Dou R, Lin Z. et al. The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int J Biol Sci. 2022;18:1415-33
68. Li LQ, Pan D, Chen H, Zhang L, Xie WJ. F-box protein FBXL2 inhibits gastric cancer proliferation by ubiquitin-mediated degradation of forkhead box M1. FEBS Lett. 2016;590:445-52
69. Wu W, Ding H, Cao J, Zhang W. FBXL5 inhibits metastasis of gastric cancer through suppressing Snail1. Cell Physiol Biochem. 2015;35:1764-72
70. Zou S, Ma C, Yang F, Xu X, Jia J, Liu Z. FBXO31 Suppresses Gastric Cancer EMT by Targeting Snail1 for Proteasomal Degradation. Mol Cancer Res. 2018;16:286-95
71. Wu P, Wang F, Wang Y, Men H, Zhu X, He G. et al. Significance of FBX8 in progression of gastric cancer. Exp Mol Pathol. 2015;98:360-6
72. Jiang Y, Liu X, Shen R, Gu X, Qian W. Fbxo21 regulates the epithelial-to-mesenchymal transition through ubiquitination of Nr2f2 in gastric cancer. J Cancer. 2021;12:1421-30
73. Yao Y, Liu Z, Huang S, Huang C, Cao Y, Li L. et al. The E3 ubiquitin ligase, FBXW5, promotes the migration and invasion of gastric cancer through the dysregulation of the Hippo pathway. Cell Death Discov. 2022;8:79
74. Yeo MS, Subhash VV, Suda K, Balcıoğlu HE, Zhou S, Thuya WL. et al. FBXW5 Promotes Tumorigenesis and Metastasis in Gastric Cancer via Activation of the FAK-Src Signaling Pathway. Cancers (Basel). 2019;11:836
75. Huang Y, Liu Y, Yu L, Chen J, Hou J, Cui L. et al. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol. 2015;36:271-8
76. Sun X, Wang T, Guan ZR, Zhang C, Chen Y, Jin J. et al. FBXO2, a novel marker for metastasis in human gastric cancer. Biochem Biophys Res Commun. 2018;495:2158-64
77. Sun C, Tao Y, Gao Y, Xia Y, Liu Y, Wang G. et al. F-box protein 11 promotes the growth and metastasis of gastric cancer via PI3K/AKT pathway-mediated EMT. Biomed Pharmacother. 2018;98:416-23
78. Miwa T, Kanda M, Tanaka H, Tanaka C, Kobayashi D, Umeda S. et al. FBXO50 Enhances the Malignant Behavior of Gastric Cancer Cells. Ann Surg Oncol. 2017;24:3771-9
79. Zhao E, Tang C, Jiang X, Weng X, Zhong X, Zhang D. et al. Inhibition of cell proliferation and induction of autophagy by KDM2B/FBXL10 knockdown in gastric cancer cells. Cell Signal. 2017;36:222-9
80. Calcagno DQ, Freitas VM, Leal MF, de Souza CR, Demachki S, Montenegro R. et al. MYC, FBXW7 and TP53 copy number variation and expression in gastric cancer. BMC Gastroenterol. 2013;13:141
81. Yokobori T, Mimori K, Iwatsuki M, Ishii H, Onoyama I, Fukagawa T. et al. p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009;69:3788-94
82. Li MR, Zhu CC, Ling TL, Zhang YQ, Xu J, Zhao EH. et al. FBXW7 expression is associated with prognosis and chemotherapeutic outcome in Chinese patients with gastric adenocarcinoma. BMC Gastroenterol. 2017;17:60
83. Kogure N, Yokobori T, Ogata K, Altan B, Mochiki E, Ohno T. et al. Low Expression of FBXO45 Is Associated with Gastric Cancer Progression and Poor Prognosis. Anticancer Res. 2017;37:191-6
84. Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31-46
85. Kuai X, Li L, Chen R, Wang K, Chen M, Cui B. et al. SCF(FBXW7)/GSK3β-Mediated GFI1 Degradation Suppresses Proliferation of Gastric Cancer Cells. Cancer Res. 2019;79:4387-98
86. Huang LY, Zhao J, Chen H, Wan L, Inuzuka H, Guo J. et al. SCF(FBW7)-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat Commun. 2018;9:3569
87. Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol Cancer. 2020;19:96
88. Wu H, Fu M, Liu J, Chong W, Fang Z, Du F. et al. The role and application of small extracellular vesicles in gastric cancer. Mol Cancer. 2021;20:71
89. Gao H, Ma J, Cheng Y, Zheng P. Exosomal Transfer of Macrophage-Derived miR-223 Confers Doxorubicin Resistance in Gastric Cancer. Onco Targets Ther. 2020;13:12169-79
90. Lin H, Zhang L, Zhang C, Liu P. Exosomal MiR-500a-3p promotes cisplatin resistance and stemness via negatively regulating FBXW7 in gastric cancer. J Cell Mol Med. 2020;24:8930-41
91. Zhang PF, Sheng LL, Wang G, Tian M, Zhu LY, Zhang R. et al. miR-363 promotes proliferation and chemo-resistance of human gastric cancer via targeting of FBW7 ubiquitin ligase expression. Oncotarget. 2016;7:35284-92
92. Zhou X, Jin W, Jia H, Yan J, Zhang G. MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J Exp Clin Cancer Res. 2015;34:28
93. Eto K, Iwatsuki M, Watanabe M, Ishimoto T, Ida S, Imamura Y. et al. The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. Int J Cancer. 2015;136:1537-45
94. Wu WD, Wang M, Ding HH, Qiu ZJ. FBXL5 attenuates RhoGDI2-induced cisplatin resistance in gastric cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:2551-7
95. Wang C, Li X, Zhang J, Ge Z, Chen H, Hu J. EZH2 contributes to 5-FU resistance in gastric cancer by epigenetically suppressing FBXO32 expression. Onco Targets Ther. 2018;11:7853-64
96. Kamran M, Long ZJ, Xu D, Lv SS, Liu B, Wang CL. et al. Aurora kinase A regulates Survivin stability through targeting FBXL7 in gastric cancer drug resistance and prognosis. Oncogenesis. 2017;6:e298
97. Masuda TA, Inoue H, Sonoda H, Mine S, Yoshikawa Y, Nakayama K. et al. Clinical and biological significance of S-phase kinase-associated protein 2 (Skp2) gene expression in gastric carcinoma: modulation of malignant phenotype by Skp2 overexpression, possibly via p27 proteolysis. Cancer Res. 2002;62:3819-25
98. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18
99. Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179:1033-55
100. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
101. Zhang X, Kong Y, Xu X, Xing H, Zhang Y, Han F. et al. F-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20a. Oncotarget. 2014;5:6178-90
102. Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y. et al. MicroRNA-25 promotes gastric cancer proliferation, invasion, and migration by directly targeting F-box and WD-40 Domain Protein 7, FBXW7. Tumour Biol. 2015;36:7831-40
103. Zhang Y, Peng Z, Zhao Y, Chen L. microRNA-25 Inhibits Cell Apoptosis of Human Gastric Adenocarcinoma Cell Line AGS via Regulating CCNE1 and MYC. Med Sci Monit. 2016;22:1415-20
104. Li J, Guo Y, Liang X, Sun M, Wang G, De W. et al. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol. 2012;138:763-74
105. Hong X, Xu Y, Qiu X, Zhu Y, Feng X, Ding Z. et al. MiR-448 promotes glycolytic metabolism of gastric cancer by downregulating KDM2B. Oncotarget. 2016;7:22092-102
106. Kong Y, Zou S, Yang F, Xu X, Bu W, Jia J. et al. RUNX3-mediated up-regulation of miR-29b suppresses the proliferation and migration of gastric cancer cells by targeting KDM2A. Cancer Lett. 2016;381:138-48
107. Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G. et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17:87
108. Zhang Y, Yang G, He X, Chen S, Zhang F, Fang X. LINC01436, regulating miR-585 and FBXO11, is an oncogenic lncRNA in the progression of gastric cancer. Cell Biol Int. 2020;44:882-93
109. Zhao Y, Chen X, Jiang J, Wan X, Wang Y, Xu P. Epigallocatechin gallate reverses gastric cancer by regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165856
110. Li Q, Tang H, Hu F, Qin C. Circular RNA SMARCA5 inhibits gastric cancer progression through targeting the miR-346/ FBXL2 axis. RSC Adv. 2019;9:18277-84
111. Liu H, Xue Q, Cai H, Jiang X, Cao G, Chen T. et al. RUNX3-mediated circDYRK1A inhibits glutamine metabolism in gastric cancer by up-regulating microRNA-889-3p-dependent FBXO4. J Transl Med. 2022;20:120
112. Liu J, Zang Y, Ma C, Wang D, Tian Z, Xu X. et al. Pseudophosphatase STYX is induced by Helicobacter pylori and promotes gastric cancer progression by inhibiting FBXO31 function. Cell Death Dis. 2022;13:268
113. Wei Z, Jiang X, Qiao H, Zhai B, Zhang L, Zhang Q. et al. STAT3 interacts with Skp2/p27/p21 pathway to regulate the motility and invasion of gastric cancer cells. Cell Signal. 2013;25:931-8
114. Zhao L, Wang X, Yang J, Jiang Q, Zhang J, Wu F. et al. MECP2 promotes the migration and invasion of gastric cancer cells by modulating the Notch1/c-Myc/mTOR signaling pathways by suppressing FBXW7 transcription. Am J Cancer Res. 2022;12:5183-204
115. Liu C, Zhang Y, Chen H, Jiang L, Xiao D. Function analysis of rs9589207 polymorphism in miR-92a in gastric cancer. Tumour Biol. 2016;37:4439-44
116. Jia Y, Tian C, Wang H, Yu F, Lv W, Duan Y. et al. Long non-coding RNA NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of esophageal squamous cell carcinoma by promoting nuclear accumulation of β-catenin. Mol Cancer. 2021;20:162
117. Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J. et al. Long Noncoding RNA GMAN, Up-regulated in Gastric Cancer Tissues, Is Associated With Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS. Gastroenterology. 2019;156:676-91.e11
118. Ji Z, Tang T, Chen M, Dong B, Sun W, Wu N. et al. C-Myc-activated long non-coding RNA LINC01050 promotes gastric cancer growth and metastasis by sponging miR-7161-3p to regulate SPZ1 expression. J Exp Clin Cancer Res. 2021;40:351
119. Wang X, Liang Q, Zhang L, Gou H, Li Z, Chen H. et al. C8orf76 Promotes Gastric Tumorigenicity and Metastasis by Directly Inducing lncRNA DUSP5P1 and Associates with Patient Outcomes. Clin Cancer Res. 2019;25:3128-40
120. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188-206
121. Pavlou MP, Diamandis EP, Blasutig IM. The long journey of cancer biomarkers from the bench to the clinic. Clin Chem. 2013;59:147-57
122. Lei KF, Liu BY, Wang YF, Chen XH, Yu BQ, Guo Y. et al. SerpinB5 interacts with KHDRBS3 and FBXO32 in gastric cancer cells. Oncol Rep. 2011;26:1115-20
123. Okines AF, Dewdney A, Chau I, Rao S, Cunningham D. Trastuzumab for gastric cancer treatment. Lancet. 2010;376:1736 author reply -7
124. Sasako M. Ramucirumab: second-line therapy for gastric cancer. Lancet Oncol. 2014;15:1182-4
125. Lordick F. Gastrointestinal cancer. Over the RAINBOW-renaissance in antiangiogenesis. Nat Rev Clin Oncol. 2015;12:7-8
126. Aoyama T, Yoshikawa T. Targeted therapy: Apatinib - new third-line option for refractory gastric or GEJ cancer. Nat Rev Clin Oncol. 2016;13:268-70
127. Zeng Y, Jin RU. Molecular pathogenesis, targeted therapies, and future perspectives for gastric cancer. Semin Cancer Biol. 2022;86:566-82
128. Ueda Y, Moriwaki K, Takeuchi T, Higuchi K, Asahi M. O-GlcNAcylation-mediated degradation of FBXL2 stabilizes FOXM1 to induce cancer progression. Biochem Biophys Res Commun. 2020;521:632-8
129. Wu Y, Qi Y, Liu H, Wang X, Zhu H, Wang Z. AMPK activator AICAR promotes 5-FU-induced apoptosis in gastric cancer cells. Mol Cell Biochem. 2016;411:299-305
130. Honjo S, Kase S, Osaki M, Ardyanto TD, Kaibara N, Ito H. COX-2 correlates with F-box protein, Skp2 expression and prognosis in human gastric carcinoma. Int J Oncol. 2005;26:353-60
131. Ailiken G, Kitamura K, Hoshino T, Satoh M, Tanaka N, Minamoto T. et al. Post-transcriptional regulation of BRG1 by FIRΔexon2 in gastric cancer. Oncogenesis. 2020;9:26
132. Lamoral-Theys D, Decaestecker C, Mathieu V, Dubois J, Kornienko A, Kiss R. et al. Lycorine and its derivatives for anticancer drug design. Mini Rev Med Chem. 2010;10:41-50
133. Cedrón JC, Gutiérrez D, Flores N, Ravelo AG, Estévez-Braun A. Synthesis and antiplasmodial activity of lycorine derivatives. Bioorg Med Chem. 2010;18:4694-701
134. Nair JJ, van Staden J. Acetylcholinesterase inhibition within the lycorine series of Amaryllidaceae alkaloids. Nat Prod Commun. 2012;7:959-62
135. Li C, Deng C, Pan G, Wang X, Zhang K, Dong Z. et al. Lycorine hydrochloride inhibits cell proliferation and induces apoptosis through promoting FBXW7-MCL1 axis in gastric cancer. J Exp Clin Cancer Res. 2020;39:230
136. Holstein SA, McCarthy PL. Immunomodulatory Drugs in Multiple Myeloma: Mechanisms of Action and Clinical Experience. Drugs. 2017;77:505-20
137. Zhang Q, Hou D, Luo Z, Chen P, Lv B, Wu L. et al. The novel protective role of P27 in MLN4924-treated gastric cancer cells. Cell Death Dis. 2015;6:e1867
Author contact
![]() Corresponding authors: Cong Luo: luocongorg.cn; Yuan Li: yuanli909909com. 1 Banshandong, Road, Hangzhou, 310005, China.
Corresponding authors: Cong Luo: luocongorg.cn; Yuan Li: yuanli909909com. 1 Banshandong, Road, Hangzhou, 310005, China.

 Global reach, higher impact
Global reach, higher impact