Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(7):1227-1240. doi:10.7150/ijms.93464 This issue Cite
Review
Potential Markers to Differentiate Uterine Leiomyosarcomas from Leiomyomas
1. Department of Obstetrics and Gynecology, Affiliated Hangzhou First People's Hospital, Westlake University School of Medicine, 310003 Hangzhou, Zhejiang Province, China.
2. Department of Gynecology, Clinical Oncology School of Fujian Medical University, Fujian Cancer Hospital, 350014 Fuzhou, Fujian Province, China.
Received 2023-12-21; Accepted 2024-5-4; Published 2024-5-13
Abstract
Uterine leiomyomas (ULM) are the most common benign tumors of the female genitalia, while uterine leiomyosarcomas (ULMS) are rare. The sarcoma is diffuse growth, prone to hematogenous metastasis, and has a poor prognosis. Due to their similar clinical symptoms and morphological features, it is sometimes difficult to distinguish them, and the final diagnosis depends on histological diagnosis. Misdiagnosis of ULM as ULMS will lead to more invasive and extensive surgery when it is not needed, while misdiagnosis of ULMS as ULM may lead to delayed treatment and poor prognosis. This review searched and studied the published articles on ULM and ULMS, and summarized the potential markers for the differential diagnosis of ULMS. These markers will facilitate differential diagnosis and personalized treatment, providing timely diagnosis and potentially better prognosis for patients.
Keywords: uterine leiomyoma, leiomyosarcoma, sarcoma, marker, diagnosis
Introduction
Uterine tumors can be divided into benign and malignant. The most common benign uterine tumors are uterine leiomyomas (ULM), and the most common malignant uterine tumors are endometrial carcinoma and uterine sarcoma. Fibroids or myomas, often known as uterine leiomyomas, are the most prevalent benign tumors of the female genitalia, which occurs in 1 out of every 4 to 5 women [1, 2]. Despite its benign nature, it has a high incidence. Because fibroids are often asymptomatic or rarely symptomatic, the reported incidence is much lower than the true incidence of fibroids [3]. Common symptoms of uterine leiomyomas include increased menstrual bleeding, anemia, lower abdominal mass, infertility, etc., so it also troubles most women with uterine leiomyomas [4]. Uterine leiomyosarcomas (ULMS) have similar clinical symptoms, but it is less common in comparison. They are the most common sarcoma of the uterine body. Three to nine percent of uterine malignancies are uterine sarcomas. ULMS have an extremely poor prognosis, are prone to hematogenous metastasis, and make up 60%-70% of all uterine sarcomas [5]. According to a study, 42% of individuals with uterine leiomyosarcomas survive for the whole five years [6].
Although uterine leiomyomas are non-malignant, studies have found that one in 498 uterine tumors has a hidden risk of undiagnosed malignant tumors, such as leiomyosarcomas [7, 8]. Uterine leiomyomas and uterine leiomyosarcomas have similar clinical symptoms and morphological features [9], and it is sometimes difficult to distinguish them, and the diagnosis is based on histological examination [10, 11]. Surgery is considered the main treatment for ULM and ULMS [12, 13], so preoperative diagnosis is very important. If the preoperative diagnosis of ULMS is misdiagnosed as ULM, using morcellators during the operation will bring the risk of sarcoma spread [14, 15]. As a result of a safety communication issued by the US Food and Drug Administration (FDA) in 2014, minimally invasive surgery in women with ULM is now severely limited due to the inability to use morcellators [16]. Preoperative diagnosis of ULMS is challenging, if the ULM is misdiagnosed as ULMS, it will lead to more invasive and extensive surgery when it is not needed, while misdiagnosis of ULMS as ULM may lead to delayed treatment and poor prognosis [17].
Importantly, diagnostic methods such as ultrasound, CT, MRI, and CA-125 detection alone cannot accurately distinguish between malignant and benign uterine fibroids. Although CA125 is frequently utilized in daily practice, it is typically only markedly increased in advanced disease. And only 35 percent of cases could be diagnosed with ULMS by endometrial biopsy, according to Sagae et al. [18]. At present, markers that can differentiate between uterine fibroids and sarcomas have not been found [19]. Therefore, this study aims to find and summarize the potential diagnostic markers between ULM and ULMS by searching and studying the articles related to ULM and ULMS published so far. These potential diagnostic markers will facilitate the identification of ULM and ULMS, which will help to identify new therapeutic targets, which will not only effectively distinguish ULM from ULMS, but also accurately treat and even monitor prognosis.
Materials and Methods
The source that we used was the PubMed database. Uterine leiomyoma, leiomyosarcoma, marker, biomarker, and diagnostic were the search phrases that were employed. 421 abstracts were assessed by title from 1122 search results that were retrieved between 1983 and 2024, 249 articles were extensively read, and 117 papers were ultimately included. Studies were not restricted by design, publication date, or number of patients reported due to the rarity of uterine leiomyosarcomas.
Potential Markers
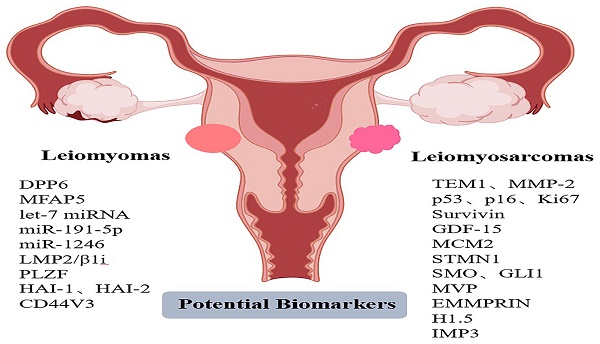
We searched the articles, summarized and described 16 potential diagnostic markers. Table 1 presents a summary of the markers. Some of the markers involved in some pathways we have added relevant graphical representation (Figures 1-6).
Tumor Endothelial Marker 1 (TEM1)
TEM1 (also known as CD248 or endosialin) is a cell membrane protein that is mainly expressed in malignant tissues or during embryonic development, but hardly expressed in benign and normal tissues [20, 21]. Its expression has been detected in skin cancer, colorectal, breast and other malignant tumors [22-24]. It has been found that TEM1 is functional in controlling the interactions between tumor cells, endothelium, and stroma, and that TEM1 expression in tumor stroma and vascular endothelial cells may support tumor progression and invasion [24, 25]. In addition, TEM1 is highly expressed in sarcomas. One study found that TEM1 was expressed in 96% of human sarcoma tissues among 19 sarcoma subtypes [26]. TEM1 is an important therapeutic target for human sarcoma. Therefore, TEM1 may influence the onset and progression of uterine leiomyosarcomas and is anticipated to be a possible target for therapy.
Wu et al. found that TEM1 promotes the invasion and migration of ULMS by promoting extracellular matrix (ECM) remodeling through up-regulation of MMP-2 [27]. MMP-2 and TEM1 are co-expressed and positively correlated in ULMS whereas they were not expressed in ULM. It was found that MMP-2 activity and expression were upregulated in TEM1-overexpressing cells, whereas the opposite was observed in TEM1-knockdown cells. Depletion of MMP-2 suppressed the invasion and migration of TEM1-overexpressing cells. MMP-2 has been shown to be associated with a number of carcinomas metastasis [28, 29]. For malignant tumors, cell-ECM adhesion is the key to their distant metastasis [30, 31]. Wu et al. suggested that TEM1 can promote the adhesion of sarcoma cells to ECM components, and TEM1 overexpression also promotes sarcoma metastasis [27]. This indicates that for the diagnosis of benign and malignant uterine fibroids, TEM1 is probably a good marker.
Potential biomarkers for differential diagnosis between ULMS and ULM.
| Biomarker | Expression in ULMS | Reference |
|---|---|---|
| TEM1, MMP-2 | Up | [26, 27] |
| DPP6, MFAP5 | Down | [34] |
| p53, p16, Ki67 | Up | [45-49, 61] |
| survivin | Up | [71] |
| GDF-15 | Up | [78] |
| MCM2 | Up | [84, 85] |
| STMN1 | Up | [60, 94] |
| SMO, GLI1 | Up | [96, 97] |
| let-7 miRNA | Down | [108] |
| miR-191-5p, miR-1246 | Down | [111] |
| MVP | Up | [114] |
| LMP2/β1i | Down | [118, 124] |
| EMMPRIN | Up | [133, 134] |
| PLZF | Down | [143] |
| H1.5 | Up | [143] |
| IMP3 | Up | [148] |
| HAI-1, HAI-2 | Down | [153] |
| CD44V3 | Down | [164] |
Graphical representation of the pathways involved in TEM1.

Graphical representation of the pathways involved in DPP6 and MFAP5.

Dipeptidyl Peptidase Like 6 (DPP6) and Microfibril Associated Protein 5 (MFAP5)
DPP6, a type II transmembrane protein derived from the ubiquitous family of serine peptidases in both prokaryotes and eukaryotes, is essential for the normal function of cells [32]. Furthermore, MFAP5, an extracellular matrix (ECM) glycoprotein, is implicated in cell survival, elastinogenesis, and signaling during microfibril construction [33]. Ke et al. found that DPP6 and MFAP5 were expressed at significantly lower levels in ULMS than in ULM, and the area under the ROC curve (AUC) determined that DPP6 and MFAP5 had the diagnostic ability to distinguish ULMS from ULM (AUC values were 0.957 and 0.899, respectively) [34]. Studies have shown that the immune cell components of ULM and ULMS are different, and DPP6 and MFAP5 are also associated to infiltrating immune cells [34, 35]. DPP6 was positively correlated with some immune cells that were more abundant in ULM and negatively correlated with some immune cells that were more abundant in ULMS. For example, the proportion of macrophage M0 was notably higher in uterine leiomyosarcomas than in leiomyomas, and DPP6 was negatively related to M0 macrophages. Additionally, MFAP5 was positively related to resting mast cells, and the ratio of resting mast cells was significantly higher in ULM than in ULMS [34]. Therefore, the association of DPP6 and MFAP5 with immune cells can speculate that they may have an impact on immune-related pathways that influence the development and incidence of ULMS. In malignant tumors, more and more studies use immune cells as a novel research direction for diagnosing diseases and prognosis [36-38]. For example, in patients with breast cancer, low DPP6 expression predicts unfavorable prognosis, which is consistent with Ke et al. 's finding that DPP6 expression level in ULMS is significantly lower than that in ULM [39].
KEGG analysis revealed that immune-related and cell cycle-related pathways, including the HTLV-1 infection pathway, were enriched in the differentially expressed genes between ULM and ULMS [34]. Related studies have shown that HTLV-1 enhances genomic instability through changing host genes expression directly, thereby affecting immune-related pathways and leading to malignant transformation [40], which again demonstrates that the regulation of immune response may be closely related to the occurrence of ULMS.
Tumor Protein p53, p16 and Ki67
P53, p16 and Ki67 have been studied more in immunohistochemistry and are often used to distinguish benign and malignant lesions [41, 42]. In human cancers, mutations in the p53 tumor suppressor gene are frequently found [43]. The most prevalent gene mutation in solid tumors, p53 mutations, can make cells resistant to the activation of intrinsic apoptotic pathways [44]. De Vos et al. first proposed that p53 gene mutations are more common in ULMS. They claim that the acquisition of p53 mutations is a difference between leiomyomas and leiomyosarcomas [45]. Nordal et al. suggested that p53 gene abnormalities possibly play an essential part in the development of uterine sarcoma [46]. Previous researches have shown that the expression of p53 between ULMS and ULM is significantly different [47, 48]. The immunohistochemical staining pattern of p53 mutation is significantly higher in ULMS than in ULM [49].
The overexpression of p16 is also described in ULMS. P16 is a tumor inhibitory protein that plays a vital role in regulating the cell cycle. It acts as a negative regulator of the cell cycle by binding to the cell cycle-dependent kinase CDK4-cyclin D[47, 50, 51]. P16 normally promotes growth arrest, but increased expression in tumor cells causes protein accumulation in their nuclei and cytoplasms, and strong and diffuse p16 positivity is observed in immunohistochemistry [50]. In uterine leiomyosarcomas, p16 gene expression appears to be upregulated, and this upregulation may also extend to p16 protein expression, which has been reported by several studies in ULMS compared with ULM [47, 52, 53]. In uterine leiomyomas, aberrant expression of p53, p16 has been described as a hallmark of malignancy [47].
Abnormal cell proliferation is the main reason for the occurrence and development of carcinogenic processes. Ki67(also known as MKI67) is a proliferation marker and a nuclear DNA binding protein [54]. Ki-67 is a late marker of cell cycle entry in normal cells and is strongly downregulated in quiescent G0 phase cells, with the highest expression of Ki-67 mRNA in G2 phase, whereas Ki-67 protein expression increases throughout the cell cycle and peaks in mitosis [54, 55]. Ki-67 is highly expressed in endometrial cancer [56], ovarian cancer [57], and cervical cancer [58], and high Ki-67 index generally indicates poor clinical prognosis [59]. Compared to uterine leiomyomas, uterine leiomyosarcomas express significantly higher Ki67 mRNA levels [60]. The increased expression of Ki67 is also considered as a diagnostic marker for the malignancy of uterine leiomyosarcoma [61], and high Ki67 is associated with poor prognosis of leiomyosarcoma [41, 42].
Survivin
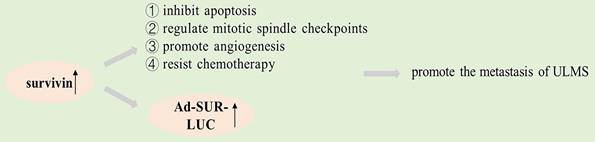
The survivin gene encodes an inhibitor of apoptosis (IAP) that is structurally unique to humans. Studies have shown that survivin is significantly expressed in many human cancers, such as lung carcinoma, breast carcinoma and colon carcinoma [62-64]. The survivin protein plays a key role in mitosis and programmed cell death, and a genome-wide search indicated that survivin expression differs in normal and tumor tissue [65]. It may be a universal characteristic of tumorigenesis that apoptosis is inhibited, thereby preventing normal homeostasis and promoting tissue tumorigenesis [66], and survivin inhibits apoptosis, regulates mitotic spindle checkpoints, promotes angiogenesis, and resists chemotherapy in cancer pathogenesis, according to studies [44, 67]. Increased expression of survivin is an adverse prognostic marker in patients, and a high expression of survivin is also linked to an increase in recurrences, lymphadenopathy, and metastasis [68, 69]. Survivin has been identified as a cancer-specific promoter in lots of researches [70].
Shalaby et al. found that survivin expressed its downstream reporter gene in ULMS merely but not in fibroids. Compared with uterine leiomyoma, the downstream reporter gene (Ad-SUR-LUC) of survivin promoter was highly expressed in uterine leiomyosarcoma cells [71]. Their study showed that survivin is a promoter that can distinguish ULMS from ULM. It will be possible to detect cancer cells by using the expression of survivin in different cells as a method for testing the promoter driving power of the downstream reporter gene, which can serve for the early detection of cancer cells [72]. Intravenous injection of Ad-SUR-LUC in the study by Shalaby et al. successfully differentiated preexisting human leiomyosarcoma from human uterine leiomyoma in a mouse model [71]. Therefore, survivin may be a promising new target for cancer therapies that are based on apoptosis.
Growth Differentiation Factor-15 (GDF-15)
GDF-15, also known as macrophage inhibitory cytokine-1 (MIC-1), is a secreted cytokine regulated by P53, which is associated with tumorigenesis and is a biomarker for ovarian and endometrial cancer [73-75]. Inflammation is considered one of the "hallmarks of cancer" and interleukin-1 and tumor necrosis factor-alpha may activate macrophages and induce GDF-15 production [76, 77]. Trovik et al. showed that uterine leiomyosarcoma patients had significantly higher circulating GDF-15 levels compared to leiomyomas patients, and the ROC curve analysis showed that GDF-15 has a certain accuracy in the diagnosis of ULMS and ULM, indicating that GDF-15 is an effective biomarker to distinguish ULM and ULMS [78]. The high expression of GDF-15 in ULMS may be due to the activation of cancer-related inflammatory processes.
GDF-15 has a certain correlation with p53 and macrophages, so it is speculated that the high expression of GDF-15 in ULMS may be due to the activation of cancer-related inflammatory process [77], or it may be the target of downstream pathways regulating cell cycle arrest and apoptosis, and has an impact on cancer proliferation, migration, invasion, etc. [79].
Graphical representation of the pathways involved in survivin.
Graphical representation of the pathways involved in GDF-15.

Graphical representation of the pathways involved in SMO.
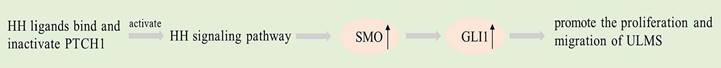
Minichromosome Maintenance Complex Component 2 (MCM2)
MCM2 belongs to the family of MCM proteins. It plays a key role in DNA replication initiation and replication fork movement and is closely associated with cell proliferation, with its protein levels increasing in G1 and peaking in S1[80, 81]. MCM2 overexpression has been found in a wide variety of malignant tumors in recent years, and as a promising proliferation marker, it is expected to become a marker for the identification of malignant tumors [80, 82, 83].
According to Quade et al., MCM2 expression was 18.2-fold higher in ULMS than in ULM, that is, MCM2 expression increased significantly in ULMS compared to ULM [84]. The population studied by Keyhanian et al., ULMS patients consistently demonstrated a high proportion of MCM2 responses, the sensitivity and specificity for diagnosing ULMS with MCM2>80% were 92% and 94%, respectively [85]. This study suggests that MCM2 is valuable in identifying ULMS. and additionally, combining MCM2 with p16 or Ki67 can better distinguish ULMS from ULM [85].
Stathmin 1 (STMN1)
STMN1, also known as oncoprotein 18, is a microtubule depolymerization-related protein that is widely expressed in the cytoplasm. It exerts regulatory control over microtubule dynamics through the inhibition of tubulin polymerization and the promotion of microtubule instability, while also facilitating tumor cell proliferation, differentiation, and invasion [86-88]. STMN1 is expressed in various carcinomatosis such as ovarian cancer [89], endometrial cancer [90], cervical cancer [91], hepatocellular carcinoma [92], and bladder cancer [93], and inhibition of STMN1 can reduce cell viability and migration potential.
The activation of the oncogenic phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway (PI3K-AKT-mTOR) has been observed in uterine smooth muscle tumors. Furthermore, the identification of STMN1 expression serves as an indicator for the activation of the PI3K-AKT-mTOR pathway [94]. Hwang et al. showed that the expression of STMN1 has some value in differentiating uterine fibroids from uterine sarcomas [95]. The research by Allen et al. found that STMN1 expression in uterine leiomyosarcomas was mainly diffusely and strongly positive, while uterine leiomyomas was mainly weakly positive. The expression of STMN1 exhibited a sensitivity of 100% in relation to leiomyosarcomas, yet its specificity was merely 55% [94]. Consequently, STMN1 emerges as a remarkably sensitive indicator for leiomyosarcomas, albeit with limited specificity for diagnostic applications. In line with the findings of Allen et al., Hu et al. also discovered that STMN1 expression in uterine leiomyosarcomas was much higher than that in uterine leiomyomas [60, 94]. Therefore, STMN1 is a gene associated with tumors and is a possible target for diagnosis and treatment.
Smoothened (SMO) and GLI Family Zinc Finger 1 (GLI1)
Dysregulation of the Hedgehog (HH) pathway has been documented in ULMS patients, with higher expression levels of GLI1 and SMO in ULMS compared to ULM. The evidence presented suggests a correlation between HH pathway dysregulation and ULMS development [96, 97]. The HH pathway signaling involves three ligands: sonic hedgehog (SHH), Indian hedgehog (IHH), and desert hedgehog (DHH), two receptors: PTCH1 and SMO, and three transcription factors: GLI1, GLI2 and GLI3 mediate. When HH ligands bind and inactivate PTCH1, the classical HH signaling pathway is activated, releasing SMO protein signals to its cytoplasmic targeting [98]. SMO triggers the translocation of GLI proteins to the nucleus, which leads to their subsequent binding to DNA [99]. GLI members are nuclear regulators situated at the pathway's end and in charge of controlling the downstream target genes' expression [100].
The HH signaling pathway has been implicated in tumorigenesis and cell differentiation in several studies [96, 101]. Analyzing the protein expression in components of Hedgehog signalling in uterine smooth muscle tumors is helpful for the diagnosis, prognosis or malignant risk prediction of uterine smooth muscle tumors [96]. Garcia et al. found activation of the Hedgehog pathway and increased GLI nuclear translocation in ULMS. SMO and GLI1 expression in ULMS is higher than that in ULM [97]. PTCH1 expression is downregulated in ULMS. SMO or GLI inhibitors applied to ULMS cells inhibit cell proliferation and migration while inducing apoptosis [97]. In the future, GLI1 and SMO may become therapeutic targets for uterine smooth muscle tumors.
Lethal-7 (let-7) miRNA, miR-191-5p and miR-1246
MicroRNAs (miRNAs) regulate the expression of a variety of target genes in cells, mainly through regulating the translation of the target genes [102]. Studies have shown that extracellular miRNAs have significant functions in cell-to-cell communication and various biological mechanisms [103]. Altered expression of miRNAs may lead to tumorigenesis, and gynecologic tumors such as ovarian cancer, endometrial cancer, cervical cancer, and uterine sarcoma are associated with unregulated expression of miRNAs [104, 105]. Blood contains miRNAs at a stable concentration, and it has been established that circulating miRNAs can be used as disease biomarkers [106].
The let-7 family of miRNAs is a key regulator of eukaryotic cell apoptosis, differentiation and pluripotency. The let-7 family is a major family of miRNAs that normally function as tumor suppressors [107]. The study's findings demonstrated that all members of the let-7 family had downregulation in ULMS, and that a worse patient prognosis was correlated with a higher degree of loss of expression [108]. This suggests that let-7 is a potential prognostic biomarker for LMS. Loss of let-7 likely result from perturbation of the signalling network involving key families of proteins, leading to an acceleration of tumor progression [109]. Downregulation of let-7 is prevalent in lots of cancers, and substitution of let-7 for normal expression has been shown to arrest tumor growth [110].
Furthermore, Yokoi et al. endeavored to ascertain diagnostic biomarkers that could effectively differentiate between ULMS and ULM through their investigation of circulating miRNAs. Their primary objective was to identify potential miRNAs that could be utilized in the development of a diagnostic model for ULMS, and seven candidate miRNAs (miR-191-5p, miR-1246, miR-4635, miR-4485-5p, miR-451a, miR-6511b-5p and miR-4430) were screened out [111]. These seven miRNAs were significantly downregulated in ULMS. The optimal model consisted of two miRNAs (miR-191-5p and miR-1246) based on the model construction, and ULMS patients could be accurately identified using this dual miRNA prediction signature [111]. Therefore, the combination of miR-191-5p and miR-1246 is a potential marker for differentiating ULMS from ULM.
Major Vault Protein (MVP)
MVP, also called lung resistance-related protein (LRP), is located on chromosome 16 and helps move various molecules in and out of signal transduction networks and toxic compounds out. Data indicate high MVP expression is related with resistance to multiple chemotherapy regimens [112]. In acute myeloid leukaemia, lung cancer, ovarian cancer and other malignant tumors, its increased expression has been found to be associated with the induction of multidrug resistance [112, 113]. By analyzing differentially expressed proteins between ULMS and ULM, Lintel et al. shown that MVP expression in ULMS was 3.05-fold higher than that in ULM. By immunohistochemistry (IHC), MVP found 50% sensitivity and 100% specificity when comparing ULMS and ULM [114]. MVP is a helpful adjunct for differentiating ULMS from ULM, although negative staining results cannot rule out malignancy, positive staining results are a strong indicator of malignancy [114].
Large Multifunctional Protease 2 (LMP2/β1i)
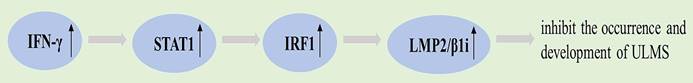
LMP2/β1i is an immunoproteasome catalytic subunit [115]. Mice with LMP2/β1i gene deficiency were found to spontaneously develop ULMS through animal models [116, 117]. LMP2/β1i is not expressed in ULMS but is present in ULM. Thus, one of the risk factors for ULMS may be defective LMP2/β1i expression [117, 118]. Tumor rejection mediated by MHC class I molecules, a process influenced by the function of the proteasome induced by interferon-γ (IFN-γ) [119, 120]. The findings confirm that IFN-γ prevents the development of primary tumors and thus exhibits a tumor suppressive effect in the immune response [119, 121].
The expression of LMP2 is significantly induced by IFN-γ, and the activation of signal transducer and activator of transcription (STAT) 1 by IFN-γ leads to the upregulation of tumor suppressors, including interferon regulatory factor 1 (IRF1). IRF1 functions as a transcriptional regulator that plays a significant role in the regulation of LMP2 expression [122, 123]. Decreased IRF1 level caused by LMP2 deficiency may be a risk factor for ULMS [118].
Graphical representation of the pathways involved in LMP2/β1i.
LMP2 immunostaining is helpful in the differential diagnosis of ULMS and ULM, as LMP2 protein expression is attenuated in 85% of ULMS samples [124]. LMP2/β1i is a promising diagnostic marker for ULMS and has the potential to become a targeted molecule for novel therapies [117].
Extracellular Matrix Metalloproteinase Inducer (EMMPRIN)
EMMPRIN, also known as CD147, is a member of the human immunoglobulin superfamily and an inducer of extracellular matrix proteolytic enzymes encoded by the BSG gene [125]. EMMPRIN, which is abundantly expressed on the surface of tumor cells, assumes a pivotal function in the advancement of numerous cancers through its stimulation of matrix metalloproteinases (MMPs) and cytokine secretion [126, 127]. MMPs are crucial in the degradation of the extracellular matrix and the progression of tumors [128, 129]. The degradation of the extracellular matrix is of utmost significance in facilitating tumor invasion, growth, and metastasis. Consequently, EMMPRIN assumes a crucial role in cancer cells by governing cell proliferation, apoptosis, migration, metastasis, and differentiation, particularly in hypoxic environments [125]. Many tumors, particularly disseminated cancer cells and those with a poor prognosis, express EMMPRIN at high levels [130-132]. Studies have shown that EMMPRIN may serve as a potential early disease diagnostic marker. For example, EMMPRIN is considered a promising therapeutic target for the treatment of hepatocellular carcinoma, and monoclonal antibodies targeting EMMPRIN have made exciting clinical progress in the treatment of hepatocellular carcinoma [125].
In the study by Kefeli et al., the degree and intensity of EMMPRIN staining and their combined score were compared between ULMS and benign uterine smooth muscle tumors, and the findings suggested that the difference was statistically significant, and the EMMPRIN expression in the LMS group was notably higher than in the ULM group [133]. The results of Ozler et al. are consistent with those of Kefeli et al., high EMMPRIN expression is predominantly observed in ULMS [134], indicating that EMMPRIN may serve as a useful immunohistochemical marker to distinguish LMS from other benign smooth muscle tumors.
Promyelocytic Leukemia Zinc Finger (PLZF) and Histone H1.5
PLZF protein is a DNA-binding transcriptional repressor that negatively regulates the progression of the cell cycle, ultimately leading to growth inhibition [135]. PLZF was found to be highly expressed in the secretory endometrium and the myometrium by IHC [136]. However, its expression has been observed to decrease in lung cancer, melanoma and hematological malignancies [137-139]. Histone H1.5 is a type of histone H1, which is a group of proteins that assist in organizing chromosomes into more complex structures [139-141]. H1.5 has been found to have an effect on the regulation of transcriptional, and this protein is strongly expressed in lung neuroendocrine tumors and prostate cancer and is associated with disease progression [139, 142].
PLZF was under-expressed in ULMS, whereas H1.5 was over-expressed in ULMS. In uterine leiomyomas, the expression of the two proteins is opposite [143]. Thus, as immunohistochemical biomarkers for ULMS and ULM, PLZF exhibits an inverse relationship with H1.5. This indicates that PLZF or H1.5 staining may serve as a useful screening test. The study by Momeni et al. showed that combining these two immunophenotypes resulted in a specificity and sensitivity of 97.5% and 90.5%, respectively, in differentiating ULM from ULMS [143].
Insulin-like Growth Factor Ⅱ mRNA Binding Protein 3 (IMP3)
IMP3 is a member of insulin-like growth factor RNA binding protein family. IMP3, an oncogenic fetal protein linked to advanced and aggressive cancers, is expressed only in malignant tumors and is not found in benign tissues. It has a role in embryogenesis and carcinogenesis of certain malignant tumors [144-146]. Research has demonstrated that IMP3 can stimulate the growth, invasion, and metastasis of tumor cells [146, 147]. The study by Cornejo et al. suggested that IMP3 is strongly expressed in ULMS but not in benign leiomyomas [148]. One extremely specific biomarker of leiomyosarcoma is IMP3, which may be involved in the pathophysiology of the disease. IMP3 is a cancer-specific biomarker linked to more aggressive tumor behavior, as demonstrated by earlier research on endometrial and renal cell carcinomas [144, 146, 149]. IMP3 immunoreactivity in leiomyosarcoma also illustrates the more aggressive behavior of the tumor and predicts a worse prognosis. Therefore, to distinguish between benign and malignant smooth muscle tumors, IMP3 staining can be a helpful adjunct. Additionally, IMP3 expression in the uterus can be utilized as a positive biomarker to raise the degree of confidence in the final diagnosis of malignant smooth muscle tumors.
Hepatocyte Growth Factor Activator Inhibitors (HAI)-1 and -2
HAI-1 and HAI-2 were originally described as endogenous inhibitors of hepatocyte growth factor activator (HGFA), matriptase, hepsin and prostasin. These proteolytic enzymes are primarily membrane-bound and may perform crucial functions in cellular homeostasis. Dysregulation of their activity and expression has been linked to the development and advancement of tumors [150-152]. In the study by Nakamura et al., HAI-1 and HAI-2 mediated cell invasion, migration and proliferation, leading to necrosis and apoptosis by reducing the expression of HGFA, hepsin and matriptase [153].
Hepatocyte growth factor (HGF) is a multifunctional growth factor that is secreted by liver mesenchymal cells and acts on the motility and morphogenesis of a variety of target cells, usually associated with the ECM [154]. Degradation of the ECM facilitates cell separation, leading to local and systemic dissemination. HAI-1 and HAI-2 regulate HGFA, which is in charge of the proteolytic activation of the precursor forms of HGF in a variety of human cancer tissues and serum [155, 156]. Reduced expression of HAI-1 and HAI-2 has been linked in the past to the advancement of ovarian and cervical cancer, according to research [157, 158]. A range of serine proteases that may be implicated in carcinogenesis, invasion, and metastasis may be efficiently inhibited by HAI-1 and HAI-2, and their overexpression decreases cell adherence and spreading [153].
The typical human uterus has high levels of expression of both HAI-1 and HAI-2 in its surface epithelium and uterine glands [159]. HAI-1 and HAI-2 expression levels in ULMS tissues were notably lower than those in ULM tissues [153]. Furthermore, compared to high expression of HAI-1 and HAI-2, reduced expression of these genes was a strong indicator of a poor prognosis [153]. Given the suggestion that HAI-1 and HAI-2 may be significant tumor suppressor genes for the detection of ULMS, both may be taken into consideration as therapeutic options for the illness.
CD44 variant 3 (CD44v3)
As a transmembrane glycoprotein, CD44 is involved in intercellular and cell-matrix interactions [160]. The simplest CD44 standard (CD44s) does not contain any additional exon products, whereas CD44 variants (CD44v) contain one or more additional exons. Studies have shown that alterations in CD44 protein are associated with tumorigenesis, local invasion, metastasis, recurrence, and poor prognosis [161-163].
CD44v3 was expressed in ULM but not in ULMS. When using CD44v3 negative staining to diagnose ULMS, the specificity, sensitivity, positive predictive value, and negative predictive value were all 100% [164]. The expression of CD44s is decreased in ULMS relapse patients. The study by Poncelet et al. suggested that CD44s immunostaining in leiomyosarcomas may be prognostic and that loss of CD44v3 expression may serve as a potential diagnostic tool for uterine leiomyosarcomas [164].
Other Biomarkers
In addition to some of the markers described above, there are other markers that are helpful in differentiating ULMS from ULM, which are briefly described as follows.
The expression of B-cell lymphoma 2 (Bcl2) and DNA fragmentation factors 40, 45 (DFF40, 45) was significantly lower in ULMS compared to ULM [165, 166], while progesterone and estrogen receptors (PR and ER) were either faintly positive or negative [167, 168]. DFF40 and DFF45 are the ultimate DNA ladder leading to apoptosis, and Bcl2 is an inhibitor of apoptosis. Bcl2 expression changes in breast cancer, endometrial cancer, ULMS and other malignant tumors [169, 170]. Low or absent expression of these markers is associated with potentially adverse outcomes [166, 171].
Mediator complex subunit 12 (MED12) mutations are the first recurrent oncogenic mechanisms identified in smooth muscle tumors [172]. These somatic mutations have demonstrated reliability as biomarkers for uterine fibroids, with MED12 mutations being present in roughly 70% of affected women [173]. MED12 mutations are rarely seen in ULMS [174, 175]. The alteration of MED12 may be related to the occurrence of smooth muscle tumors, and its expression may be inhibited in malignant tumors [176]. Furthermore, the Wilms' tumor 1 (WT1) that is expressed in various gynecologic tumors, such as endometrial stromal tumors and ovarian cancer [177, 178]. It has been found that uterine leiomyosarcomas is more likely to have a loss of WT1 expression, and WT1 is expressed in most uterine leiomyomas, so WT1 may also have a certain value in distinguishing benign and malignant uterine leiomyosarcomas [49, 178].
Besides, immunohistochemistry showed that the expression of hyperthermia inhibited histone acetyltransferase 1 (HAT1) in ULMS was higher than that in ULM, and was associated with poor prognosis. Studies suggest that further preclinical investigation of HAT1 as a promising drug target for treating ULMS is warranted, particularly when combined with hyperthermia [179]. And there was a notable contrast in lactate dehydrogenase (LDH) levels observed among patients with benign uterine masses and sarcomas. Serum LDH levels may be elevated in patients with ULMS, but the sensitivity is low, and it is difficult to identify ULMS by LDH detection alone [180, 181]. Di Cello et al. found that LDH3, LDH4, and LDH5 isoenzymes were notably higher in uterine sarcoma patients compared to those with uterine fibroids, while LDH1 and LDH2 were significantly lower [182]. It has been found that in addition to LDH, D-dimer may be elevated in ULMS and C-reactive protein is highly positive, and the combined detection of these three is useful in the differential diagnosis of ULMS and ULM [183].
Studies have demonstrated the involvement of carbonic anhydrases (CAs) in relation to different types of cancer, whereby they aid in regulating pH homeostasis in the microenvironment of tumor cells [184]. CA isoenzymes can be used as a histopathological biomarker for the differential diagnosis of ULMS and ULM. CA isoenzymes are absent in most fibroids, whereas all uterine leiomyosarcomas show positive staining [185].
Discussion
ULMS is a disease that exhibits a high mortality rate, a high recurrence rate, and the prognosis is poor. The pathogenesis of ULMS is still unclear, and there is no specific biomarker that can be used for differentiating it from other analogues. Consequently, the preoperative detection of ULM and ULMS presents significant challenges. ULM are diagnosed by means of pelvic examination, ultrasonography, and, if necessary, enhanced magnetic resonance imaging (MRI), but it is not straightforward to use contrast-enhanced magnetic resonance imaging or other clinical tests to determine the degree of malignancy of uterine smooth muscle tissue or to diagnose a mass [9]. If the above examination is not very definite as ULM, or if the patient's mass is rapidly growing or is postmenopausal, then the possibility of a malignant uterine tumor such as ULMS should also be considered [186]. Clinically, ULMS may be found more incidentally by performing histopathological examination of specimens [10]. Therefore, ULMS and ULM cannot be reliably differentiated due to the absence of specific symptoms or diagnostic imaging studies. Currently, surgery remains the sole viable approach for diagnosis and treatment. Myomectomy is usually performed with minimally invasive procedures such as laparoscopic surgery [10, 11]. Minimally invasive surgery for a presumed benign uterine leiomyoma may result in unexpected intra-abdominal spread of sarcoma, resulting in poor survival [13, 187]. For the safety of the patient with regard to tumors, alternative invasive methods, including procedures based on open surgery, lead to higher morbidity, mortality, and costs for both the patient and the healthcare system [187]. Given these challenges, it is crucial to prioritize the development of highly accurate biomarkers and non-invasive diagnostic methods in fields like gynecology and oncology. The purpose of this review is to analyze the available literature on the genes or proteins that differ between ULM and ULMS to provide valuable insights for the differential diagnosis of ULM and ULMS. It aims to aid in identifying specific diagnostic markers and therapeutic targets for ULMS and ULM, with the potential to establish a foundational basis for future assessment of malignant risk.
The search for a reliable and easily accessible method to distinguish ULMS from ULM has been the subject of much research. By summarizing the previous literature, we found many candidate molecules for the differential diagnosis between ULMS and ULM, and we selected some of them to describe. Compared with ULM, TEM1, MMP-2, p53, p16, Ki67, survivin, GDF-15, MCM2, STMN1, SMO, GLI1, MVP, EMMPRIN, H1.5 and IMP3 were up-regulated in ULMS. DPP6, MFAP5, let-7 miRNA, miR-191-5p, miR-1246, LMP2, PLZF, HAI-1, HAI-2 and CD44V3 were down-regulated in ULMS. Table 1 shows an overview of the biomarkers. In total, 489 genes with differential expression between ULMS and ULM were identified, wherein 416 were notably upregulated while 73 were notably downregulated. These discrepancies primarily participate in the mechanism of genes related to the cell cycle [188]. For instance, ULMS has been linked to numerous rearrangements that target the chromatin remodeling protein ATRX [188]. ATRX expression loss is linked to a telomere phenotype that is selectively prolonged, enabling tumor cells to evade planned cell death. ULMS has a correlation between this mechanism and poor prognosis and overall survival [189, 190]. In addition, the cyclin AURKA also appears to be critical for ULMS pathogenesis, as it inhibits cell-cycle arresting and apoptosis of ULMS cell lines [191]. AURKA is also overexpressed in ovarian and cervical cancer [192, 193].
PD-L1 expression and cytotoxic T-cell infiltration were significantly higher in ULMS compared to ULM, suggesting a possible role for PD-1/PD-L1 checkpoint inhibition in the leiomyosarcoma patient population [194]. The expression of TOP2A, which is significantly elevated in ULMS but not in nonmalignant smooth muscle diseases, may also be an important diagnostic tool for difficult cases in which the diagnosis of ULMS is not clear [195]. The expression of cellular retinol binding protein-1 (CRBP-1) in ULMS is higher than that in ULM, and CRBP-1 overexpression is linked to alterations in signaling molecules for cell proliferation and apoptosis [196]. It has also been suggested that methylation level can be used to distinguish ULMS from ULM. Brany et al. reported that methylation events in benign fibroids and sarcomas should have different patterns, and the KLF4 and DLEC1 genes can be considered as potential methylation biomarkers for uterine fibroids [197].
In recent years, more studies tend to combine multiple indicators to distinguish uterine leiomyoma and uterine leiomyosarcoma. The detection of LDH, D-dimer, and C-reactive protein in combination is beneficial for distinguishing between ULMS and benign lesions. Specificity and positive predictive value were found to be 100% when all three of these markers were combined. In addition, applying these three markers in combination with MRI will allow more accurate diagnosis of ULMS, and prospective studies of these markers and MRI results are warranted in the future [183]. The combined detection of MCM2, Ki67 and p16 immunohistochemistry has good diagnostic value in the differential diagnosis of ULMS [85]. Furthermore, the study of collagen in ULM may also help to distinguish ULM from ULMS. In ULM, there is excessive deposition of ECM, the major component being collagen, which is involved in keeping the tissue's structural integrity [198]. Collagens have the ability to alone or in conjunction with integrins and growth factor-mediated mitogenic pathways to modify the behavior and function of cells [199]. It has been proposed that aberrant collagen fiber orientation and structure are present in ULM, and that changes in collagen genes may contribute to the pathophysiology of leiomyomas [200]. Thus, the microstructure collagen properties of ULM can be characterized using an inventive multidisciplinary method based on Phase-Contrast MicroComputed Tomography, Transmission Electron Microscopy, and Fourier Transform Infrared Imaging Spectroscopy [198].
The treatment and prognosis of ULM and ULMS are different. Accurate differentiation of ULM and ULMS is of great significance for formulating appropriate treatment plans, evaluating prognosis and improving patients' quality of life. While some studies can serve as a basis for future research on the pathophysiology and diagnosis of ULMS, the findings of studies on candidate molecules for the differential diagnosis of ULM and ULMS cannot yet be applied clinically. Based on the molecular properties of ULMS tissue, differentiated diagnosis and tailored treatment can result in an earlier diagnosis and a better prognosis.
Abbreviations
ULM: uterine leiomyomas; ULMS: uterine leiomyosarcomas; TEM1: tumor endothelial marker 1; ECM: extracellular matrix; DPP6: dipeptidyl peptidase like 6; MFAP5: microfibril associated protein 5; GDF-15: growth differentiation factor-15; MIC-1: macrophage inhibitory cytokine-1; MCM2: minichromosome maintenance complex component 2; STMN1: stathmin 1; HH: hedgehog; SMO: smoothened; GLI1: GLI family zinc finger 1; miRNAs: microRNAs; let-7: lethal-7; MVP: major vault protein; LRP: lung resistance-related protein; IHC: immunohistochemistry; LMP2/β1i: large multifunctional protease 2; IFN-γ: interferon-γ; STAT: signal transducer and activator of transcription; IRF1: interferon regulatory factor 1; EMMPRINE: extracellular matrix metalloproteinase Inducer; MMPs: matrix metalloproteinases; PLZF: promyelocytic leukemia zinc finger; IMP3: insulin-like growth factor Ⅱ mRNA binding protein 3; HAI: hepatocyte growth factor activator inhibitors; HGFA: hepatocyte growth factor activator; HGF: hepatocyte growth factor; CD44s: CD44 standard; CD44v: CD44 variants; DFF40: DNA fragmentation factors 40; Bcl2: B-cell lymphoma 2; MED12: mediator complex subunit 12; PR: progesterone receptor; ER: estrogen receptor; WT1: Wilms' tumor 1; HAT1: hyperthermia inhibited histone acetyltransferase 1; LDH: lactate dehydrogenase; CAs: carbonic anhydrases; MRI: magnetic resonance imaging; CRBP-1: cellular retinol binding protein-1.
Author Contributions
JT designed the study. JG drafted the manuscript. JZ revised the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Buttram VC Jr, Reiter RC. Uterine leiomyomata: etiology, symptomatology, and management. Fertil Steril. 1981;36:433-45
2. Stewart EA. Uterine fibroids. Lancet. 2001;357:293-8
3. Walker CL, Stewart EA. Uterine fibroids: the elephant in the room. Science. 2005;308:1589-92
4. Lumsden MA, Hamoodi I, Gupta J, Hickey M. Fibroids: diagnosis and management. Bmj. 2015;351:h4887
5. Mallmann P. Uterine Sarcoma - Difficult to Diagnose, Hard to Treat. Oncol Res Treat. 2018;41:674
6. Hosh M, Antar S, Nazzal A, Warda M, Gibreel A, Refky B. Uterine Sarcoma: Analysis of 13,089 Cases Based on Surveillance, Epidemiology, and End Results Database. Int J Gynecol Cancer. 2016;26:1098-104
7. Mas A, Simón C. Molecular differential diagnosis of uterine leiomyomas and leiomyosarcomas. Biol Reprod. 2019;101:1115-23
8. Halaska MJ, Haidopoulos D, Guyon F, Morice P, Zapardiel I, Kesic V. European Society of Gynecological Oncology Statement on Fibroid and Uterine Morcellation. Int J Gynecol Cancer. 2017;27:189-92
9. Seagle BL, Alexander AL, Strohl AE, Shahabi S. Discussing sarcoma risks during informed consent for nonhysterectomy management of fibroids: an unmet need. Am J Obstet Gynecol. 2018;218:103.e1-e5
10. Bhave Chittawar P, Franik S, Pouwer AW, Farquhar C. Minimally invasive surgical techniques versus open myomectomy for uterine fibroids. Cochrane Database Syst Rev. 2014: Cd004638.
11. Siedhoff MT, Doll KM, Clarke-Pearson DL, Rutstein SE. Laparoscopic hysterectomy with morcellation vs abdominal hysterectomy for presumed fibroids: an updated decision analysis following the 2014 Food and Drug Administration safety communications. Am J Obstet Gynecol. 2017;216:259.e1-e6
12. George S, Serrano C, Hensley ML, Ray-Coquard I. Soft Tissue and Uterine Leiomyosarcoma. J Clin Oncol. 2018;36:144-50
13. Ricci S, Stone RL, Fader AN. Uterine leiomyosarcoma: Epidemiology, contemporary treatment strategies and the impact of uterine morcellation. Gynecol Oncol. 2017;145:208-16
14. AAGL practice report. Morcellation during uterine tissue extraction. J Minim Invasive Gynecol. 2014;21:517-30
15. Mori KM, Abaid LN, Mendivil AA, Brown JV 3rd, Beck TL, Micha JP. et al. The incidence of occult malignancy following uterine morcellation: A ten-year single institution experience retrospective cohort study. Int J Surg. 2018;53:239-42
16. Ottarsdottir H, Cohen SL, Cox M, Vitonis A, Einarsson JI. Trends in Mode of Hysterectomy After the U.S. Food and Drug Administration Power Morcellation Advisory. Obstet Gynecol. 2017;129:1014-21
17. Major FJ, Blessing JA, Silverberg SG, Morrow CP, Creasman WT, Currie JL. et al. Prognostic factors in early-stage uterine sarcoma. A Gynecologic Oncology Group study. Cancer. 1993;71:1702-9
18. Sagae S, Yamashita K, Ishioka S, Nishioka Y, Terasawa K, Mori M. et al. Preoperative diagnosis and treatment results in 106 patients with uterine sarcoma in Hokkaido, Japan. Oncology. 2004;67:33-9
19. Chen I, Firth B, Hopkins L, Bougie O, Xie RH, Singh S. Clinical Characteristics Differentiating Uterine Sarcoma and Fibroids. Jsls. 2018 22
20. Bagley RG, Honma N, Weber W, Boutin P, Rouleau C, Shankara S. et al. Endosialin/TEM 1/CD248 is a pericyte marker of embryonic and tumor neovascularization. Microvasc Res. 2008;76:180-8
21. MacFadyen JR, Haworth O, Roberston D, Hardie D, Webster MT, Morris HR. et al. Endosialin (TEM1, CD248) is a marker of stromal fibroblasts and is not selectively expressed on tumour endothelium. FEBS Lett. 2005;579:2569-75
22. Huber MA, Kraut N, Schweifer N, Dolznig H, Peter RU, Schubert RD. et al. Expression of stromal cell markers in distinct compartments of human skin cancers. J Cutan Pathol. 2006;33:145-55
23. Rmali KA, Puntis MC, Jiang WG. Prognostic values of tumor endothelial markers in patients with colorectal cancer. World J Gastroenterol. 2005;11:1283-6
24. Rouleau C, Gianolio DA, Smale R, Roth SD, Krumbholz R, Harper J. et al. Anti-Endosialin Antibody-Drug Conjugate: Potential in Sarcoma and Other Malignancies. Mol Cancer Ther. 2015;14:2081-9
25. Tomkowicz B, Rybinski K, Foley B, Ebel W, Kline B, Routhier E. et al. Interaction of endosialin/TEM1 with extracellular matrix proteins mediates cell adhesion and migration. Proc Natl Acad Sci U S A. 2007;104:17965-70
26. Guo Y, Hu J, Wang Y, Peng X, Min J, Wang J. et al. Tumour endothelial marker 1/endosialin-mediated targeting of human sarcoma. Eur J Cancer. 2018;90:111-21
27. Wu C, Sun W, Shen D, Li H, Tong X, Guo Y. TEM1 up-regulates MMP-2 and promotes ECM remodeling for facilitating invasion and migration of uterine sarcoma. Discov Oncol. 2023;14:5
28. Song Z, Wang J, Su Q, Luan M, Chen X, Xu X. The role of MMP-2 and MMP-9 in the metastasis and development of hypopharyngeal carcinoma. Braz J Otorhinolaryngol. 2021;87:521-8
29. Middleton JD, Sivakumar S, Hai T. Chemotherapy-Induced Changes in the Lung Microenvironment: The Role of MMP-2 in Facilitating Intravascular Arrest of Breast Cancer Cells. Int J Mol Sci. 2021 22
30. Aktary Z, Alaee M, Pasdar M. Beyond cell-cell adhesion: Plakoglobin and the regulation of tumorigenesis and metastasis. Oncotarget. 2017;8:32270-91
31. Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18:533-48
32. Strop P, Bankovich AJ, Hansen KC, Garcia KC, Brunger AT. Structure of a human A-type potassium channel interacting protein DPPX, a member of the dipeptidyl aminopeptidase family. J Mol Biol. 2004;343:1055-65
33. Li Q, Zhang Y, Jiang Q. MFAP5 suppression inhibits migration/invasion, regulates cell cycle and induces apoptosis via promoting ROS production in cervical cancer. Biochem Biophys Res Commun. 2018;507:51-8
34. Ke Y, You L, Xu Y, Wu D, Lin Q, Wu Z. DPP6 and MFAP5 are associated with immune infiltration as diagnostic biomarkers in distinguishing uterine leiomyosarcoma from leiomyoma. Front Oncol. 2022;12:1084192
35. Shen X, Yang Z, Feng S, Li Y. Identification of uterine leiomyosarcoma-associated hub genes and immune cell infiltration pattern using weighted co-expression network analysis and CIBERSORT algorithm. World J Surg Oncol. 2021;19:223
36. Zhang C, Zheng JH, Lin ZH, Lv HY, Ye ZM, Chen YP. et al. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of osteosarcoma. Aging (Albany NY). 2020;12:3486-501
37. Yao S, Cheng TD, Elkhanany A, Yan L, Omilian A, Abrams SI. et al. Breast Tumor Microenvironment in Black Women: A Distinct Signature of CD8+ T-Cell Exhaustion. J Natl Cancer Inst. 2021;113:1036-43
38. Xiang A, Lin X, Xu L, Chen H, Guo J, Zhou F. PCOLCE Is Potent Prognostic Biomarker and Associates With Immune Infiltration in Gastric Cancer. Front Mol Biosci. 2020;7:544895
39. Choy TK, Wang CY, Phan NN, Khoa Ta HD, Anuraga G, Liu YH. et al. Identification of Dipeptidyl Peptidase (DPP) Family Genes in Clinical Breast Cancer Patients via an Integrated Bioinformatics Approach. Diagnostics (Basel). 2021 11
40. Cook L, Melamed A, Yaguchi H, Bangham CR. The impact of HTLV-1 on the cellular genome. Curr Opin Virol. 2017;26:125-31
41. Rubisz P, Ciebiera M, Hirnle L, Zgliczyńska M, Łoziński T, Dzięgiel P. et al. The Usefulness of Immunohistochemistry in the Differential Diagnosis of Lesions Originating from the Myometrium. Int J Mol Sci. 2019 20
42. Lee CH, Turbin DA, Sung YC, Espinosa I, Montgomery K, van de Rijn M. et al. A panel of antibodies to determine site of origin and malignancy in smooth muscle tumors. Mod Pathol. 2009;22:1519-31
43. Köbel M, Ronnett BM, Singh N, Soslow RA, Gilks CB, McCluggage WG. Interpretation of P53 Immunohistochemistry in Endometrial Carcinomas: Toward Increased Reproducibility. Int J Gynecol Pathol. 2019;38(Suppl 1):S123-s31
44. Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000-5
45. de Vos S, Wilczynski SP, Fleischhacker M, Koeffler P. p53 alterations in uterine leiomyosarcomas versus leiomyomas. Gynecol Oncol. 1994;54:205-8
46. Nordal RR, Kristensen GB, Stenwig AE, Tropé CG, Nesland JM. Immunohistochemical analysis of p53 protein in uterine sarcomas. Gynecol Oncol. 1998;70:45-8
47. O'Neill CJ, McBride HA, Connolly LE, McCluggage WG. Uterine leiomyosarcomas are characterized by high p16, p53 and MIB1 expression in comparison with usual leiomyomas, leiomyoma variants and smooth muscle tumours of uncertain malignant potential. Histopathology. 2007;50:851-8
48. Niemann TH, Raab SS, Lenel JC, Rodgers JR, Robinson RA. p53 protein overexpression in smooth muscle tumors of the uterus. Hum Pathol. 1995;26:375-9
49. Delgado B, Dreiher J, Braiman D, Meirovitz M, Shaco-Levy R. P16, Ki67, P53, and WT1 Expression in Uterine Smooth Muscle Tumors: An Adjunct in Confirming the Diagnosis of Malignancy in Ambiguous Cases. Int J Gynecol Pathol. 2021;40:257-62
50. Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753-6
51. Atkins KA, Arronte N, Darus CJ, Rice LW. The Use of p16 in enhancing the histologic classification of uterine smooth muscle tumors. Am J Surg Pathol. 2008;32:98-102
52. Skubitz KM, Skubitz AP. Differential gene expression in leiomyosarcoma. Cancer. 2003;98:1029-38
53. Ünver NU, Acikalin MF, Öner Ü, Ciftci E, Ozalp SS, Colak E. Differential expression of P16 and P21 in benign and malignant uterine smooth muscle tumors. Arch Gynecol Obstet. 2011;284:483-90
54. Sobecki M, Mrouj K, Colinge J, Gerbe F, Jay P, Krasinska L. et al. Cell-Cycle Regulation Accounts for Variability in Ki-67 Expression Levels. Cancer Res. 2017;77:2722-34
55. Rudolph P, Peters J, Lorenz D, Schmidt D, Parwaresch R. Correlation between mitotic and Ki-67 labeling indices in paraffin-embedded carcinoma specimens. Hum Pathol. 1998;29:1216-22
56. Di Donato V, Iacobelli V, Schiavi MC, Colagiovanni V, Pecorella I, Palaia I. et al. Impact of Hormone Receptor Status and Ki-67 Expression on Disease-Free Survival in Patients Affected by High-risk Endometrial Cancer. Int J Gynecol Cancer. 2018;28:505-13
57. Liu P, Sun YL, Du J, Hou XS, Meng H. CD105/Ki67 coexpression correlates with tumor progression and poor prognosis in epithelial ovarian cancer. Int J Gynecol Cancer. 2012;22:586-92
58. Ovestad IT, Dalen I, Hansen E, Loge JL, Dybdahl BM, Dirdal MB. et al. Clinical value of fully automated p16/Ki-67 dual staining in the triage of HPV-positive women in the Norwegian Cervical Cancer Screening Program. Cancer Cytopathol. 2017;125:283-91
59. Pezzilli R, Partelli S, Cannizzaro R, Pagano N, Crippa S, Pagnanelli M. et al. Ki-67 prognostic and therapeutic decision driven marker for pancreatic neuroendocrine neoplasms (PNENs): A systematic review. Adv Med Sci. 2016;61:147-53
60. Hu X, Zhang H, Zheng X, Lin Z, Feng G, Chen Y. et al. STMN1 and MKI67 Are Upregulated in Uterine Leiomyosarcoma and Are Potential Biomarkers for its Diagnosis. Med Sci Monit. 2020;26:e923749
61. Chen L, Yang B. Immunohistochemical analysis of p16, p53, and Ki-67 expression in uterine smooth muscle tumors. Int J Gynecol Pathol. 2008;27:326-32
62. Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917-21
63. Gianani R, Jarboe E, Orlicky D, Frost M, Bobak J, Lehner R. et al. Expression of survivin in normal, hyperplastic, and neoplastic colonic mucosa. Hum Pathol. 2001;32:119-25
64. Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127-34
65. Islam A, Kageyama H, Takada N, Kawamoto T, Takayasu H, Isogai E. et al. High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617-23
66. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70
67. Deveraux QL, Reed JC. IAP family proteins-suppressors of apoptosis. Genes Dev. 1999;13:239-52
68. Osaka E, Suzuki T, Osaka S, Yoshida Y, Sugita H, Asami S. et al. Survivin as a prognostic factor for osteosarcoma patients. Acta Histochem Cytochem. 2006;39:95-100
69. Marioni G, Bertolin A, Giacomelli L, Marchese-Ragona R, Savastano M, Calgaro N. et al. Expression of the apoptosis inhibitor protein Survivin in primary laryngeal carcinoma and cervical lymph node metastasis. Anticancer Res. 2006;26:3813-7
70. Zhu ZB, Makhija SK, Lu B, Wang M, Kaliberova L, Liu B. et al. Transcriptional targeting of tumors with a novel tumor-specific survivin promoter. Cancer Gene Ther. 2004;11:256-62
71. Shalaby S, Khater M, Laknaur A, Arbab A, Al-Hendy A. Molecular Bio-Imaging Probe for Non-Invasive Differentiation Between Human Leiomyoma Versus Leiomyosarcoma. Reprod Sci. 2020;27:644-54
72. Bao R, Connolly DC, Murphy M, Green J, Weinstein JK, Pisarcik DA. et al. Activation of cancer-specific gene expression by the survivin promoter. J Natl Cancer Inst. 2002;94:522-8
73. Bauskin AR, Brown DA, Kuffner T, Johnen H, Luo XW, Hunter M. et al. Role of macrophage inhibitory cytokine-1 in tumorigenesis and diagnosis of cancer. Cancer Res. 2006;66:4983-6
74. Staff AC, Bock AJ, Becker C, Kempf T, Wollert KC, Davidson B. Growth differentiation factor-15 as a prognostic biomarker in ovarian cancer. Gynecol Oncol. 2010;118:237-43
75. Staff AC, Trovik J, Eriksson AG, Wik E, Wollert KC, Kempf T. et al. Elevated plasma growth differentiation factor-15 correlates with lymph node metastases and poor survival in endometrial cancer. Clin Cancer Res. 2011;17:4825-33
76. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
77. Bootcov MR, Bauskin AR, Valenzuela SM, Moore AG, Bansal M, He XY. et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci U S A. 1997;94:11514-9
78. Trovik J, Salvesen HB, Cuppens T, Amant F, Staff AC. Growth differentiation factor-15 as biomarker in uterine sarcomas. Int J Gynecol Cancer. 2014;24:252-9
79. Agarwal MK, Hastak K, Jackson MW, Breit SN, Stark GR, Agarwal ML. Macrophage inhibitory cytokine 1 mediates a p53-dependent protective arrest in S phase in response to starvation for DNA precursors. Proc Natl Acad Sci U S A. 2006;103:16278-83
80. Yang J, Ramnath N, Moysich KB, Asch HL, Swede H, Alrawi SJ. et al. Prognostic significance of MCM2, Ki-67 and gelsolin in non-small cell lung cancer. BMC Cancer. 2006;6:203
81. Tsuruga H, Yabuta N, Hashizume K, Ikeda M, Endo Y, Nojima H. Expression, nuclear localization and interactions of human MCM/P1 proteins. Biochem Biophys Res Commun. 1997;236:118-25
82. Cho Mar K, Eimoto T, Nagaya S, Tateyama H. Cell proliferation marker MCM2, but not Ki67, is helpful for distinguishing between minimally invasive follicular carcinoma and follicular adenoma of the thyroid. Histopathology. 2006;48:801-7
83. Yousef EM, Furrer D, Laperriere DL, Tahir MR, Mader S, Diorio C. et al. MCM2: An alternative to Ki-67 for measuring breast cancer cell proliferation. Mod Pathol. 2017;30:682-97
84. Quade BJ, Wang TY, Sornberger K, Dal Cin P, Mutter GL, Morton CC. Molecular pathogenesis of uterine smooth muscle tumors from transcriptional profiling. Genes Chromosomes Cancer. 2004;40:97-108
85. Keyhanian K, Lage JM, Chernetsova E, Sekhon H, Eslami Z, Islam S. Combination of MCM2 With Ki67 and p16 Immunohistochemistry Can Distinguish Uterine Leiomyosarcomas. Int J Gynecol Pathol. 2020;39:354-61
86. Rana S, Maples PB, Senzer N, Nemunaitis J. Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev Anticancer Ther. 2008;8:1461-70
87. Wang J, Yao Y, Ming Y, Shen S, Wu N, Liu J. et al. Downregulation of stathmin 1 in human gallbladder carcinoma inhibits tumor growth in vitro and in vivo. Sci Rep. 2016;6:28833
88. Suzuki K, Watanabe A, Araki K, Yokobori T, Harimoto N, Gantumur D. et al. High STMN1 Expression Is Associated with Tumor Differentiation and Metastasis in Clinical Patients with Pancreatic Cancer. Anticancer Res. 2018;38:939-44
89. Wei SH, Lin F, Wang X, Gao P, Zhang HZ. Prognostic significance of stathmin expression in correlation with metastasis and clinicopathological characteristics in human ovarian carcinoma. Acta Histochem. 2008;110:59-65
90. Werner HM, Trovik J, Halle MK, Wik E, Akslen LA, Birkeland E. et al. Stathmin protein level, a potential predictive marker for taxane treatment response in endometrial cancer. PLoS One. 2014;9:e90141
91. Wang X, Ren JH, Lin F, Wei JX, Long M, Yan L. et al. Stathmin is involved in arsenic trioxide-induced apoptosis in human cervical cancer cell lines via PI3K linked signal pathway. Cancer Biol Ther. 2010;10:632-43
92. Hsieh SY, Huang SF, Yu MC, Yeh TS, Chen TC, Lin YJ. et al. Stathmin1 overexpression associated with polyploidy, tumor-cell invasion, early recurrence, and poor prognosis in human hepatoma. Mol Carcinog. 2010;49:476-87
93. Hemdan T, Lindén M, Lind SB, Namuduri AV, Sjöstedt E, de Ståhl TD. et al. The prognostic value and therapeutic target role of stathmin-1 in urinary bladder cancer. Br J Cancer. 2014;111:1180-7
94. Allen MM, Douds JJ, Liang SX, Desouki MM, Parkash V, Fadare O. An immunohistochemical analysis of stathmin 1 expression in uterine smooth muscle tumors: differential expression in leiomyosarcomas and leiomyomas. Int J Clin Exp Pathol. 2015;8:2795-801
95. Hwang H, Matsuo K, Duncan K, Pakzamir E, Pham HQ, Correa A. et al. Immunohistochemical panel to differentiate endometrial stromal sarcoma, uterine leiomyosarcoma and leiomyoma: something old and something new. J Clin Pathol. 2015;68:710-7
96. Garcia N, Bozzini N, Baiocchi G, da Cunha IW, Maciel GA, Soares Junior JM. et al. May Sonic Hedgehog proteins be markers for malignancy in uterine smooth muscle tumors? Hum Pathol. 2016;50:43-50
97. Garcia N, Al-Hendy A, Baracat EC, Carvalho KC, Yang Q. Targeting Hedgehog Pathway and DNA Methyltransferases in Uterine Leiomyosarcoma Cells. Cells. 2020 10
98. Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn J Basic Med Sci. 2018;18:8-20
99. Carballo GB, Honorato JR, de Lopes GPF, Spohr T. A highlight on Sonic hedgehog pathway. Cell Commun Signal. 2018;16:11
100. Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer. 2006;42:437-45
101. Yin VT, Esmaeli B. Targeting the Hedgehog Pathway for Locally Advanced and Metastatic Basal Cell Carcinoma. Curr Pharm Des. 2017;23:655-9
102. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126-39
103. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376-85
104. Torres A, Torres K, Maciejewski R, Harvey WH. MicroRNAs and their role in gynecological tumors. Med Res Rev. 2011;31:895-923
105. Gonzalez Dos Anjos L, de Almeida BC, Gomes de Almeida T, Mourão Lavorato Rocha A, De Nardo Maffazioli G, Soares FA. et al. Could miRNA Signatures be Useful for Predicting Uterine Sarcoma and Carcinosarcoma Prognosis and Treatment? Cancers (Basel). 2018 10
106. Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126:1163-72
107. Wang X, Cao L, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (Review). Oncol Lett. 2012;3:955-60
108. de Almeida BC, Dos Anjos LG, Uno M, Cunha IWD, Soares FA, Baiocchi G. et al. Let-7 miRNA's Expression Profile and Its Potential Prognostic Role in Uterine Leiomyosarcoma. Cells. 2019 8
109. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
110. Barh D, Malhotra R, Ravi B, Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol. 2010;17:70-80
111. Yokoi A, Matsuzaki J, Yamamoto Y, Tate K, Yoneoka Y, Shimizu H. et al. Serum microRNA profile enables preoperative diagnosis of uterine leiomyosarcoma. Cancer Sci. 2019;110:3718-26
112. Scheffer GL, Wijngaard PL, Flens MJ, Izquierdo MA, Slovak ML, Pinedo HM. et al. The drug resistance-related protein LRP is the human major vault protein. Nat Med. 1995;1:578-82
113. Chen YL, Yang TY, Wu CL, Chen KC, Hsu SL, Hsueh CM. Mechanisms underlying lung resistance-related protein (LRP)-mediated doxorubicin resistance of non-small cell lung cancer cells. Chin J Physiol. 2016;59:331-47
114. Lintel NJ, Luebker SA, Lele SM, Koepsell SA. MVP immunohistochemistry is a useful adjunct in distinguishing leiomyosarcoma from leiomyoma and leiomyoma with bizarre nuclei. Hum Pathol. 2018;73:122-7
115. Chen X, Mao Y, Guo Y, Xiao D, Lin Z, Huang Y. et al. LMP2 deficiency causes abnormal metabolism, oxidative stress, neuroinflammation, myelin loss and neurobehavioral dysfunctions. J Transl Med. 2023;21:226
116. Hayashi T, Faustman DL. Development of spontaneous uterine tumors in low molecular mass polypeptide-2 knockout mice. Cancer Res. 2002;62:24-7
117. Hayashi T, Horiuchi A, Sano K, Hiraoka N, Ichimura T, Sudo T. et al. Potential diagnostic biomarkers: differential expression of LMP2/β1i and cyclin B1 in human uterine leiomyosarcoma. Tumori. 2014;100:99e-106e
118. Hayashi T, Horiuchi A, Sano K, Hiraoka N, Kanai Y, Shiozawa T. et al. Molecular Approach to Uterine Leiomyosarcoma: LMP2-Deficient Mice as an Animal Model of Spontaneous Uterine Leiomyosarcoma. Sarcoma. 2011;2011:476498
119. Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ. et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107-11
120. Delp K, Momburg F, Hilmes C, Huber C, Seliger B. Functional deficiencies of components of the MHC class I antigen pathway in human tumors of epithelial origin. Bone Marrow Transplant. 2000;25(Suppl 2):S88-95
121. Nakajima C, Uekusa Y, Iwasaki M, Yamaguchi N, Mukai T, Gao P. et al. A role of interferon-gamma (IFN-gamma) in tumor immunity: T cells with the capacity to reject tumor cells are generated but fail to migrate to tumor sites in IFN-gamma-deficient mice. Cancer Res. 2001;61:3399-405
122. Brucet M, Marqués L, Sebastián C, Lloberas J, Celada A. Regulation of murine Tap1 and Lmp2 genes in macrophages by interferon gamma is mediated by STAT1 and IRF-1. Genes Immun. 2004;5:26-35
123. Hayashi T, Kobayashi Y, Kohsaka S, Sano K. The mutation in the ATP-binding region of JAK1, identified in human uterine leiomyosarcomas, results in defective interferon-gamma inducibility of TAP1 and LMP2. Oncogene. 2006;25:4016-26
124. Hayashi T, Horiuchi A, Sano K, Hiraoka N, Kasai M, Ichimura T. et al. Potential role of LMP2 as tumor-suppressor defines new targets for uterine leiomyosarcoma therapy. Sci Rep. 2011;1:180
125. Xiong L, Edwards CK 3rd, Zhou L. The biological function and clinical utilization of CD147 in human diseases: a review of the current scientific literature. Int J Mol Sci. 2014;15:17411-41
126. Muramatsu T, Miyauchi T. Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol Histopathol. 2003;18:981-7
127. Taylor PM, Woodfield RJ, Hodgkin MN, Pettitt TR, Martin A, Kerr DJ. et al. Breast cancer cell-derived EMMPRIN stimulates fibroblast MMP2 release through a phospholipase A(2) and 5-lipoxygenase catalyzed pathway. Oncogene. 2002;21:5765-72
128. Shapiro SD. Matrix metalloproteinase degradation of extracellular matrix: biological consequences. Curr Opin Cell Biol. 1998;10:602-8
129. Riethdorf S, Reimers N, Assmann V, Kornfeld JW, Terracciano L, Sauter G. et al. High incidence of EMMPRIN expression in human tumors. Int J Cancer. 2006;119:1800-10
130. Tsai WC, Chao YC, Lee WH, Chen A, Sheu LF, Jin JS. Increasing EMMPRIN and matriptase expression in hepatocellular carcinoma: tissue microarray analysis of immunohistochemical scores with clinicopathological parameters. Histopathology. 2006;49:388-95
131. Tsai WC, Chao YC, Sheu LF, Lin YF, Nieh S, Chen A. et al. EMMPRIN and fascin overexpression associated with clinicopathologic parameters of pancreatobiliary adenocarcinoma in Chinese people. Apmis. 2007;115:929-38
132. Zheng HC, Takahashi H, Murai Y, Cui ZG, Nomoto K, Miwa S. et al. Upregulated EMMPRIN/CD147 might contribute to growth and angiogenesis of gastric carcinoma: a good marker for local invasion and prognosis. Br J Cancer. 2006;95:1371-8
133. Kefeli M, Yildiz L, Gun S, Ozen FZ, Karagoz F. EMMPRIN (CD147) Expression in Smooth Muscle Tumors of the Uterus. Int J Gynecol Pathol. 2016;35:1-7
134. Ozler A, Evsen MS, Turgut A, Sak ME, Tunc SY, Agacayak E. et al. CD147 expression in uterine smooth muscle tumors, and its potential role as a diagnostic and prognostic marker in patients with leiomyosarcoma. J Exp Ther Oncol. 2014;10:325-30
135. Yeyati PL, Shaknovich R, Boterashvili S, Li J, Ball HJ, Waxman S. et al. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18:925-34
136. Fahnenstich J, Nandy A, Milde-Langosch K, Schneider-Merck T, Walther N, Gellersen B. Promyelocytic leukaemia zinc finger protein (PLZF) is a glucocorticoid- and progesterone-induced transcription factor in human endometrial stromal cells and myometrial smooth muscle cells. Mol Hum Reprod. 2003;9:611-23
137. Koken MH, Reid A, Quignon F, Chelbi-Alix MK, Davies JM, Kabarowski JH. et al. Leukemia-associated retinoic acid receptor alpha fusion partners, PML and PLZF, heterodimerize and colocalize to nuclear bodies. Proc Natl Acad Sci U S A. 1997;94:10255-60
138. Felicetti F, Bottero L, Felli N, Mattia G, Labbaye C, Alvino E. et al. Role of PLZF in melanoma progression. Oncogene. 2004;23:4567-76
139. Hechtman JF, Beasley MB, Kinoshita Y, Ko HM, Hao K, Burstein DE. Promyelocytic leukemia zinc finger and histone H1.5 differentially stain low- and high-grade pulmonary neuroendocrine tumors: a pilot immunohistochemical study. Hum Pathol. 2013;44:1400-5
140. Hizume K, Yoshimura SH, Takeyasu K. Linker histone H1 per se can induce three-dimensional folding of chromatin fiber. Biochemistry. 2005;44:12978-89
141. Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1-12
142. Sato S, Takahashi S, Asamoto M, Nakanishi M, Wakita T, Ogura Y. et al. Histone H1 expression in human prostate cancer tissues and cell lines. Pathol Int. 2012;62:84-92
143. Momeni M, Kalir T, Farag S, Kinoshita Y, Roman TY, Chuang L. et al. Immunohistochemical detection of promyelocytic leukemia zinc finger and histone 1.5 in uterine leiomyosarcoma and leiomyoma. Reprod Sci. 2014;21:1171-6
144. Jiang Z, Chu PG, Woda BA, Rock KL, Liu Q, Hsieh CC. et al. Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: a retrospective study. Lancet Oncol. 2006;7:556-64
145. Jiang Z, Lohse CM, Chu PG, Wu CL, Woda BA, Rock KL. et al. Oncofetal protein IMP3: a novel molecular marker that predicts metastasis of papillary and chromophobe renal cell carcinomas. Cancer. 2008;112:2676-82
146. Zheng W, Yi X, Fadare O, Liang SX, Martel M, Schwartz PE. et al. The oncofetal protein IMP3: a novel biomarker for endometrial serous carcinoma. Am J Surg Pathol. 2008;32:304-15
147. Vikesaa J, Hansen TV, Jønson L, Borup R, Wewer UM, Christiansen J. et al. RNA-binding IMPs promote cell adhesion and invadopodia formation. Embo j. 2006;25:1456-68
148. Cornejo K, Shi M, Jiang Z. Oncofetal protein IMP3: a useful diagnostic biomarker for leiomyosarcoma. Hum Pathol. 2012;43:1567-72
149. Li C, Zota V, Woda BA, Rock KL, Fraire AE, Jiang Z. et al. Expression of a novel oncofetal mRNA-binding protein IMP3 in endometrial carcinomas: diagnostic significance and clinicopathologic correlations. Mod Pathol. 2007;20:1263-8
150. Kataoka H, Itoh H, Nuki Y, Hamasuna R, Naganuma S, Kitamura N. et al. Mouse hepatocyte growth factor (HGF) activator inhibitor type 2 lacking the first Kunitz domain potently inhibits the HGF activator. Biochem Biophys Res Commun. 2002;290:1096-100
151. Shimomura T, Denda K, Kitamura A, Kawaguchi T, Kito M, Kondo J. et al. Hepatocyte growth factor activator inhibitor, a novel Kunitz-type serine protease inhibitor. J Biol Chem. 1997;272:6370-6
152. Netzel-Arnett S, Hooper JD, Szabo R, Madison EL, Quigley JP, Bugge TH. et al. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237-58
153. Nakamura K, Abarzua F, Hongo A, Kodama J, Nasu Y, Kumon H. et al. Hepatocyte growth factor activator inhibitors (HAI-1 and HAI-2) are potential targets in uterine leiomyosarcoma. Int J Oncol. 2010;37:605-14
154. Jeffers M, Rong S, Vande Woude GF. Enhanced tumorigenicity and invasion-metastasis by hepatocyte growth factor/scatter factor-met signalling in human cells concomitant with induction of the urokinase proteolysis network. Mol Cell Biol. 1996;16:1115-25
155. Nagakawa O, Yamagishi T, Fujiuchi Y, Junicho A, Akashi T, Nagaike K. et al. Serum hepatocyte growth factor activator (HGFA) in benign prostatic hyperplasia and prostate cancer. Eur Urol. 2005;48:686-90
156. Parr C, Watkins G, Mansel RE, Jiang WG. The hepatocyte growth factor regulatory factors in human breast cancer. Clin Cancer Res. 2004;10:202-11
157. Nakamura K, Abarzua F, Kodama J, Hongo A, Nasu Y, Kumon H. et al. Expression of hepatocyte growth factor activator inhibitors (HAI-1 and HAI-2) in ovarian cancer. Int J Oncol. 2009;34:345-53
158. Nakamura K, Abarzua F, Hongo A, Kodama J, Nasu Y, Kumon H. et al. The role of hepatocyte growth factor activator inhibitor-1 (HAI-1) as a prognostic indicator in cervical cancer. Int J Oncol. 2009;35:239-48
159. Szabo R, Hobson JP, List K, Molinolo A, Lin CY, Bugge TH. Potent inhibition and global co-localization implicate the transmembrane Kunitz-type serine protease inhibitor hepatocyte growth factor activator inhibitor-2 in the regulation of epithelial matriptase activity. J Biol Chem. 2008;283:29495-504
160. Günthert U. CD44: a multitude of isoforms with diverse functions. Curr Top Microbiol Immunol. 1993;184:47-63
161. Matsumura Y, Tarin D. Significance of CD44 gene products for cancer diagnosis and disease evaluation. Lancet. 1992;340:1053-8
162. Fox SB, Gatter KC, Jackson DG, Screaton GR, Bell MV, Bell JI. et al. CD44 and cancer screening. Lancet. 1993;342:548-9
163. Saegusa M, Hashimura M, Machida D, Okayasu I. Down-regulation of CD44 standard and variant isoforms during the development and progression of uterine cervical tumours. J Pathol. 1999;187:173-83
164. Poncelet C, Walker F, Madelenat P, Bringuier AF, Scoazec JY, Feldmann G. et al. Expression of CD44 standard and isoforms V3 and V6 in uterine smooth muscle tumors: a possible diagnostic tool for the diagnosis of leiomyosarcoma. Hum Pathol. 2001;32:1190-6
165. Zhai YL, Nikaido T, Toki T, Shiozawa A, Orii A, Fujii S. Prognostic significance of bcl-2 expression in leiomyosarcoma of the uterus. Br J Cancer. 1999;80:1658-64
166. Banas T, Pitynski K, Okon K, Czerw A. DNA fragmentation factors 40 and 45 (DFF40/DFF45) and B-cell lymphoma 2 (Bcl-2) protein are underexpressed in uterine leiomyosarcomas and may predict survival. Onco Targets Ther. 2017;10:4579-89
167. Soltan MM, Albasry AM, Eldosouky MK, Abdelhamid HS. Immunoexpression of progesterone receptor, epithelial growth factor receptor and galectin-3 in uterine smooth muscle tumors. Cell Mol Biol (Noisy-le-grand). 2018;64:7-12
168. Maltese G, Fontanella C, Lepori S, Scaffa C, Fucà G, Bogani G. et al. Atypical Uterine Smooth Muscle Tumors: A Retrospective Evaluation of Clinical and Pathologic Features. Oncology. 2018;94:1-6
169. Bodner K, Bodner-Adler B, Kimberger O, Czerwenka K, Mayerhofer K. Bcl-2 expression and other clinicopathologic parameters in uterine leiomyosarcoma. Wien Klin Wochenschr. 2004;116:135-9
170. Cekanova M, Fernando RI, Siriwardhana N, Sukhthankar M, Parra C, Woraratphoka J. et al. BCL-2 family protein, BAD is down-regulated in breast cancer and inhibits cell invasion. Exp Cell Res. 2015;331:1-10
171. Clement PB, Young RH. Non-endometrioid carcinomas of the uterine corpus: a review of their pathology with emphasis on recent advances and problematic aspects. Adv Anat Pathol. 2004;11:117-42
172. Ravegnini G, Mariño-Enriquez A, Slater J, Eilers G, Wang Y, Zhu M. et al. MED12 mutations in leiomyosarcoma and extrauterine leiomyoma. Mod Pathol. 2013;26:743-9
173. Mäkinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ. et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252-5
174. Zhang Q, Ubago J, Li L, Guo H, Liu Y, Qiang W. et al. Molecular analyses of 6 different types of uterine smooth muscle tumors: Emphasis in atypical leiomyoma. Cancer. 2014;120:3165-77
175. Bertsch E, Qiang W, Zhang Q, Espona-Fiedler M, Druschitz S, Liu Y. et al. MED12 and HMGA2 mutations: two independent genetic events in uterine leiomyoma and leiomyosarcoma. Mod Pathol. 2014;27:1144-53
176. Pérot G, Croce S, Ribeiro A, Lagarde P, Velasco V, Neuville A. et al. MED12 alterations in both human benign and malignant uterine soft tissue tumors. PLoS One. 2012;7:e40015
177. Shimizu M, Toki T, Takagi Y, Konishi I, Fujii S. Immunohistochemical detection of the Wilms' tumor gene (WT1) in epithelial ovarian tumors. Int J Gynecol Pathol. 2000;19:158-63
178. Sumathi VP, Al-Hussaini M, Connolly LE, Fullerton L, McCluggage WG. Endometrial stromal neoplasms are immunoreactive with WT-1 antibody. Int J Gynecol Pathol. 2004;23:241-7
179. Lin CY, Chao A, Wu RC, Lee LY, Ueng SH, Tsai CL. et al. Synergistic effects of pazopanib and hyperthermia against uterine leiomyosarcoma growth mediated by downregulation of histone acetyltransferase 1. J Mol Med (Berl). 2020;98:1175-88
180. Seki K, Hoshihara T, Nagata I. Leiomyosarcoma of the uterus: ultrasonography and serum lactate dehydrogenase level. Gynecol Obstet Invest. 1992;33:114-8
181. Goto A, Takeuchi S, Sugimura K, Maruo T. Usefulness of Gd-DTPA contrast-enhanced dynamic MRI and serum determination of LDH and its isozymes in the differential diagnosis of leiomyosarcoma from degenerated leiomyoma of the uterus. Int J Gynecol Cancer. 2002;12:354-61
182. Di Cello A, Borelli M, Marra ML, Franzon M, D'Alessandro P, Di Carlo C. et al. A more accurate method to interpret lactate dehydrogenase (LDH) isoenzymes' results in patients with uterine masses. Eur J Obstet Gynecol Reprod Biol. 2019;236:143-7
183. Nishigaya Y, Kobayashi Y, Matsuzawa Y, Hasegawa K, Fukasawa I, Watanabe Y. et al. Diagnostic value of combination serum assay of lactate dehydrogenase, D-dimer, and C-reactive protein for uterine leiomyosarcoma. J Obstet Gynaecol Res. 2019;45:189-94
184. Pastorekova S, Parkkila S, Zavada J. Tumor-associated carbonic anhydrases and their clinical significance. Adv Clin Chem. 2006;42:167-216
185. Hynninen P, Parkkila S, Huhtala H, Pastorekova S, Pastorek J, Waheed A. et al. Carbonic anhydrase isozymes II, IX, and XII in uterine tumors. Apmis. 2012;120:117-29
186. Stukan M, Rutkowski P, Smadja J, Bonvalot S. Ultrasound-Guided Trans-Uterine Cavity Core Needle Biopsy of Uterine Myometrial Tumors to Differentiate Sarcoma from a Benign Lesion-Description of the Method and Review of the Literature. Diagnostics (Basel). 2022 12
187. Amant F, Van den Bosch T, Vergote I, Timmerman D. Morcellation of uterine leiomyomas: a plea for patient triage. Lancet Oncol. 2015;16:1454-6
188. Machado-Lopez A, Alonso R, Lago V, Jimenez-Almazan J, Garcia M, Monleon J. et al. Integrative Genomic and Transcriptomic Profiling Reveals a Differential Molecular Signature in Uterine Leiomyoma versus Leiomyosarcoma. Int J Mol Sci. 2022 23
189. Liau JY, Tsai JH, Jeng YM, Lee JC, Hsu HH, Yang CY. Leiomyosarcoma with alternative lengthening of telomeres is associated with aggressive histologic features, loss of ATRX expression, and poor clinical outcome. Am J Surg Pathol. 2015;39:236-44
190. Ahvenainen TV, Mäkinen NM, von Nandelstadh P, Vahteristo MEA, Pasanen AM, Bützow RC. et al. Loss of ATRX/DAXX expression and alternative lengthening of telomeres in uterine leiomyomas. Cancer. 2018;124:4650-6
191. Shan W, Akinfenwa PY, Savannah KB, Kolomeyevskaya N, Laucirica R, Thomas DG. et al. A small-molecule inhibitor targeting the mitotic spindle checkpoint impairs the growth of uterine leiomyosarcoma. Clin Cancer Res. 2012;18:3352-65
192. Gritsko TM, Coppola D, Paciga JE, Yang L, Sun M, Shelley SA. et al. Activation and overexpression of centrosome kinase BTAK/Aurora-A in human ovarian cancer. Clin Cancer Res. 2003;9:1420-6
193. Twu NF, Yuan CC, Yen MS, Lai CR, Chao KC, Wang PH. et al. Expression of Aurora kinase A and B in normal and malignant cervical tissue: high Aurora A kinase expression in squamous cervical cancer. Eur J Obstet Gynecol Reprod Biol. 2009;142:57-63
194. Shanes ED, Friedman LA, Mills AM. PD-L1 Expression and Tumor-infiltrating Lymphocytes in Uterine Smooth Muscle Tumors: Implications for Immunotherapy. Am J Surg Pathol. 2019;43:792-801
195. Baiocchi G, Poliseli FL, De Brot L, Mantoan H, Schiavon BN, Faloppa CC. et al. TOP2A copy number and TOP2A expression in uterine benign smooth muscle tumours and leiomyosarcoma. J Clin Pathol. 2016;69:884-9
196. Orlandi A, Francesconi A, Clément S, Ropraz P, Spagnoli LG, Gabbiani G. High levels of cellular retinol binding protein-1 expression in leiomyosarcoma: possible implications for diagnostic evaluation. Virchows Arch. 2002;441:31-40
197. Braný D, Dvorská D, Grendár M, Ňachajová M, Szépe P, Lasabová Z. et al. Different methylation levels in the KLF4, ATF3 and DLEC1 genes in the myometrium and in corpus uteri mesenchymal tumours as assessed by MS-HRM. Pathol Res Pract. 2019;215:152465
198. Belloni A, Furlani M, Greco S, Notarstefano V, Pro C, Randazzo B. et al. Uterine leiomyoma as useful model to unveil morphometric and macromolecular collagen state and impairment in fibrotic diseases: An ex-vivo human study. Biochim Biophys Acta Mol Basis Dis. 2022;1868:166494
199. Koohestani F, Braundmeier AG, Mahdian A, Seo J, Bi J, Nowak RA. Extracellular matrix collagen alters cell proliferation and cell cycle progression of human uterine leiomyoma smooth muscle cells. PLoS One. 2013;8:e75844
200. Leppert PC, Baginski T, Prupas C, Catherino WH, Pletcher S, Segars JH. Comparative ultrastructure of collagen fibrils in uterine leiomyomas and normal myometrium. Fertil Steril. 2004;82(Suppl 3):1182-7
Author contact
![]() Corresponding author: Jinyi Tong; Email: tong-jinyicom.
Corresponding author: Jinyi Tong; Email: tong-jinyicom.

 Global reach, higher impact
Global reach, higher impact