Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(7):1307-1320. doi:10.7150/ijms.96274 This issue Cite
Review
Cross-Talk between the TGF-β and Cell Adhesion Signaling Pathways in Cancer
1. Guangdong Provincial Key Laboratory of Medical Molecular Diagnostics, Dongguan Key Laboratory of Medical Bioactive Molecular Developmental and Translational Research, The First Dongguan Affiliated Hospital, Guangdong Medical University, Dongguan, Guangdong, 523808, China.
2. Institute of Laboratory Medicine, School of Medical Technology, Guangdong Medical University, Dongguan, Guangdong, 523808, China.
3. Department of Pathology, Binhaiwan Central Hospital of Dongguan, Dongguan, Guangdong, 523905, China.
4. School of Biomedical Engineering, Guangdong Medical University, Dongguan, Guangdong, 523808, China.
† These authors have contributed equally to this work.
Received 2024-3-14; Accepted 2024-4-30; Published 2024-5-13
Abstract
Transforming growth factor-β (TGF-β) is strongly associated with the cell adhesion signaling pathway in cell differentiation, migration, etc. Mechanistically, TGF-β is secreted in an inactive form and localizes to the extracellular matrix (ECM) via the latent TGF-β binding protein (LTBP). However, it is the release of mature TGF-β that is essential for the activation of the TGF-β signaling pathway. This progress requires specific integrins (one of the main groups of cell adhesion molecules (CAMs)) to recognize and activate the dormant TGF-β. In addition, TGF-β regulates cell adhesion ability through modulating CAMs expression. The aberrant activation of the TGF-β signaling pathway, caused by abnormal expression of key regulatory molecules (such as Smad proteins, certain transcription factors, and non-coding RNAs), promotes tumor invasive and metastasis ability via epithelial-mesenchymal transition (EMT) during the late stages of tumorigenesis. In this paper, we summarize the crosstalk between TGF-β and cell adhesion signaling pathway in cancer and its underlying molecular mechanisms.
Keywords: TGF-β signaling pathway, cell adhesion signaling pathway, cancer
Introduction
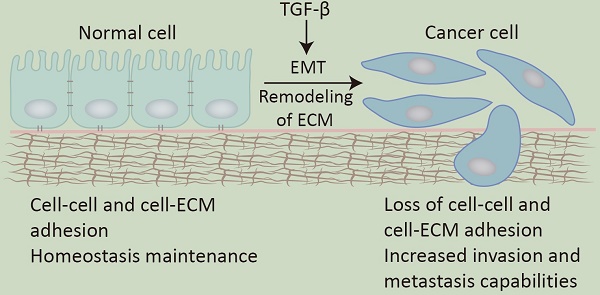
Transforming growth factor-β (TGF-β) was initially isolated as a tumor factor from mouse sarcoma cells and was found to induce certain non-tumor cells to form growth colonies on soft agar [1, 2]. With the in-depth study of it, researchers gradually find that TGF-β has a dual role: as a tumor suppressor, TGF-β inhibits cell proliferation and promotes cell apoptosis and aging to maintain the homeostasis of tissues and organs; on the other hand, as a tumor promoter, TGF-β promotes tumor cell proliferation, transformation, invasion, and metastasis of tumor cells, which plays a key role in tumorigenesis and development [3]. As the common characteristic of malignant tumors, invasion and metastasis often involve changes in cell adhesion mechanisms [4], which include EMT, remodeling of the ECM, and loss of cell-cell adhesion [5].
It is reported that TGF-β is highly relevant to the cell adhesion mechanisms [6]. TGF-β is stored in an inactive form on the ECM and can be activated by CAMs through specific binding. The release of TGF-β from ECM sites greatly enhances the TGF-β signaling pathway [7]. Additionally, TGF-β can regulate the synthesis, deposition, and remodeling of ECM components [8] and the expression of certain CAMs in cells and the extracellular matrix [9].
Cell adhesion is a fundamental mechanism shared by all multicellular organisms, allowing individual cells to assemble into a three-dimensional structure. A variety of cell adhesion mechanisms determine the diversity and specificity of tissues, which are characterized by cell-cell and cell-extracellular matrix connections. Normally, cell adhesion systems are in a dynamic process, not only maintaining the homeostasis of the tissue structure, but also forming a complex signaling network that regulates various aspects of cellular activities, including proliferation, differentiation, motion, etc. [11]. The cross-regulation of three systems, namely, the extracellular matrix, cell adhesion molecules, and cell-secreted cytokines or growth factors, plays an essential role in signaling networks [12].
Current studies have highlighted the interplay between the TGF-β and cell adhesion signaling pathway in tumors. The functional integrity and crosstalk between these two signaling systems are essential for maintaining tissue and organ homeostasis. In this paper, we review the extensive crosstalk between these pathways and the potential underlying molecule mechanism and discussed the role of the TGF-β signaling pathway in tumor development.
The TGF-β signaling pathway
The synthesis, secretion, and localization of TGF-β
TGF-β is a member of the TGF-β superfamily, and there are three TGF-β isoforms in mammals: TGF-β1, TGF-β2, and TGF-β3. These isoforms exhibit a high level of protein sequence conservation. Interestingly, knockout mice of each isoform display distinct phenotypes, indicating the functional diversity of these three isoforms [13-15].
As a multifunctional cytokine that is ubiquitously expressed in almost every tissue and cell type, TGF-β is involved in various processes including immune regulation, wound healing, and the development of tissues and organs. It is also crucial in maintaining tissue homeostasis and plays a role in the pathogenesis of diseases like cancer, by regulating cell growth, proliferation, differentiation, and apoptosis.
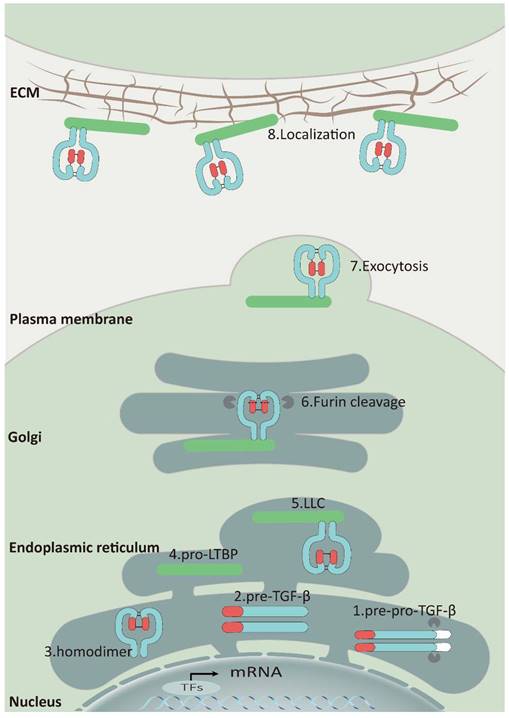
As a secreted protein, TGF-β is synthesized as a pre-pro-TGF-β form in ribosomes. This precursor consists of a small N-terminal signal peptide, a prodomain known as the latency-associated polypeptide, and a C-terminal mature polypeptide. Guided by the signal peptide, the nascent protein is transferred to the endoplasmic reticulum and cleaved, generating pro-TGF-β monomers. These monomers then fold and form a homodimer through disulfide bonds, creating a pro-TGF-β structure. This complex is transported to the cis-Golgi apparatus, where furin or furin-like proteinases cleave the prodomain, releasing the mature TGF-β cytokine [16]. Nevertheless, latency-associated protein (LAP) can still form a complex with TGF-β through non-covalent binding, preventing it from binding to the TGF-β receptors known as the small latent complex (SLC) (Figure 1) [17].
Furthermore, SLC can form a large latent complex (LLC) by covalently attaching to the latent TGF-β binding protein 1,3,4 (LTBP1,3,4) through a pair of disulfide bonds in the endoplasmic reticulum. Most cells release LLC outside of the cell as an inactive form, which is attached to ECM by LTBPs binding to fibrillin, fibulins, fibronectin, and other ECM proteins (Figure 1) [18-20]. The interaction between LAP and LTBPs facilitates the correct folding of TGF-β precursor proteins and is crucial for TGF-β activation [21]. When a specific cysteine residue (C33) in the binding site of LAP to LTBPs is substituted with serine in vivo, mice exhibit inflammatory response and tumor characteristics that resemble those observed in TGF-β1-deficient mice [22].
The TGF-β signaling pathway
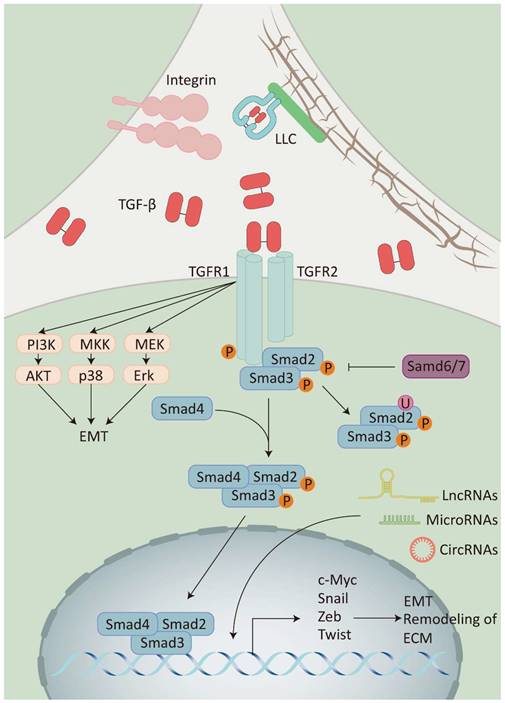
Classical TGF-β signaling enables cell membrane-to-nucleus message transduction through the Smad protein family [23]. At first, the activated TGF-β binds to the TGF-β type II receptor homodimer (TβRII) located on the cell membrane. This interaction leads to the recruitment and activation of the TGF-β type I receptor homodimer (TβRI) and then forms a heterotetramer. Subsequently, the activated TβRI phosphorylates the receptor-regulated Smad (R-Smad), which facilitates binding the R-Smad complex to co-mediator Smad (Co-Smad) to form a trimeric complex. Finally, the trimeric complex is transported and accumulates in the nucleus, acting as a transcription factor which enhances or inhibits the production of TGF-β target genes (Figure 2) [24].
Furthermore, activated TGF-β transmits signals through various signaling pathways, including phosphatidylinositol-3 kinase (PI3K)/AKT pathway, Rho/Rho-kinase pathway, and mitogen-activated protein kinase (MAPK) pathway, which includes extracellular signal-regulated kinase 1/2 (Erk1/2) pathway, p38 mitogen-activated protein kinase (p38 MAPK) pathway, and c-Jun amino-terminal kinase (JNK). These pathways are collectively known as non-classical TGF-β signaling pathway [25, 26]. It has been shown that the diversity of TGF-β signaling depends on the combinatorial utilization of core pathway components such as ligands, receptors, Smads, and transcription factors. This diversity is further modulated by crosstalk with other signaling pathways and the integration of multiple signaling modules beyond TGF-β receptor-activated Smads, collectively regulating the transcription of target genes.
The Synthesis, Secretion, and Localization of TGF-β: Upon entry into the endoplasmic reticulum, the signal peptide of pre-pro-TGF-β (1) is rapidly cleaved to generate pro-TGF-β monomers (2), and then these monomers fold and form a homodimer (3) through disulfide bonds. Subsequently, the homodimer binds to LTBP (4) to form a ternary complex (5) and is translocated to the Golgi. Although TGF-β is released by furin, latency-associated protein (LAP) can still connect with TGF-β via non-covalent binding and form LLC that would be secreted (6) (7). Finally, LLC binds to fibronectin, fibronectin, etc. via LTBP and attaches to the ECM (8).
The PI3K/AKT pathway is initiated by the activation of PI3K, which phosphorylates phosphatidylinositol 4,5-bisphosphate (PIP2) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 serves as a second messenger that recruits and activates various downstream effectors, including the serine/threonine kinase AKT (also known as protein kinase B, PKB) [27]. The crosstalk between the TGF-β signaling pathway and the PI3K/AKT signaling pathway frequently promotes EMT in epithelial malignant cancer. For instance, AKT-mediated Twist1 phosphorylation promotes breast cancer EMT and metastasis by modulating its transcriptional target TGF-β2, leading to enhanced TGF-β receptors signaling, which in turn maintains hyperactive PI3K/AKT signaling [28]. In BxPC-3 pancreatic cancer cells, TGF-β1 promotes TGF-β1-induced EMT by enhancing the phosphorylation of AKT [29]. The MAPK pathway is activated by a range of input signals, such as cytokines, chemokines, growth factors, and stress signals. For instance, TGF-β can stimulate the p38 MAPK signaling pathway, thereby augmenting the migratory and invasive capacities of cancer cells in non-small cell lung cancer [30]. Within the liver cancer cells' tumor microenvironment, the cancer-associated fibroblasts (CAFs)-derived cardiotrophin-like cytokine factor 1 (CLCF1) increases TGF-β secretion in tumor cells. This, in turn, activates Erk 1/2 signaling in CAFs, leading to the production of additional CLCF1, creating a positive feedback loop that accelerates the development of liver cancer cells [31].
The TGF-β signaling pathway: Integrins release TGF-β by binding to LLC on ECM. TGF-β activates receptor-regulated Smad (Smad2/3) by recruiting the receptor(TβRI/II) and then binds to co-mediator Smad (Smad4) to activate the expression of target genes, while inhibitory Smad (Smad6/7) inhibits this process. TGF-β can also activate different signaling pathways to participate in the signaling pathway modification, including the PI3K/AKT pathway, the p38 MAPK signaling pathway, and the Erk1/2 pathway. Additionally, non-coding RNAs (LncRNAs, MicroRNAs, and CircRNAs) can also participate in the TGF-β signaling pathway by regulating the expression of TGF-β transcription factors.
The crosstalk between CAMs and TGF-β signaling pathway
The cell adhesion mechanism is regulated by the adhesion functional unit, CAMs, which play a crucial role in the interaction of cells to ECM and other cells [32]. Characterized by their protein structures, CAMs can be divided into four groups: integrins, selectins, cadherins, and members of the immunoglobulin superfamily (IgSF) [33, 34]. Among them, integrins are mainly related to cell-ECM interaction, while selectins, cadherins, and IgSF members are mainly involved in cell-cell adhesion [35].
Cell adhesion involves the interaction of cells with each other and with the ECM. It is fundamental to the organization of cells into tissues and organs, cell migration, and cellular signal transduction. CAMs function as transmembrane proteins, the extracellular domain stabilizes cells to bind to adjacent cells and ECM, while the intracellular domain interacts with the actin and/or cytoskeleton, allowing signals to be transmitted from outside to cells (outside-in signaling) [36]. The outside-in signaling regulates cell adhesion ability and responds to the microenvironment [37]. Additionally, CAMs can interact with transformed growth factors, transcription factors, and other signal proteins that participate in more extensive signaling pathways to mediate cell fate [38, 39]. In the following discussion, we primarily focus on how integrins, cadherins, and IgSF are involved and regulate the TGF-β signaling pathway (Table 1).
The association of CAMs and TGF-β signaling pathways
| CAMs | Relationship with TGF-β signaling pathways | Ref. |
|---|---|---|
| Integrins | ||
| αvβ6 | Activate TGF-β and closely related to tumor invasion, metastasis, and poor prognosis in several human epithelial-derived malignancies | [50, 51] |
| αvβ8 | Activate TGF-β and regulate the development and maturation of vascular and microglial cells | [64, 65] |
| Cadherins | ||
| E-cadherin | TGF-β downregulates E-cadherin expression and disrupts cell-cell adhesion and epithelial integrity, which promotes the process of EMT | [176] |
| N-cadherin | TGF-β upregulates N-cadherin expression, which promotes the process of EMT | [176] |
| Cadherin-11 | Promote TGF-β expression to induce the differentiation of MSCs into SMCs and regulate the ECM | [71] |
| FAT1 | Promote TGF-β expression and form an immunosuppressive microenvironment, which facilitates the immune evasion of cancer | [72] |
| IgSF | ||
| ICAMs | TGF-β promotes ICAMs expression and leads to neutrophil-mediated damage in lung injury | [74] |
| ALCAM | TGF-β induces the expression and shedding of ALCAM and promotes bone metastasis of prostate cancer | [75] |
| VCAM-1 | TGF-β inhibits VCAM-1 expression in colorectal cancer vasculature, which allows tumor cells to avoid immunosurveillance | [76] |
| Jam-A | TGF-β induces the lysosomal pathway degradation of Jam-A, which allows breast cancer cells to acquire invasive cell properties | [77, 78] |
The crosstalk between TGF-β and integrins
Although latent TGF-β is widespread in many tissues and most cells have TGF-β receptors, TGF-β signaling requires mature TGF-β to bind to the receptors. Thus, a key element in controlling the TGF-β signaling pathway is knowing how to release mature TGF-β from the latent form of TGF-β [40]. In vitro, it is relatively easy to induce conformational changes of latent TGF-β that lead to the release of mature TGF-β (i.e., TGF-β activation). For instance, purified latent TGF-β can be activated by acidic or alkaline pH, thermal denaturation, ionizing radiation, and oxidation of reactive oxygen species (ROS) [41-43]. In contrast, the role of these activation pathways in vivo has not yet been elucidated. Current research shows that the essential activation mechanisms of TGF-β in vivo are primarily triggered by interaction with integrins (Figure 2) [43].
The integrins family are transmembrane heterodimer receptors on the cell surface and consist of 18 α-subunits and 8 β-subunits that can combine to generate 24 distinct integrins, 18 of which have ECM receptors activities. Integrins can be categorized into three primary groups based on ligand specificity: recognize the Arg-Gly-Asp (RGD) peptide receptors, collagen receptors, and laminin receptors [44, 45]. Among these, integrins that bind to RGD have been extensively researched. The αvβ1, αvβ3, αvβ5, αvβ6, and αvβ8 receptors could attach to the LAP. Nevertheless, the activation of TGF-β is mainly mediated by integrins αvβ6 and αvβ8 [16].
The crosstalk between TGF-β and integrin αvβ6
αvβ6 is an integrin that is mainly found in epithelial cells and upregulated following epithelial damage. Integrin αvβ6, unlike other RGD integrins, can recognize not only the RGD sequence, but also a specific LXXL/I motif. This motif forms an amphipathic α-helix that fits into a hydrophobic pocket made up entirely of residues from the β6 subunit. Thus, integrin αvβ6 exhibits a significantly strong affinity to pro-TGF-β [46]. Integrin αvβ6 selectively triggers the activation of TGF-β1 and TGF-β3 while excluding TGF-β2. The integrin αvβ6 is absent in mice lacking the gene responsible for generating the β6 integrin subunit (Itgβ6 -/-) because β6 exclusively binds to αv [47]. There was no significant difference in the overall quantity of TGF-β protein in the lung tissue of Itgβ6 -/- and Itgβ6 +/+ mice. However, in the bleomycin-induced lung fibrosis model, which is known to be heavily dependent on TGF-β activity, Itgβ6 -/- mice did not experience any significant effects following bleomycin treatment. Another study shows that the activation of TGF-β was observed by cell lines co-culture that express integrin αvβ6 and TGF-β receptors [48]. Furthermore, the activation mechanism requires the association of integrin αvβ6 with the actin cytoskeleton through the cytoplasmic tail of the β6 subunit. In a mouse model of tendinopathy created based on excessive tensile strain, the presence of integrin αvβ6 triggers the activation of TGF-β1 in response to mechanical force. This activation leads to the degeneration of tendons, the formation of cartilage, and the growth of blood vessels, all of which contribute to the development of tendinopathy. External stretch or increased intracellular stress in vitro also directly activates TGF-β [49]. These results indicate that the interaction between latent TGF-β and integrin αvβ6 is insufficient for activation. Activation of latent TGF-β requires the additional binding of integrin αvβ6 to the actin cytoskeleton.
The increased expression of integrin αvβ6 is strongly linked to tumor invasion, metastasis, and a poor prognosis in several human epithelial-derived malignancies [50, 51]. In triple-negative breast cancer (TNBC), tumor cells could activate the TGF-β signaling pathway by αvβ6 integrin. This activation results in the increase of SOX4, which is an important regulator of immune evasion pathways [52]. Colorectal cancer (CRC) cells that express integrin αvβ6 activate cancer-associated fibroblasts (CAFs) by TGF-β activation. CAF cells promote the migration of colorectal cancer cells to distant locations by activating the SDF-1/C-X-C chemokine receptor type 4 (CXCR4) axis [53]. The co-expression of integrin αvβ6 with eukaryotic translation initiation factor 4E (eIF4E) or Ets-1 is regarded as a reliable prognostic marker of CRC [54, 55]. Conversely, the concurrent presence of integrin αvβ6 and Matrix Metallopeptidase 9 (MMP-9) is a reliable predictive marker in individuals diagnosed with gastric cancer [56].
The crosstalk between TGF-β and integrin αvβ8
While integrin αvβ6 has been extensively studied, the adhesion and signaling roles of integrin αvβ8 in development, physiology, and disease are still yet to be investigated. It is suggested that integrin αvβ6 and integrin αvβ8 trigger TGF-β through distinct pathways. The activation of TGF-β by integrin αvβ8 occurs in a unique structural domain called β8. This domain lacks a cytoplasmic tail and only has a differentiated cytoplasmic structural domain [57]. This domain does not interact with the actin cytoskeleton and exists in an extended closed structure [58, 59]. The activation of TGF-β by integrin αvβ8 does not depend on actin cytoskeletal interactions and is continuous activation [60]. In addition, integrin αvβ8 could interact with LAP in a local binding way and form a protein complex. This process does not rely on the release and spread of mature TGF-β but rather directly activates the TGF-β signaling pathway in the local area [61], differentiating it from other activation modes which involve the release and dispersion of mature TGF-β [62, 63].
The current studies indicate that developmental cerebral hemorrhage and postnatal microglia maturation arrest and activation occur in both Itgβ8 mutant and TGFβR1-/- mice. These findings align with the characteristics observed in microglial cell lineage-specific conditional deletion of TGFβR2, implying that the central nervous system (CNS) microenvironment relies on the same molecular mechanism, namely the αvβ8-TGF-β signaling, to regulate the development and maturation of vascular and microglial cells [64, 65].
Integrins Participation in the TGF-β Signaling Pathway via Integrin Adhesome
Kindlin-1 is a protein that binds to the tail of integrin and enhances the activation of TGF-β through αvβ6 integrin. Kindler syndrome is caused by mutations or deletions of Kindlin-1 in human and mouse keratinocytes. The disease is characterized by skin blistering, premature skin aging, and an elevated risk of malignancy [66]. Mouse keratinocytes lacking the Kindlin-1 gene experience significant disruptions in β6 integrins-dependent cell adhesion, spreading, assembly of F-actin stress fibers, and release of TGF-β. This leads to an increased risk of tumor development [67]. Kindlin-2, a member of the kindlin family, also plays a role in the activation of TGF-β by integrins. In breast cancer, Kindlin-2 stabilizes the β1 integrin-TβRI complexes and facilitates the TGF-β signaling pathway [68]. Kindlin-2 is also established as a requirement for BC tumor development and progression in transgenic mice [69].
The crosstalk between TGF-β and Cadherins
Cadherins feature cadherin repeat sequences in their extracellular domain which are stabilized by calcium ions. Cadherins can be categorized into classical cadherins, protocadherins, desmosomal cadherins, and atypical cadherins [70]. Several of them have a strong link with the TGF-β signaling pathway. Classic Cadherins, such as E-Cadherin and N-Cadherin, have a significant function in TGF-β-mediated EMT, as will be discussed later. Cadherin-11 is also a classical adhesion molecule expressed by mesenchymal stem cells (MSCs) and is necessary to induce the differentiation of MSCs. Cadherin-11 can regulate the expression of TGF-β1 and induce the differentiation of MSCs into contractile smooth muscle cells (SMCs) through TGF-β receptor II pathway. At the same time, Cadherin-11 can influence MSCs differentiation indirectly by regulating the ECM via the TGF-β1 pathway [71]. Furthermore, in glioma tumors, FAT atypical cadherin 1 (FAT1) promotes the translation of TGF-β1/2 by inhibiting the level of miR-663a, forming an immunosuppressive microenvironment, which in turn facilitates the immune evasion of GBM [72].
The crosstalk between TGF-β and IgSF
The IgSF is one of the largest and most diverse protein families [73]. All members of the IgSF typically contain one or more immunoglobulin or immunoglobulin-like domains. Many IgSF proteins function as cell adhesion molecules that are regulated by TGF-β. For example, TGF-β induces lung injury by the overexpression of intercellular adhesion molecules (ICAMs) on endothelial cells, leading to neutrophil-mediated damage [74]. TGF-β can also induce the expression and shedding of activated leukocyte cell adhesion molecule (ALCAM) and thereby promote bone metastasis of prostate cancer [75]. In colorectal cancer, the high expression of TGF-β may inhibit the expression of vascular cell adhesion molecule 1 (VCAM-1) in tumor vasculature, which allows tumor cells to avoid immunosurveillance by circulating lymphocytes [76]. TGF-β can induce the lysosomal pathway degradation of Junctional adhesion molecule A (Jam-A), a tight junction component facilitating epithelial cell-cell adhesion. This allows breast cancer cells to acquire invasive cell properties during the EMT process [77, 78].
The crosstalk between ECM and TGF-β signaling pathway
The ECM is a complex network that provides structural and biochemical support to cells within tissues and organs [79]. It consists of various structural proteins, polysaccharides, adhesion proteins, and other molecules secreted by cells and organized into a three-dimensional network surrounding cells [80, 81]. TGF-β is mainly secreted and stored in the ECM as a latent complex [82]. TGF-β activation in normal cells causes the transformation of myofibroblasts into fibroblasts, which promotes pro-healing homeostasis [83]. TGF-β can regulate the remodeling of ECM components that facilitate tumor migration and invasion in the majority of tumor tissues. This remodeling is frequently accompanied by alterations in the expression of base proteinases (MMPs) [84, 85]. By inhibiting Grhl2 expression, TGF-β can promote MMP-2, MMP-7, and MMP-9 expression in gastric cancer, thereby facilitating the invasion and migration of gastric malignant cells [86]. TGF-β upregulates MMP-2 and MMP-9 expression in p38 MAPK signaling pathway that leads to the invasive and migratory phenotypes in MCF10A human breast epithelial cells [87]. Specifically, TGF-β promotes the expression of MMP-2 by inhibiting the transcription factor ATF2 [88]. Additionally, it has been demonstrated that MT1-MMP can cleave LAP and is essential for αvβ8-mediated TGF-β activation [89]. In human prostate and breast cancer cells, MT1-MMP promotes cancer cell EMT and migration by activating TGF-β [90].
Regulation of TGF-β and EMT
The epithelial-mesenchymal transition is a crucial process that has a significant impact on both physiological and pathological events, including embryogenesis, tissue damage repair, and cancer progression [91]. It is well known acknowledged that EMT is closely related to cell adhesion mechanism: alterations in various cell adhesion molecules, such as claudins and E-cadherin [92], induce the detachment of epithelial cells from each other and the underlying basement membrane [93]. During this process, epithelial cells gradually acquire mesenchymal phenotypes (such as reduced intercellular adhesion and increased motility) in response to the microenvironment [94].
The initiation and development of EMT are affected by several signaling pathways, among which the TGF-β signaling pathway plays an important role [95]. Under physiological conditions, TGF-β is involved in mesenchymal phenotypic transitions in epithelial and endothelial progenitor cells during development [23], and in wound healing, it promotes macrophage-mediated inflammation and tissue debridement [96]. However, when the TGF-β signaling pathway is disturbed, it can promote tissue fibrosis and facilitate cancer cells' migration, invasion, and metastasis through EMT [3, 97, 98].
Smads signaling in TGF-β-mediated EMT
TGF-β induces EMT by promoting the expression of transcriptional repressors of E-cadherin, such as Snail, Zeb, and Twist [99]. As a prominent effector protein of the TGF-β signaling pathway, Smad is a central mediator of EMT in many contexts. In keratinocyte-specific Smad2-deficient mice, a decrease in E-cadherin and the activation of Snail were observed in the epidermis. Furthermore, Smad2 deficiency resulted in a notable increase in Smad4 binding to the Snail promoter. This led to an acceleration of chemically induced skin tumor formation and malignant progression [100]. In primary renal tubular epithelial cells from Smad3-deficient mice, exogenous TGF-β1 treatment was unable to alter epithelial cell phenotypic characteristics, which resulted in Smad3-deficient mice being protected from tubulointerstitial fibrosis after unilateral ureteral obstruction (UUO) [101]. Consistent with lung carcinoma cells [102], Smad3 deletion similarly inhibited the process of EMT. However, in gastric cancer cells, Smad3-mediated TGF-β signaling induces carcinoembryonic antigen (CEA)-related cell adhesion molecule 6 (CEACAM6) expression and promotes EMT in gastric cancer cells [103]. In bronchial epithelial cells chronically exposed to toluene diisocyanate (TDI), TGF-β1 secretion was increased, and activation of the TGF-β signaling pathway through massive activation of Smad2/3. This led to a decrease in cell adhesion molecules and facilitated the process of EMT and the cancerous transformation of bronchial epithelial cells [104]. Knockdown of Smad4 effectively inhibits TGF-β-induced EMT in normal mammary cells and strongly suppresses bone metastasis of breast cancer cells in nude mice [105]. The Smad7-mediated negative feedback loop of TGF-β signaling was disturbed in some breast and lung cancer cell lines, while TGF-β-induced EMT and cancer cell invasion were reversed when Smad7 transcription was upregulated [106].
c-Myc in TGF-β-mediated EMT
Myc is a group of proto-oncogenes that encode transcription factors, specifically c-Myc (MYC), I-Myc (MYCL), and n-Myc (MYCN) [107]. Dysregulation of c-Myc activity is a common occurrence in many malignancies, resulting in the development of tumors and the perpetuation of the disease [108, 109]. C-Myc is a crucial downstream target gene of the TGF-β signaling pathway and is widely recognized for its significant involvement in TGF-β-mediated EMT [110]. For example, in pancreatic cancer cells, TGF-β regulates the expression of RAP2 via the transcription factor c-Myc. Promotes N-cadherin, and vimentin, and decreases E-cadherin in pancreatic cancer [111]. Furthermore, quercetin also inhibits EMT by blocking the TGF-β signaling pathway, which reduces the migration and invasion of pancreatic cancer cells. Quercetin decreases TGF-β expression, inhibiting the phosphorylation and nuclear translocation of Smad2 and Smad3, and reducing the expression of Snail1, Zeb2, and c-Myc [112]. In esophageal squamous cell carcinoma (ESCC), Chaperone-containing TCP1 subunit 6A (CCT6A) stimulates the TGF-β/Smad/c-Myc pathway, thereby promoting ESCC EMT and cell invasiveness [113].
Transcription factors in TGF-β-mediated EMT
In addition to the important role of Smads signaling for EMT, many transcription factors have been widely reported to be associated with EMT. This regulation mainly involves three transcription factor families: Snail, Zeb, and bHLH family [93, 114]. They regulate each other's expression, cooperate functionally on target genes, and play a central role in development, fibrosis, and cancer [115]. Among them, Snail1/2, Zeb1/2, and Twist1/2 are considered to be implicated in the EMT process as master drivers [116].
Snail transcription factors
The Snail family are zinc finger proteins, including SNAIL (Snail1), SLUG (Snail2) and SMUC (Snail3). Snail promotes the EMT process by directly suppressing E-cadherin transcription through binding to the promoter sequence of the E-cadherin gene, E-box (5'-CACCTG) [117]. In epithelial cells, the mRNA levels of E-cadherin and Snail are significantly negatively correlated [117]. When the TGF-β signaling pathway is over-activated, Smad2 and Smad3 bound and activate Snail1/Snail2 [118, 119], which suppresses E-cadherin, and Snail1/Snail2 continues to bind to the Snail1/Snail2 gene respectively and further suppresses E-cadherin expression [120]. Additionally, Snail1/Snail2 can suppress the expression of other epithelial markers associated with epithelial phenotypes, such as Claudin, Occludin, and Cytokeratin [121, 122], while simultaneously increasing the expression of mesenchymal markers associated with mesenchymal phenotypes, such as fibronectin and vimentin [123]. Studies have demonstrated that Snail family members have high expression in various cancer types, such as lung, breast, and colorectal malignancies [124-126]. Increased levels of Snail impair cancer cell adhesion and facilitate tumor invasion and metastasis.
Zeb transcription factors
Zeb1 and Zeb2 belong to the human Zeb family of zinc finger proteins, which inhibit E-cadherin transcription by binding to regulatory gene sequences on the E-box [127]. In addition, Zeb1 could interact with DNA methyltransferase 1 (DNMT1) through the Smad-binding domain and promote high methylation of the E-cadherin promoter region, which indirectly inhibits E-cadherin expression at the epigenetic level [128]. Zeb1 and Zeb2 play a role in the TGF-β signaling pathway by interacting with Smad. However, they have contrasting regulatory functions in osteoblast development, implying that maintaining a balance between Zeb1 and Zeb2 may help regulate the TGF-β signaling pathway [129]. Nevertheless, TGF-β triggers EMT in the majority of cancer cells and epithelial cells by activating Zeb1 and Zeb2 [130]. As an illustration, Zeb1 enhances the invasion of colorectal cancer by suppressing basement membrane (BM) gene expression [131].
Twist transcription factors
The bHLH family is a large family of transcription factors that control a wide range of developmental and pathological processes. Twist1 and Twist2 are highly conserved members of the Twist subfamily of bHLH transcription factors [132]. Twist1 inhibits E-cadherin and promotes N-cadherin levels through E-box cis-acting elements, respectively [133], and also promotes EMT through up-regulation of vimentin [134]. Twist1 directly activates Galectin-3 transcription and induces macrophage polarization towards the M2 phenotype, leading to the promotion of renal fibrosis [135]. In prostate cancer, Twist1 regulates the transcriptional activity of CLU by binding to the distal promoter region of induces clusterin (CLU), which promotes TGF-β-mediated EMT and distant metastasis of the tumor [136]. In breast cancer, TGF-β-induced EMT in HER2 cells is coordinated with the activation of the Wnt/β-catenin pathway. Under the action of TGF-β, Twist can directly promote the EMT process and also bind to the promoter of Wnt3 to activate the Wnt/β-catenin pathway and participate in Wnt/β-catenin pathway-mediated EMT [137].
Non-coding RNAs in TGF-β-mediated EMT
In addition to the above-mentioned transcription factors that can regulate TGF-β-mediated EMT, non-coding RNAs (ncRNAs) are also involved in regulating the progression of EMT. Although ncRNAs do not directly encode proteins, they can regulate gene transcription and translation by interacting with specific messenger RNAs (mRNAs) [138, 139]. Recent research has demonstrated that ncRNAs have a crucial function in tumor migration, invasion, and metastasis [140, 141]. Specifically, some microRNAs, lncRNAs, and circRNAs have been identified as effectors of TGF-β-mediated EMT (Table 2) [142].
MicroRNAs
MicroRNAs (miRNAs) are ~22 nt small noncoding RNAs that can bind to 3′ UTR of their target mRNAs to suppress expression [143]. It has been shown that miRNAs control the expression of EMT transcription factors, which affects TGF-β-mediated EMT. For example, miR-22, miR-30a, and miR-199a can directly target and inhibit Snail expression [144-146], while miR-141 targets NRP-1 and indirectly inhibits Snail expression [147], both of which inhibit the TGF-β-mediated EMT phenotype. In addition, miR-34a/b/c and miR-203 inhibited Snail expression, while Snail could bind miR-34a/b/c and miR-203 promoters to suppress its transcription, thus forming a dual negative feedback mechanism. However, frequent inactivation of miR-34a/b/c and miR-203 might disrupt the balance of these reciprocal regulations and promote TGF-β-mediated EMT [148, 149]. Research shows that miRNA-200 significantly inhibits Zeb1 expression, and the enforced expression of the miR-200 alone was sufficient to prevent TGF-β-induced EMT. In contrast, ectopic expression of these microRNAs in mesenchymal cells initiates mesenchymal-to-epithelial transition (MET). Furthermore, in invasive breast cancer, lack of miR-200 expression is positively correlated with E-cadherin deficiency [150]. In addition, miR-300 negatively regulates EMT by directly targeting Twist, inhibiting epithelial cancer cell invasion and metastasis [151]. And miR-15a/16 inhibits prostate cancer cell invasion by targeting endogenous Smad3 and Acvr2a proteins to suppress Snail and Twist expression.
List of ncRNAs and their function in TGF-β-mediated EMT
| Non-coding RNA | Targets gene | Cancer type | Role in TGF-β-mediated EMT | Regulation pathway | Ref. |
|---|---|---|---|---|---|
| MiR-15a/16 | Smad3, ACVR2A | Prostate cancer | Suppressor | MiR-15a/16 targets and inhibits Smad3 and ACVR2A expression | [177] |
| MiR-22 | Snail | Lung cancer | Suppressor | MiR-22 targets and inhibits the expression of Snail | [144] |
| MiR-23a | E-cadherin | Lung cancer | Promoter | MiR-23a inhibited E-cadherin expression and stimulated TGF-β-induced EMT | [154] |
| MiR-30a | Snail | Peritoneal fibrosis, liver fibrosis | Suppressor | MiR-30a targets and inhibits the expression of Snail | [145, 146] |
| MiR-34 a/b/c | Snail | Colorectal cancer | Suppressor | Snail and miR-34 form a dual negative feedback loop, inhibiting each other | [148] |
| MiR-141 | Snail | Pancreatic cancer | Suppressor | MiR-141 targets NRP-1 to inhibit Snail expression | [147] |
| MiR-145 and miR-497 | MTDH | Non-small cell lung cancer | Suppressor | MiR-145 and miR-497 attenuated MTDH expression by directly binding 3′-UTR of MTDH mRNA and exerting the tumor-suppression role | [153] |
| MiR-194 | N-cadherin | Gastric cancer | Suppressor | MiR-194 inhibits N-cadherin expression | [156] |
| MiR-199a | Snail1, E-cadherin | Lung cancer | Suppressor | MiR-199a targeted and inhibited the expression of Snail and E-cadherin | [178] |
| MiR-199b-5p | N-cadherin | Hepatocellular carcinoma | Suppressor | MiR-23a inhibited the migration and invasion of HCC cells and suppressed tumor metastasis of xenografts in nude mice by targeting binding to N-cadherin | [155] |
| MiR-200 | Zeb1, Zeb2 | Breast cancer | Suppressor | MiR-200 targeted and inhibited the expression of Zeb | [150] |
| MiR-203 | Snail2 | Breast cancer | Suppressor | SNAI2 and miR-203 form a dual negative feedback loop, inhibiting each other's expression and thus controlling EMT | [149] |
| MiR-300 | Twist | Breast cancer | Suppressor | MiR-300 directly targets Twist | [151] |
| LncRNA LETS1 | TβRI | Breast cancer, lung cancer | Promoter | LncRNA LETS1 enhances TGF-β-Smad signaling by stabilizing TβRI on the cell surface | [160] |
| LncRNA Smyca | Smad3, Smad4 | Breast cancer | Promoter | LncRNA Smyca acts as a scaffold, providing an additional binding surface for enhanced binding of Smad3 to Smad4 | [161] |
| LncRNA LINC00941 | Smad4 | Colorectal cancer | Promoter | LncRNA LINC00941 inhibits Smad4 protein degradation by directly binding to Smad4 protein and inhibiting Smad4 protein degradation | [162] |
| LncRNA TUG1 | Twist1 | Colorectal cancer | Promoter | LncRNA TUG1 promotes Twist1 expression | [163] |
| LncRNA LITATS1 | TβRI, SMURF2 | Breast cancer, non-small cell lung cancer | Suppressor | LncRNA LITATS1 enhances polyubiquitination and proteasomal degradation of TGF-β type I receptor (TβRI) | [164] |
| LncRNA LINP1 | — | Lung Cancer | Suppressor | Smad4 binds and inhibits lncRNA LINP1 expression and induces EMT and cell invasion in lung cancer cells | [165] |
| LncRNA ANCR | RUNX2 | Breast cancer | Suppressor | LncRNA ANCR inhibits TGF-β1-induced EMT and breast cancer cell migration and metastasis of breast cancer cells by decreasing the expression of RUNX2 | [166] |
| CircITGB6 | PDPN | Liver cancer | Promoter | CircITGB6 promotes TGF-β-induced EMT and tumor metastasis through enhancing IGF2BP3-mediated PDPN mRNA stability | [170] |
| CircPTK2 | TIF1γ | Non-small cell lung cancer | Suppressor | CircPTK2 overexpression augmented TIF1γ expression, inhibited TGF-β-induced EMT and NSCLC cell invasion | [171] |
| CircVANGL1 | MiR-150-5p | Melanoma | Promoter | CircVANGL1 directly binds to miR‐150‐5p and promotes EMT of TGF‐β‐treated melanoma cells | [172] |
| Circ-DOCK5 | MiR-627-3p | Esophageal squamous cell carcinoma | Suppressor | Circ-DOCK5 increased the stability of miR-627-3p resulting in downregulation of ZEB1 and suppression of TGF-β-induced EMT | [173] |
MiRNAs can also regulate TGF-β-mediated EMT by modulating the expression of epithelial or mesenchymal proteins [152]. For instance, miR-145 and miR-497 promote E-cadherin and inhibit vimentin expression by inhibiting MTDH expression, suppressing the process of TGF-β-induced EMT in non-small cell lung cancer [153]. In A549 lung cancer cells, overexpression of miR-23a inhibited E-cadherin expression and stimulated TGF-β-induced EMT [154]. In addition, miR-199b-5p bound to the 3'-UTR of N-cadherin mRNA, which reduced N-cadherin expression in HCC cells and attenuated TGF-β1-induced EMT progression, thereby inhibiting metastasis and invasion of hepatocellular carcinoma cells [155]. Another study showed that miR-194 could directly interact with the 3'-UTR of N-cadherin mRNA and reduce its expression in advanced gastric cancer cells [156].
LncRNAs
Long non-coding RNAs (lncRNAs) is a lengthy sequence of more than 200 nucleotides that controls the expression of specific genes by interacting with RNA, DNA, and proteins to form complexes. It is essential in various biological phenomena, including cell growth, differentiation, and metastasis [157, 158]. Furthermore, lncRNAs are abnormally expressed in nearly all types of cancer and are linked to oncogenesis, metastasis, and tumor staging [159]. And many reports have confirmed that lncRNAs are closely associated with TGF-β-mediated tumor EMT.
On the one hand, overexpression of lncRNAs in a variety of tumors induces EMT and promotes tumor metastasis. For example, the lncRNA LETS1 improves TGF-β-Smad signaling by stabilizing cell-surface TGF-β type I receptor (TβRI) to promote the migration of breast and lung cancer cells [160]. LncRNA Smyca functions as a scaffold and facilitates the binding of Smad3 and Smad4, which promotes TGF-β/Smad signaling-mediated EMT and lung cancer cell migration [161]. In a mouse colorectal cancer model, lncRNA LINC00941 and lncRNA Tug1 promote the migratory and invasive ability of colorectal cancer, thereby expediting lung metastasis. Mechanistically, LINC00941 directly binds to Smad4 protein and inhibits its degradation, thereby activating EMT [162], while the knockdown of Tug1 reduces Twist1 expression in CRC cells [163].
On the other hand, overexpression of certain lncRNAs in tumors inhibits TGF-β-induced EMT and tumor metastasis. For example, lncRNA LITATS1 inhibits EMT in breast cancer and non-small cell lung cancer cells by promoting polyubiquitination and proteasomal degradation of TβRI [164]; lncRNA LINP1 inhibits TGF-β1/Smad4-induced EMT and cell invasion in lung cancer cells [165]. In addition, lncRNA ANCR inhibited TGF-β1-induced EMT and breast cancer cell migration and metastasis by suppressing Runx2 [166].
CircRNAs
Circular RNAs (circRNAs) are noncoding RNAs with tissue-specific and cell-specific expression patterns that form a covalently closed loop between the 5′ and 3′ ends [167]. Aberrant expression of many circRNAs is observed in a wide range of cancers, suggesting that they play a crucial role in tumorigenesis and progression [168]. This role may involve many different molecular mechanisms. The most common function of circRNAs is the ability to act as miRNA sponges, binding directly to the corresponding miRNAs and thereby regulating the expression of target genes. In addition, circRNAs are involved in transcriptional regulation, splicing, or even translating proteins [169]. In recent years, many reports have shown that circRNAs are closely associated with TGF-β-mediated tumor EMT. For example, circITGB6 enhances IGF2BP3-mediated PDPN mRNA stability, which promotes TGF-β-induced EMT process and tumor metastasis [170]. In addition, circPTK2 overexpression augmented TIF1γ expression and inhibited TGF-β-induced EMT and NSCLC cell invasion [171]. CircRNAs not only function by regulating gene transcription and translation levels, but also modulate miRNA activity. In melanoma, circVANGL1 directly binds to miR-150-5p and promotes melanoma cell proliferation and invasion [172]. In esophageal squamous cell carcinoma, circ-DOCK5 increased the stability of miR-627-3p, resulting in downregulation of Zeb1 and suppression of TGF-β-induced EMT [173].
Conclusion
This review provides an overview of the crosstalk between cell adhesion mechanisms and TGF-β signaling pathway. Cell adhesion mechanisms not only facilitate the formation of tissue structures by promoting cell-cell or cell-ECM adhesions, but also participate in numerous non-adhesion signaling pathways. For example, CAMs can activate the TGF-β signaling pathway by binding to latent TGF-β and releasing mature TGF-β factors from ECM. This activation results in the transcription of downstream target genes, including Smad, c-Myc, Snail, Zeb, Twist, and non-coding RNAs. As a result, it in turn regulates the expression of CAMs or ECM remodeling, thus changing the cell adhesion ability. Currently, numerous studies have demonstrated a strong correlation between the TGF-β signaling pathway and the cell adhesion signaling pathway in cancer proliferation, migration, invasion, and immune evasion. Consequently, a range of drugs have been developed to specifically target crucial proteins involved in the TGF-β signaling pathway and cell adhesion signaling network. Various medications block the TGF-β signaling pathway by directly or indirectly inhibiting the production or function of TGF-β or TGF-β receptors. This includes small molecule inhibitors of TGF-β receptor kinase activity, TGF-β-directed antibodies, TGF-β ligand traps, antisense oligonucleotides (ASOs) targeting the TGF-β pathway, and vaccine-based approaches to modulate TGF-β signaling (these are well described in reviews [97, 174]). Furthermore, there have been drugs specifically designed to target integrins [175]. However, the development of drug research is not smooth sailing. The main shortcoming of antagonists targeting the TGF-β signaling pathway is the multifaceted function of TGF-β within the human body. In other words, TGF-β is necessary for healthy cells to control numerous essential physiological functions. Furthermore, there is a limited understanding of the dual opposite effects of TGF-β, which acts both as a tumor suppressor and a tumor promoter. Therefore, it is crucial to maintain rigorous control in clinical applications. Drugs that target integrins have a limited market due to the specificity of their target class, their pharmacokinetics, and the absence of clinical pharmacodynamic biomarkers. Hence, it is imperative to obtain further crosstalk between the TGF-β signaling pathway and the cell-adhesive signaling pathways. Hence, it is imperative to obtain further crosstalk between the TGF-β signaling pathway and the cell-adhesive signaling pathways. This will find novel regulatory factors that can be targeted for cancer treatment in the foreseeable future.
Acknowledgements
Funding
This study was supported by grants from Guangdong Basic and Applied Basic Research Foundation (2021A1515110494, 2021B1515140066, 2023A1515140057), Dongguan Social Science and Technology Development Project (20211800904532), Discipline Construction Project of Guangdong Medical University (4SG24013G), Youth Research Projects of Guangdong Medical University (GDMUD2022003, GDMUD2022007), Talent Development Foundation of The First Dongguan Affiliated Hospital of Guangdong Medical University (PF100-2-05).
Author contributions
All authors contributed significantly to the drafting and editing of this manuscript. JL, RC, and XQ conceived the manuscript idea and wrote the manuscript. XQ, SM, JZ and YL revised the manuscript. JL, RD, and BL created the manuscript tables and figures.
Competing Interests
The authors have declared that no competing interest exists.
References
1. de Larco JE, Todaro GJ. Growth factors from murine sarcoma virus-transformed cells. Proc Natl Acad Sci U S A. 1978;75:4001-5
2. Roberts AB, Anzano MA, Lamb LC, Smith JM, Frolik CA, Marquardt H. et al. Isolation from murine sarcoma cells of novel transforming growth factors potentiated by EGF. Nature. 1982;295:417-9
3. David CJ, Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nature Reviews Molecular Cell Biology. 2018;19:419-35
4. Janiszewska M, Primi MC, Izard T. Cell adhesion in cancer: Beyond the migration of single cells. The Journal of Biological Chemistry. 2020;295:2495-505
5. Wu J-S, Jiang J, Chen B-J, Wang K, Tang Y-L, Liang X-H. Plasticity of cancer cell invasion: Patterns and mechanisms. Transl Oncol. 2021;14:100899
6. Munger JS, Sheppard D. Cross talk among TGF-β signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb Perspect Biol. 2011;3:a005017
7. Nolte M, Margadant C. Controlling Immunity and Inflammation through Integrin-Dependent Regulation of TGF-β. Trends Cell Biol. 2020;30:49-59
8. Vasiukov G, Menshikh A, Owens P, Novitskaya T, Hurley P, Blackwell T. et al. Adenosine/TGFβ axis in regulation of mammary fibroblast functions. PLoS One. 2021;16:e0252424
9. McEntee CP, Gunaltay S, Travis MA. Regulation of barrier immunity and homeostasis by integrin-mediated transforming growth factor β activation. Immunology. 2020;160:139-48
10. Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345-57
11. Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol. 2011 3
12. Hynes RO. The extracellular matrix: not just pretty fibrils. Science (New York, NY). 2009;326:1216-9
13. Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M. et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693-9
14. Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP. et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development (Cambridge, England). 1997;124:2659-70
15. Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N. et al. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415-21
16. Robertson IB, Rifkin DB. Regulation of the Bioavailability of TGF-β and TGF-β-Related Proteins. Cold Spring Harb Perspect Biol. 2016 8
17. Gray AM, Mason AJ. Requirement for activin A and transforming growth factor-beta 1 pro-regions in homodimer assembly. Science (New York, NY). 1990;247:1328-30
18. Robertson IB, Horiguchi M, Zilberberg L, Dabovic B, Hadjiolova K, Rifkin DB. Latent TGF-β-binding proteins. Matrix Biology: Journal of the International Society For Matrix Biology. 2015;47:44-53
19. Tzavlaki K, Moustakas A. TGF-β Signaling. Biomolecules. 2020 10
20. Li Y, Fan W, Link F, Wang S, Dooley S. Transforming growth factor β latency: A mechanism of cytokine storage and signalling regulation in liver homeostasis and disease. JHEP Reports: Innovation In Hepatology. 2022;4:100397
21. Brunner AM, Marquardt H, Malacko AR, Lioubin MN, Purchio AF. Site-directed mutagenesis of cysteine residues in the pro region of the transforming growth factor beta 1 precursor. Expression and characterization of mutant proteins. The Journal of Biological Chemistry. 1989;264:13660-4
22. Yoshinaga K, Obata H, Jurukovski V, Mazzieri R, Chen Y, Zilberberg L. et al. Perturbation of transforming growth factor (TGF)-beta1 association with latent TGF-beta binding protein yields inflammation and tumors. Proc Natl Acad Sci U S A. 2008;105:18758-63
23. Massagué J, Sheppard D. TGF-β signaling in health and disease. Cell. 2023;186:4007-37
24. Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577-84
25. Zhang YE. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb Perspect Biol. 2017 9
26. Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128-39
27. Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71-88
28. Xue G, Restuccia DF, Lan Q, Hynx D, Dirnhofer S, Hess D. et al. Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-β signaling axes. Cancer Discov. 2012;2:248-59
29. Yoshida J, Ishikawa T, Endo Y, Matsumura S, Ota T, Mizushima K. et al. Metformin inhibits TGF-β1-induced epithelial-mesenchymal transition and liver metastasis of pancreatic cancer cells. Oncol Rep. 2020;44:371-81
30. Zhu N, Zhang XJ, Zou H, Zhang YY, Xia JW, Zhang P. et al. PTPL1 suppresses lung cancer cell migration via inhibiting TGF-β1-induced activation of p38 MAPK and Smad 2/3 pathways and EMT. Acta Pharmacol Sin. 2021;42:1280-7
31. Song M, He J, Pan QZ, Yang J, Zhao J, Zhang YJ. et al. Cancer-Associated Fibroblast-Mediated Cellular Crosstalk Supports Hepatocellular Carcinoma Progression. Hepatology. 2021;73:1717-35
32. Cavallaro U, Dejana E. Adhesion molecule signalling: not always a sticky business. Nature Reviews Molecular Cell Biology. 2011;12:189-97
33. Aricescu AR, Jones EY. Immunoglobulin superfamily cell adhesion molecules: zippers and signals. Curr Opin Cell Biol. 2007;19:543-50
34. Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. The Journal of Cell Biology. 2011;192:907-17
35. Harjunpää H, Llort Asens M, Guenther C, Fagerholm SC. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front Immunol. 2019;10:1078
36. Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nature Reviews Cancer. 2018;18:533-48
37. Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nature Reviews Molecular Cell Biology. 2009;10:445-57
38. Hintermann E, Christen U. The Many Roles of Cell Adhesion Molecules in Hepatic Fibrosis. Cells. 2019 8
39. Dejana E. Endothelial cell-cell junctions: happy together. Nature Reviews Molecular Cell Biology. 2004;5:261-70
40. Batlle E, Massagué J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity. 2019;50:924-40
41. Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO. et al. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93:1159-70
42. Munger JS, Harpel JG, Gleizes PE, Mazzieri R, Nunes I, Rifkin DB. Latent transforming growth factor-beta: structural features and mechanisms of activation. Kidney Int. 1997;51:1376-82
43. Hinck AP, Mueller TD, Springer TA. Structural Biology and Evolution of the TGF-β Family. Cold Spring Harb Perspect Biol. 2016 8
44. Bachmann M, Kukkurainen S, Hytönen VP, Wehrle-Haller B. Cell Adhesion by Integrins. Physiol Rev. 2019;99:1655-99
45. Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901-3
46. Dong X, Hudson NE, Lu C, Springer TA. Structural determinants of integrin β-subunit specificity for latent TGF-β. Nat Struct Mol Biol. 2014;21:1091-6
47. Bandyopadhyay A, Raghavan S. Defining the role of integrin alphavbeta6 in cancer. Curr Drug Targets. 2009;10:645-52
48. Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J. et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319-28
49. Wang X, Liu S, Yu T, An S, Deng R, Tan X. et al. Inhibition of Integrin αvβ6 Activation of TGF-β Attenuates Tendinopathy. Advanced Science (Weinheim, Baden-Wurttemberg, Germany). 2022;9:e2104469
50. Niu J, Li Z. The roles of integrin αvβ6 in cancer. Cancer Lett. 2017;403:128-37
51. Brzozowska E, Deshmukh S. Integrin Alpha v Beta 6 (αvβ6) and Its Implications in Cancer Treatment. Int J Mol Sci. 2022 23
52. Bagati A, Kumar S, Jiang P, Pyrdol J, Zou AE, Godicelj A. et al. Integrin αvβ6-TGFβ-SOX4 Pathway Drives Immune Evasion in Triple-Negative Breast Cancer. Cancer Cell. 2021 39
53. Peng C, Zou X, Xia W, Gao H, Li Z, Liu N. et al. Integrin αvβ6 plays a bi-directional regulation role between colon cancer cells and cancer-associated fibroblasts. Biosci Rep. 2018 38
54. Peng C, Gao H, Niu Z, Wang B, Tan Z, Niu W. et al. Integrin αvβ6 and transcriptional factor Ets-1 act as prognostic indicators in colorectal cancer. Cell & Bioscience. 2014;4:53
55. Niu Z, Wang J, Muhammad S, Niu W, Liu E, Peng C. et al. Protein expression of eIF4E and integrin αvβ6 in colon cancer can predict clinical significance, reveal their correlation and imply possible mechanism of interaction. Cell & Bioscience. 2014;4:23
56. Lian P-L, Liu Z, Yang G-Y, Zhao R, Zhang Z-Y, Chen Y-G. et al. Integrin αvβ6 and matrix metalloproteinase 9 correlate with survival in gastric cancer. World J Gastroenterol. 2016;22:3852-9
57. Dong X, Zhao B, Iacob RE, Zhu J, Koksal AC, Lu C. et al. Force interacts with macromolecular structure in activation of TGF-β. Nature. 2017;542:55-9
58. Cormier A, Campbell MG, Ito S, Wu S, Lou J, Marks J. et al. Cryo-EM structure of the αvβ8 integrin reveals a mechanism for stabilizing integrin extension. Nat Struct Mol Biol. 2018;25:698-704
59. Minagawa S, Lou J, Seed RI, Cormier A, Wu S, Cheng Y. et al. Selective targeting of TGF-β activation to treat fibroinflammatory airway disease. Sci Transl Med. 2014;6:241ra79
60. Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H. et al. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. The Journal of Cell Biology. 2002;157:493-507
61. Campbell MG, Cormier A, Ito S, Seed RI, Bondesson AJ, Lou J. et al. Cryo-EM Reveals Integrin-Mediated TGF-β Activation without Release from Latent TGF-β. Cell. 2020 180
62. Moses HL, Roberts AB, Derynck R. The Discovery and Early Days of TGF-β: A Historical Perspective. Cold Spring Harb Perspect Biol. 2016 8
63. Massagué J. TGFβ signalling in context. Nature Reviews Molecular Cell Biology. 2012;13:616-30
64. Arnold TD, Niaudet C, Pang M-F, Siegenthaler J, Gaengel K, Jung B. et al. Excessive vascular sprouting underlies cerebral hemorrhage in mice lacking αVβ8-TGFβ signaling in the brain. Development (Cambridge, England). 2014;141:4489-99
65. Arnold TD, Lizama CO, Cautivo KM, Santander N, Lin L, Qiu H. et al. Impaired αVβ8 and TGFβ signaling lead to microglial dysmaturation and neuromotor dysfunction. The Journal of Experimental Medicine. 2019;216:900-15
66. Rognoni E, Widmaier M, Jakobson M, Ruppert R, Ussar S, Katsougkri D. et al. Kindlin-1 controls Wnt and TGF-β availability to regulate cutaneous stem cell proliferation. Nat Med. 2014;20:350-9
67. Sin S, Bonin F, Petit V, Meseure D, Lallemand F, Bièche I. et al. Role of the focal adhesion protein kindlin-1 in breast cancer growth and lung metastasis. J Natl Cancer Inst. 2011;103:1323-37
68. Yousafzai NA, El Khalki L, Wang W, Szpendyk J, Sossey-Alaoui K. Kindlin-2 Regulates the Oncogenic Activities of Integrins and TGF-β In Triple Negative Breast Cancer Progression and Metastasis. Res Sq. 2024
69. Li B, Chi X, Song J, Tang Y, Du J, He X. et al. Integrin-interacting protein Kindlin-2 induces mammary tumors in transgenic mice. Sci China Life Sci. 2019;62:225-34
70. Patel SD, Chen CP, Bahna F, Honig B, Shapiro L. Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr Opin Struct Biol. 2003;13:690-8
71. Passanha FR, Geuens T, LaPointe VLS. Cadherin-11 Influences Differentiation in Human Mesenchymal Stem Cells by Regulating the Extracellular Matrix Via the TGFβ1 Pathway. Stem Cells. 2022;40:669-77
72. Irshad K, Srivastava C, Malik N, Arora M, Gupta Y, Goswami S. et al. Upregulation of Atypical Cadherin FAT1 Promotes an Immunosuppressive Tumor Microenvironment via TGF-β. Front Immunol. 2022;13:813888
73. Wai Wong C, Dye DE, Coombe DR. The role of immunoglobulin superfamily cell adhesion molecules in cancer metastasis. Int J Cell Biol. 2012;2012:340296
74. Suzuki Y, Tanigaki T, Heimer D, Wang W, Ross WG, Murphy GA. et al. TGF-beta 1 causes increased endothelial ICAM-1 expression and lung injury. J Appl Physiol (1985). 1994;77:1281-7
75. Hansen AG, Arnold SA, Jiang M, Palmer TD, Ketova T, Merkel A. et al. ALCAM/CD166 is a TGF-β-responsive marker and functional regulator of prostate cancer metastasis to bone. Cancer Res. 2014;74:1404-15
76. Bessa X, Elizalde JI, Mitjans F, Piñol V, Miquel R, Panés J. et al. Leukocyte recruitment in colon cancer: role of cell adhesion molecules, nitric oxide, and transforming growth factor beta1. Gastroenterology. 2002;122:1122-32
77. Kern U, Wischnewski V, Biniossek ML, Schilling O, Reinheckel T. Lysosomal protein turnover contributes to the acquisition of TGFβ-1 induced invasive properties of mammary cancer cells. Mol Cancer. 2015;14:39
78. Wang Y, Lui WY. Transforming growth factor-β1 attenuates junctional adhesion molecule-A and contributes to breast cancer cell invasion. Eur J Cancer. 2012;48:3475-87
79. Barallobre-Barreiro J, Loeys B, Mayr M, Rienks M, Verstraeten A, Kovacic JC. Extracellular Matrix in Vascular Disease, Part 2/4: JACC Focus Seminar. J Am Coll Cardiol. 2020;75:2189-203
80. Ambade AS, Hassoun PM, Damico RL. Basement Membrane Extracellular Matrix Proteins in Pulmonary Vascular and Right Ventricular Remodeling in Pulmonary Hypertension. Am J Respir Cell Mol Biol. 2021;65:245-58
81. Manou D, Caon I, Bouris P, Triantaphyllidou IE, Giaroni C, Passi A. et al. The Complex Interplay Between Extracellular Matrix and Cells in Tissues. Methods Mol Biol. 2019;1952:1-20
82. Chen Y, Zhao H, Feng Y, Ye Q, Hu J, Guo Y. et al. Pan-Cancer Analysis of the Associations of TGFBI Expression With Prognosis and Immune Characteristics. Front Mol Biosci. 2021;8:745649
83. Kollmannsberger P, Bidan CM, Dunlop JWC, Fratzl P, Vogel V. Tensile forces drive a reversible fibroblast-to-myofibroblast transition during tissue growth in engineered clefts. Sci Adv. 2018;4:eaao4881
84. Burgess JK, Mauad T, Tjin G, Karlsson JC, Westergren-Thorsson G. The extracellular matrix - the under-recognized element in lung disease? J Pathol. 2016;240:397-409
85. Cawston TE, Young DA. Proteinases involved in matrix turnover during cartilage and bone breakdown. Cell Tissue Res. 2010;339:221-35
86. Xiang J, Fu X, Ran W, Wang Z. Grhl2 reduces invasion and migration through inhibition of TGFβ-induced EMT in gastric cancer. Oncogenesis. 2017;6(1):e284
87. Kim ES, Kim MS, Moon A. TGF-beta-induced upregulation of MMP-2 and MMP-9 depends on p38 MAPK, but not ERK signaling in MCF10A human breast epithelial cells. Int J Oncol. 2004;25:1375-82
88. Kim ES, Sohn YW, Moon A. TGF-beta-induced transcriptional activation of MMP-2 is mediated by activating transcription factor (ATF)2 in human breast epithelial cells. Cancer Lett. 2007;252:147-56
89. Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H. et al. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J Cell Biol. 2002;157:493-507
90. Nguyen HL, Kadam P, Helkin A, Cao K, Wu S, Samara GJ. et al. MT1-MMP Activation of TGF-β Signaling Enables Intercellular Activation of an Epithelial-mesenchymal Transition Program in Cancer. Curr Cancer Drug Targets. 2016;16:618-30
91. Marconi GD, Fonticoli L, Rajan TS, Pierdomenico SD, Trubiani O, Pizzicannella J. et al. Epithelial-Mesenchymal Transition (EMT): The Type-2 EMT in Wound Healing, Tissue Regeneration and Organ Fibrosis. Cells. 2021 10
92. Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Science Signaling. 2014;7:re8
93. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69-84
94. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-8
95. Ang HL, Mohan CD, Shanmugam MK, Leong HC, Makvandi P, Rangappa KS. et al. Mechanism of epithelial-mesenchymal transition in cancer and its regulation by natural compounds. Med Res Rev. 2023;43:1141-200
96. Vannella KM, Wynn TA. Mechanisms of Organ Injury and Repair by Macrophages. Annu Rev Physiol. 2017;79:593-617
97. Peng D, Fu M, Wang M, Wei Y, Wei X. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol Cancer. 2022;21:104
98. David CJ, Huang Y-H, Chen M, Su J, Zou Y, Bardeesy N. et al. TGF-β Tumor Suppression through a Lethal EMT. Cell. 2016;164:1015-30
99. Hao Y, Baker D, Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci. 2019 20
100. Hoot KE, Lighthall J, Han G, Lu SL, Li A, Ju W. et al. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008;118:2722-32
101. Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest. 2003;112:1486-94
102. Motizuki M, Yokoyama T, Saitoh M, Miyazawa K. The Snail signaling branch downstream of the TGF-β/Smad3 pathway mediates Rho activation and subsequent stress fiber formation. J Biol Chem. 2023;300:105580
103. Wu G, Wang D, Xiong F, Wang Q, Liu W, Chen J. et al. The emerging roles of CEACAM6 in human cancer (Review). Int J Oncol. 2024 64
104. Han DH, Shin MK, Oh JW, Lee J, Sung JS, Kim M. Chronic Exposure to TDI Induces Cell Migration and Invasion via TGF-β1 Signal Transduction. Int J Mol Sci. 2023 24
105. Deckers M, van Dinther M, Buijs J, Que I, Löwik C, van der Pluijm G. et al. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66:2202-9
106. Yu J, Lei R, Zhuang X, Li X, Li G, Lev S. et al. MicroRNA-182 targets SMAD7 to potentiate TGFβ-induced epithelial-mesenchymal transition and metastasis of cancer cells. Nat Commun. 2016;7:13884
107. Meškytė EM, Keskas S, Ciribilli Y. MYC as a Multifaceted Regulator of Tumor Microenvironment Leading to Metastasis. Int J Mol Sci. 2020 21
108. Kalkat M, De Melo J, Hickman KA, Lourenco C, Redel C, Resetca D. et al. MYC Deregulation in Primary Human Cancers. Genes (Basel). 2017 8
109. Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5
110. Moses HL. TGF-beta regulation of epithelial cell proliferation. Mol Reprod Dev. 1992;32:179-84
111. Jin K, Liu C, Cheng H, Fei Q, Huang Q, Xiao Z. et al. TGF-β1-induced RAP2 regulates invasion in pancreatic cancer. Acta Biochim Biophys Sin (Shanghai). 2022;54:361-9
112. Guo Y, Tong Y, Zhu H, Xiao Y, Guo H, Shang L. et al. Quercetin suppresses pancreatic ductal adenocarcinoma progression via inhibition of SHH and TGF-β/Smad signaling pathways. Cell Biol Toxicol. 2021;37:479-96
113. Xia X, Zhao S, Chen W, Xu C, Zhao D. CCT6A promotes esophageal squamous cell carcinoma cell proliferation, invasion and epithelial-mesenchymal transition by activating TGF-β/Smad/c-Myc pathway. Ir J Med Sci. 2023;192:2653-60
114. Saitoh M. Transcriptional regulation of EMT transcription factors in cancer. Semin Cancer Biol. 2023;97:21-9
115. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature Reviews Molecular Cell Biology. 2014;15:178-96
116. Xu J, Lamouille S, Derynck R. TGF-β-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156-72
117. Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J. et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84-9
118. Xu Z, Jiang Y, Steed H, Davidge S, Fu Y. TGFβ and EGF synergistically induce a more invasive phenotype of epithelial ovarian cancer cells. Biochem Biophys Res Commun. 2010;401:376-81
119. Kumar B, Uppuladinne MV, Jani V, Sonavane U, Joshi RR, Bapat SA. Auto-regulation of Slug mediates its activity during epithelial to mesenchymal transition. Biochim Biophys Acta. 2015;1849:1209-18
120. Assani G, Zhou Y. Effect of modulation of epithelial-mesenchymal transition regulators Snail1 and Snail2 on cancer cell radiosensitivity by targeting of the cell cycle, cell apoptosis and cell migration/invasion. Oncol Lett. 2019;17:23-30
121. Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959-67
122. Tripathi MK, Misra S, Chaudhuri G. Negative regulation of the expressions of cytokeratins 8 and 19 by SLUG repressor protein in human breast cells. Biochem Biophys Res Commun. 2005;329:508-15
123. Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG. et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76-83
124. Kim BN, Ahn DH, Kang N, Yeo CD, Kim YK, Lee KY. et al. TGF-β induced EMT and stemness characteristics are associated with epigenetic regulation in lung cancer. Sci Rep. 2020;10:10597
125. Liu YY, Liu HY, Yu TJ, Lu Q, Zhang FL, Liu GY. et al. O-GlcNAcylation of MORC2 at threonine 556 by OGT couples TGF-β signaling to breast cancer progression. Cell Death Differ. 2022;29:861-73
126. Zhao G-X, Xu Y-Y, Weng S-Q, Zhang S, Chen Y, Shen X-Z. et al. CAPS1 promotes colorectal cancer metastasis via Snail mediated epithelial mesenchymal transformation. Oncogene. 2019;38:4574-89
127. Gheldof A, Hulpiau P, van Roy F, De Craene B, Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell Mol Life Sci. 2012;69:2527-41
128. Fukagawa A, Ishii H, Miyazawa K, Saitoh M. δEF1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells. Cancer Med. 2015;4:125-35
129. Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J. 2003;22:2443-52
130. Tsubakihara Y, Moustakas A. Epithelial-Mesenchymal Transition and Metastasis under the Control of Transforming Growth Factor β. Int J Mol Sci. 2018 19
131. Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S. et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830-40
132. Merindol N, Riquet A, Szablewski V, Eliaou JF, Puisieux A, Bonnefoy N. The emerging role of Twist proteins in hematopoietic cells and hematological malignancies. Blood Cancer J. 2014;4:e206
133. Alexander NR, Tran NL, Rekapally H, Summers CE, Glackin C, Heimark RL. N-cadherin gene expression in prostate carcinoma is modulated by integrin-dependent nuclear translocation of Twist1. Cancer Res. 2006;66:3365-9
134. Khales SA, Mozaffari-Jovin S, Geerts D, Abbaszadegan MR. TWIST1 activates cancer stem cell marker genes to promote epithelial-mesenchymal transition and tumorigenesis in esophageal squamous cell carcinoma. BMC Cancer. 2022;22:1272
135. Wu Q, Sun S, Wei L, Liu M, Liu H, Liu T. et al. Twist1 regulates macrophage plasticity to promote renal fibrosis through galectin-3. Cell Mol Life Sci. 2022;79:137
136. Shiota M, Zardan A, Takeuchi A, Kumano M, Beraldi E, Naito S. et al. Clusterin mediates TGF-β-induced epithelial-mesenchymal transition and metastasis via Twist1 in prostate cancer cells. Cancer Res. 2012;72:5261-72
137. Wu Y, Tran T, Dwabe S, Sarkissyan M, Kim J, Nava M. et al. A83-01 inhibits TGF-β-induced upregulation of Wnt3 and epithelial to mesenchymal transition in HER2-overexpressing breast cancer cells. Breast Cancer Res Treat. 2017;163:449-60
138. Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177-86
139. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610
140. Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65:625-39
141. Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179:1033-55
142. Grelet S, McShane A, Geslain R, Howe PH. Pleiotropic Roles of Non-Coding RNAs in TGF-β-Mediated Epithelial-Mesenchymal Transition and Their Functions in Tumor Progression. Cancers (Basel). 2017 9
143. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci. 2020 21
144. Zhang K, Li XY, Wang ZM, Han ZF, Zhao YH. MiR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharmacol Sci. 2017;21:3598-604
145. Zhou Q, Yang M, Lan H, Yu X. miR-30a negatively regulates TGF-β1-induced epithelial-mesenchymal transition and peritoneal fibrosis by targeting Snai1. Am J Pathol. 2013;183:808-19
146. Zheng J, Wang W, Yu F, Dong P, Chen B, Zhou MT. MicroRNA-30a Suppresses the Activation of Hepatic Stellate Cells by Inhibiting Epithelial-to-Mesenchymal Transition. Cell Physiol Biochem. 2018;46:82-92
147. Ma L, Zhai B, Zhu H, Li W, Jiang W, Lei L. et al. The miR-141/neuropilin-1 axis is associated with the clinicopathology and contributes to the growth and metastasis of pancreatic cancer. Cancer Cell Int. 2019;19:248
148. Siemens H, Jackstadt R, Hünten S, Kaller M, Menssen A, Götz U. et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256-71
149. Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J Biol Chem. 2013;288:10241-53
150. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593-601
151. Yu J, Xie F, Bao X, Chen W, Xu Q. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol Cancer. 2014;13:121
152. Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol. 2022;15:129
153. Yin Q, Han Y, Zhu D, Li Z, Shan S, Jin W. et al. miR-145 and miR-497 suppress TGF-β-induced epithelial-mesenchymal transition of non-small cell lung cancer by targeting MTDH. Cancer Cell Int. 2018;18:105
154. Cao M, Seike M, Soeno C, Mizutani H, Kitamura K, Minegishi Y. et al. MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int J Oncol. 2012;41:869-75
155. Zhou SJ, Liu FY, Zhang AH, Liang HF, Wang Y, Ma R. et al. MicroRNA-199b-5p attenuates TGF-β1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Br J Cancer. 2017;117:233-44
156. Song Y, Zhao F, Wang Z, Liu Z, Chiang Y, Xu Y. et al. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Ann Surg Oncol. 2012;19(Suppl 3):S509-17
157. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. 2020;21:102-17
158. Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9:1932-56
159. Nandwani A, Rathore S, Datta M. LncRNAs in cancer: Regulatory and therapeutic implications. Cancer Lett. 2021;501:162-71
160. Fan C, González-Prieto R, Kuipers TB, Vertegaal ACO, van Veelen PA, Mei H. et al. The lncRNA LETS1 promotes TGF-β-induced EMT and cancer cell migration by transcriptionally activating a TβR1-stabilizing mechanism. Sci Signal. 2023;16:eadf1947
161. Chen HY, Chan SJ, Liu X, Wei AC, Jian RI, Huang KW. et al. Long noncoding RNA Smyca coactivates TGF-β/Smad and Myc pathways to drive tumor progression. J Hematol Oncol. 2022;15:85
162. Wu N, Jiang M, Liu H, Chu Y, Wang D, Cao J. et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-β/SMAD2/3 signaling pathway. Cell Death Differ. 2021;28:219-32
163. Shen X, Hu X, Mao J, Wu Y, Liu H, Shen J. et al. The long noncoding RNA TUG1 is required for TGF-β/TWIST1/EMT-mediated metastasis in colorectal cancer cells. Cell Death Dis. 2020;11:65
164. Fan C, Wang Q, Kuipers TB, Cats D, Iyengar PV, Hagenaars SC. et al. LncRNA LITATS1 suppresses TGF-β-induced EMT and cancer cell plasticity by potentiating TβRI degradation. EMBO J. 2023;42:e112806
165. Zhang C, Hao Y, Wang Y, Xu J, Teng Y, Yang X. TGF-β/SMAD4-Regulated LncRNA-LINP1 Inhibits Epithelial-Mesenchymal Transition in Lung Cancer. Int J Biol Sci. 2018;14:1715-23
166. Li Z, Dong M, Fan D, Hou P, Li H, Liu L. et al. LncRNA ANCR down-regulation promotes TGF-β-induced EMT and metastasis in breast cancer. Oncotarget. 2017;8:67329-43
167. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-91
168. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188-206
169. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer. 2020;6:319-36
170. Li K, Guo J, Ming Y, Chen S, Zhang T, Ma H. et al. A circular RNA activated by TGFβ promotes tumor metastasis through enhancing IGF2BP3-mediated PDPN mRNA stability. Nat Commun. 2023;14:6876
171. Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T. et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17:140
172. Zhou H, Wu J, Leng S, Hou C, Mo L, Xie X. et al. Knockdown of circular RNA VANGL1 inhibits TGF-β-induced epithelial-mesenchymal transition in melanoma cells by sponging miR-150-5p. J Cell Mol Med. 2021;25:10837-45
173. Meng L, Zheng Y, Liu S, Ju Y, Ren S, Sang Y. et al. ZEB1 represses biogenesis of circ-DOCK5 to facilitate metastasis in esophageal squamous cell carcinoma via a positive feedback loop with TGF-β. Cancer Lett. 2021;519:117-29
174. Kim BG, Malek E, Choi SH, Ignatz-Hoover JJ, Driscoll JJ. Novel therapies emerging in oncology to target the TGF-β pathway. J Hematol Oncol. 2021;14:55
175. Slack RJ, Macdonald SJF, Roper JA, Jenkins RG, Hatley RJD. Emerging therapeutic opportunities for integrin inhibitors. Nat Rev Drug Discov. 2022;21:60-78
176. Lu J, Kornmann M, Traub B. Role of Epithelial to Mesenchymal Transition in Colorectal Cancer. Int J Mol Sci. 2023 24
177. Jin W, Chen F, Wang K, Song Y, Fei X, Wu B. miR-15a/miR-16 cluster inhibits invasion of prostate cancer cells by suppressing TGF-β signaling pathway. Biomed Pharmacother. 2018;104:637-44
178. Suzuki T, Mizutani K, Minami A, Nobutani K, Kurita S, Nagino M. et al. Suppression of the TGF-β1-induced protein expression of SNAI1 and N-cadherin by miR-199a. Genes Cells. 2014;19:667-75
Author contact
![]() Corresponding authors: Xianxiu Qiu: bmsqiuedu.cn; Sha Ma: smaedu.cn; Jincheng Zeng: zengjcedu.cn.
Corresponding authors: Xianxiu Qiu: bmsqiuedu.cn; Sha Ma: smaedu.cn; Jincheng Zeng: zengjcedu.cn.

 Global reach, higher impact
Global reach, higher impact