Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(6):1176-1186. doi:10.7150/ijms.95490 This issue Cite
Research Paper
The Role of JAK-STAT-SOCS1 Axis in Tumorigenesis, Malignant Progression and Lymphatic Metastasis of Penile Cancer
1. State Key Laboratory of Oncology in South China, Guangdong Provincial Clinical Research Center for Cancer, Sun Yat-sen University Cancer Center, Guangzhou 510060, P. R. China.
2. Department of Urology, Sun Yat-sen University Cancer Center, Guangzhou 510060, P.R. China.
3. Department of Urology, Xiangya Hospital, Central South University, Changsha 410008, P.R. China.
4. Department of Anesthesiology, Second Affiliated Hospital of Shantou University Medical College, Shantou 515041, P. R. China.
*These authors contributed to the work equally and should be regarded as co-first authors. (Da-Ming Xu; Ling-Xiao Chen; Xiao-Yu Zhuang).
Received 2024-2-19; Accepted 2024-4-24; Published 2024-4-29
Abstract
Background: To uncover the potential significance of JAK-STAT-SOCS1 axis in penile cancer, our study was the pioneer in exploring the altered expression processes of JAK-STAT-SOCS1 axis in tumorigenesis, malignant progression and lymphatic metastasis of penile cancer.
Methods: In current study, the comprehensive analysis of JAK-STAT-SOCS1 axis in penile cancer was analyzed via multiple analysis approaches based on GSE196978 data, single-cell data (6 cancer samples) and bulk RNA data (7 cancer samples and 7 metastasis lymph nodes).
Results: Our study observed an altered molecular expression of JAK-STAT-SOCS1 axis during three different stages of penile cancer, from tumorigenesis to malignant progression to lymphatic metastasis. STAT4 was an important dominant molecule in penile cancer, which mediated the immunosuppressive tumor microenvironment by driving the apoptosis of cytotoxic T cell and was also a valuable biomarker of immune checkpoint inhibitor treatment response.
Conclusions: Our findings revealed that the complexity of JAK-STAT-SOCS1 axis and the predominant role of STAT4 in penile cancer, which can mediate tumorigenesis, malignant progression, and lymphatic metastasis. This insight provided valuable information for developing precise treatment strategies for patients with penile cancer.
Keywords: JAK-STAT-SOCS1 Axis, Penile Cancer
Introduction
Penile cancer is an exceedingly rare malignant urologic neoplasm with an overall cancer incidence of 0.5-0.94/100 000 males in the United States and Europe [1]. Surgical treatment may be appropriate only for local disease, but patients with advanced penile cancer are often associated with inguinal or pelvic lymph node involvement. Limited by treatment options, survival outcomes among patients with advanced cancer substantially are very poor [2]. In fact, patients with positive inguinal or pelvic lymph node metastasis have an overall survival of approximately 15% at 5 years [3]. Established risk factors for penile cancer include phimosis, chronic inflammation in penis, lichen sclerosus, UV-A phototherapy, habit of smoking and poor socioeconomic status [4]. Over 95% of penile cancers are diagnosed as penile squamous cell carcinoma (PSCC). The histological subtypes of PSCC include human papillomavirus (HPV)-independent PSCC (usual, pseudohyperplastic, pseudoglandular, verrucous, cuniculatum, papillary, sarcomatoid and mixed), HPV-associated PSCC (basaloid, warty, warty-basaloid, lymphoepitheliomalike and mixed) and other [1,5]. In HPV-associated PSCC, high-risk HPV infection leads to uncontrolled cell proliferation by infecting penile epithelial cells and integrating viral genes (such as E6 and E7) [6,7]. By contrast, in HPV-independent PSCC, chronic inflammation and TP53 mutation are the main causes of carcinogenesis of penile cancer, which can promote the progression of malignant phenotypes such as invasion and angiogenesis [8,9]. Additionally, the oncogenic function of the PI3K-AKT-mTOR signaling pathway in penile cancer has been demonstrated [10]. Notably, this rare malignancy makes a great therapeutic challenge. In penile cancer, understanding the underlying biological processes, cancer microenvironment and significant biomarkers that drive the cancer is the foundation for the development of effective treatment. Recently, more and more scattered evidence suggests the significance of JAK-STAT pathway in penile cancer [11,12]. Simultaneously, JAK-STAT signaling axis has also been confirmed to be associated with cancer drug resistance [13]. JAK-STAT pathway signal transduction is mainly mediated by extracellular and intracellular stepwise phosphorylation, which eventually binds to DNA regulatory elements and induces gene transcription [14]. As mentioned, the research significance of JAK-STAT pathway in penile cancer has gradually emerged. Therefore, more sophisticated understanding of JAKs family, STATs family and their negative feedback regulators (SOCS1) will offer more opportunities for specificity therapy.
The novelty of our study lied in applying single-cell RNA sequencing experiment, bulk RNA sequencing experiment and other approaches to delineate the altered expression processes of JAK-STAT-SOCS1 axis in tumorigenesis, malignant progression and lymphatic metastasis of penile cancer. Collectively, our study was the first to make a comprehensive analysis of JAK-STAT-SOCS1 axis in penile cancer, thereby offering the new drug target for penile cancer treatment.
Materials and Methods
Differential expression analysis via Gene Expression Omnibus (GEO)
GSE196978 (6 tumor sample: GSM5906201, GSM5906202, GSM5906203, GSM5906204, GSM5906205, GSM5906206 and 6 normal penile tissue: GSM5906195, GSM5906196, GSM5906197, GSM5906198, GSM5906199, GSM5906200) were used to performed a comparation between penile cancer and normal penile tissue (p < 0.05) via R (4.2.2) and limma (3.54.0) packages.
Single-cell RNA sequencing experiments
Patients: A total of 6 penile cancer patients, classified according to the cancer staging criteria of the 8th American Joint Committee on Cancer, were included in our study for Single-cell RNA-seq analysis, which were divided into two group: advanced cancer group (Stage IV malignancy, n = 4) and early cancer group (Stage I malignancy, n = 2).
Cell quality control: An equal volume of 0.4% trypan blue staining solution was added into a small amount of single cell suspension, and cell counting was performed utilizing Countess® II Automated Cell Counter. Finally, the viable cells concentration was need to adjust to an ideal concentration.
10X labeled cDNA fragments: Gel beads including the barcode information associate with cells and enzymes, get into the reservoir and are allowed to be separated by oil to make a formation of gel-beads-in-emulsions (GEMs). Subsequently, gel bead lysis released capture sequences with barcode information. The cDNA fragments were reverse transcribed, and samples were labeled. PCR amplification was allowed to perform based on cDNA template. Combining all the GEMs products to construct a standard sequencing library.
Library construction of standard sequencing library: The cDNA fragments were cut into 200 to 300 bp fragments through enzyme digestion. The routine steps of next-generation sequencing library construction were performed. PCR was used to amplify the DNA library
Sequencing of libraries: The constructed library was sequenced using PE150 mode of Illumina sequencing platform.
Bulk RNA sequencing experiments
Patients: A total of 7 penile cancer patients with lymphatic metastasis were included in our study for Bulk RNA-seq analysis, which were divided into two group: Primary tumor site group (Penile cancer, n = 7) and their paired lymphatic metastasis group (Metastatic lymph node, n = 7).
RNA Sample Preparation: After total RNA extraction from the sample, the mRNA was enriched by removing rRNA using a conventional kit. The enriched mRNA was reverse transcribed to form double-stranded cDNA. After repairing the double ends of cDNA, the adaptor was added and the library was constructed by PCR amplification.
Library preparation: Eukaryotic mRNA with polyA tails was enriched using magnetic beads with Oligo and disrupted by ultrasound. In the M-MuLV reverse transcriptase system, cDNA double-strands are synthesized, followed by end repair, A-tail addition, and connection of an A sequencing adapter. Approximately 200bp cDNA fragments were screened using AMPure XP beads, and PCR products were allowed to purify again using AMPure XP beads to obtain the final library.
Molecular interaction analysis
InAct (https://www.ebi.ac.uk/intact/home) is a database for molecular interaction tool based on previous literature curation. We made a molecular interaction analysis of STAT4 via InAct to reveal its important accessory molecules. Then. STAT4 and its 26 accessory protein molecules in homo sapiens were selected to perform an immune infiltration analysis through GSEA platform [15].
Prediction of immune checkpoint inhibitor treatment response
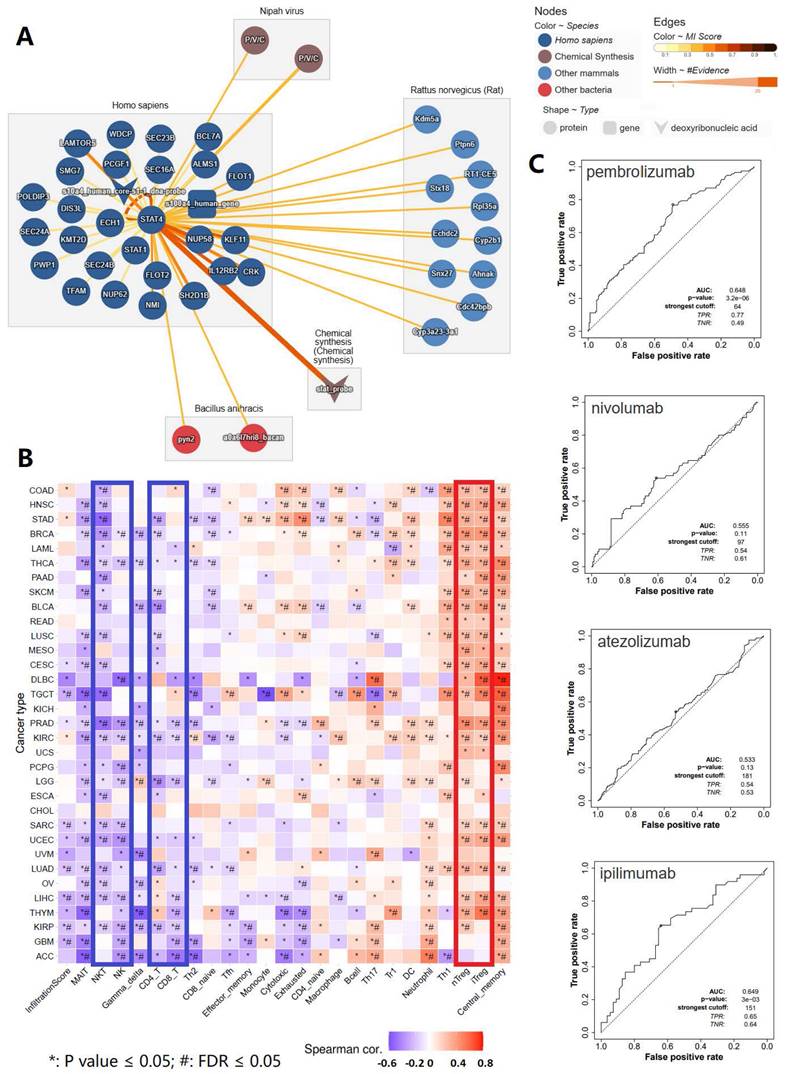
ROC Plotter is a database which has the ability to evaluate the efficacy of immunotherapy based on gene expression [16]. We conducted an exploratory investigation to gather the relationship data between STAT4 expression and the therapy efficacy of anti PD-1 therapy, containing pembrolizumab and nivolumab regimens, anti PD-L1 therapy, such as atezolizumab regimen and anti CTLA4 therapy, like ipilimumab regimen, via ROC Plotter.
Single cell trajectories analysis
Single cell trajectory in T cell subpopulation was measured by Monocle tool (version2.10.1) to reveal the expression abundance and expression status of target genes based on pseudotime variation [17,18].
Function enrichment analysis
Gene ontology (GO) is an international authoritative database for the analysis of gene function which has three different analysis levels to reveal the gene properties [19]. KEGG is a pathways database for the analysis of gene signaling pathways [20]. Our study made a GO and KEGG enrichment analysis of upregulated genes in cytotoxic T cell which was the T cell subgroup with the strongest expression of STAT4.
Results
Differential expression between penile cancer and normal penile tissue
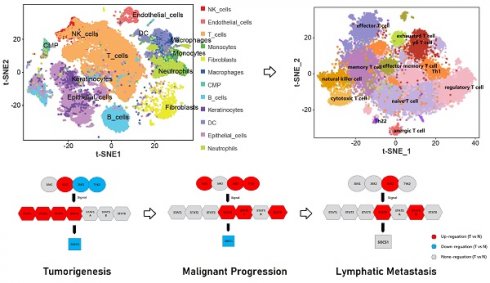
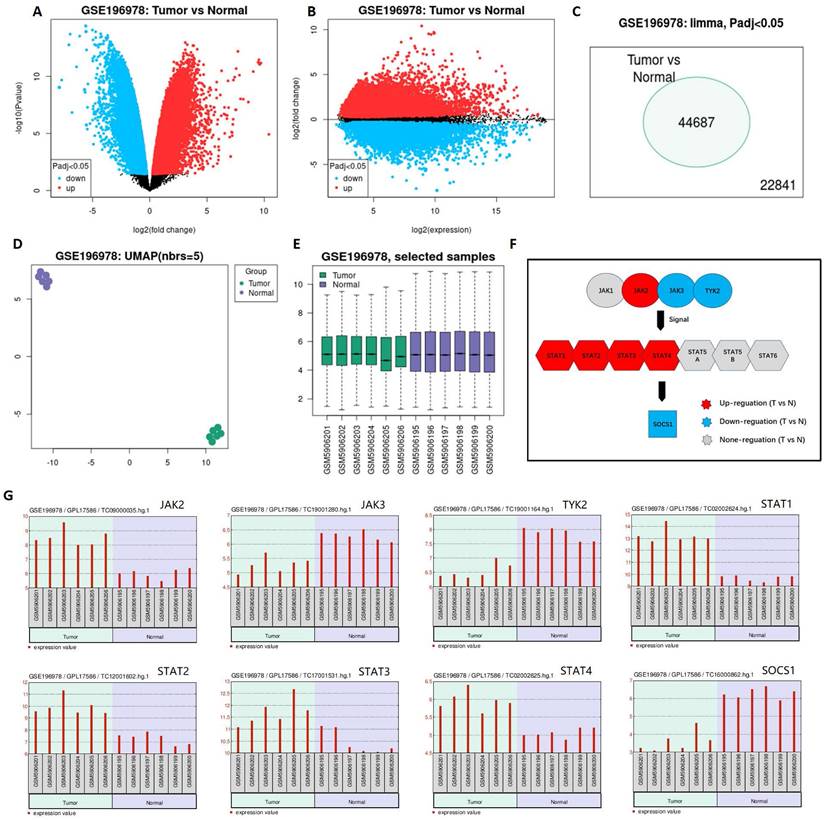
The result demonstrated that the differential expression between penile cancer and normal penile tissue (Figure 1). Figure 1A and 1B indicated the differences in gene expression via volcano plot and mean-difference plot. Figure 1C draw a Venn diagram to compare two samples, showing 44687 significantly different genes (p <0.05). Figure 1D and 1E indicated 12 samples' UMAP plot and box plot. Figure 1F displayed that the up-regulation expression of JAK2, STAT1, STAT2, STAT3, STAT4 and the down-expression of JAK3 TYK2, SOCS1 in penile cancer comparing with normal penile tissue, which had a statistical significance. Figure 1G revealed the detail gene expression of JAK2, STAT1, STAT2, STAT3, STAT4, JAK3 TYK2 and SOCS1 in each sample.
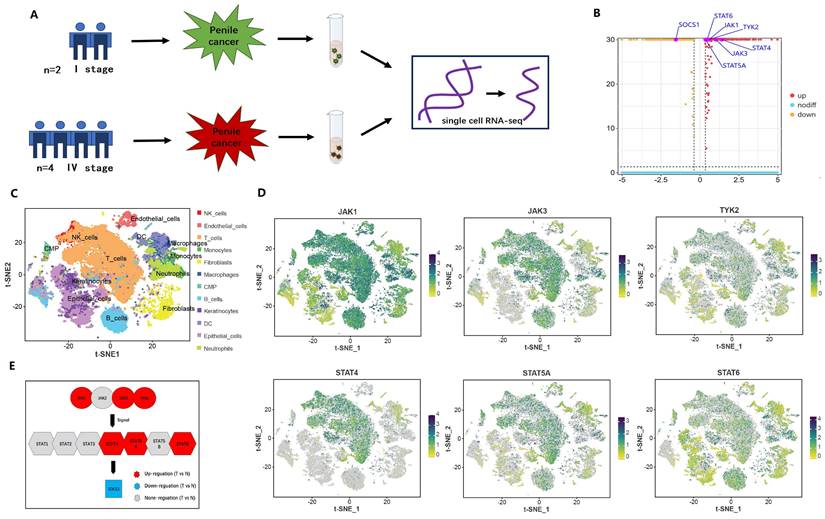
Single-cell RNA sequencing was used to reveal the difference between advanced penile cancer and early penile cancer
The result indicated that the differential expression between advanced penile cancer (IV stage) and early penile cancer (I stage) (Figure 2). Figure 2A showed the experimental procedure of Single-cell RNA sequencing. Figure 2B displayed that the upregulated expression of JAK1, JAK3, TYK2, STAT4, STAT5A, STAT6 and the down-expression of SOCS1 in advanced penile cancer comparing with early penile cancer, which had a statistical significance. Figure 2C mapped the tumor microenvironment of six penile cancer specimens, mainly including T cells, B cells, NK cells, Endothelial cells, DC, Macrophages, Monocytes, Neutrophils, Fibroblasts, Keratinocytes, Epithelial cells and CMP. Figure 2D indicated that the expression degree of JAK1, JAK3, TYK2, STAT4, STAT5A and STAT6 in tumor microenvironment. Figure 2E displayed the differences in JAK-STAT-SOCS1 Axis expression between early and advanced penile cancer through a schematic diagram.
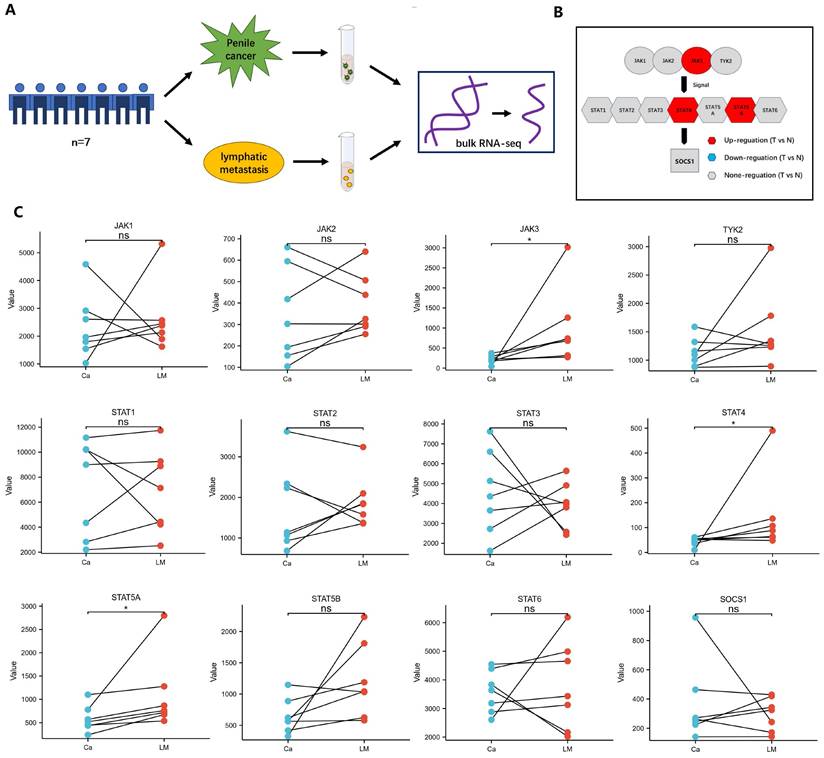
Bulk RNA sequencing was used to reveal the difference between penile cancer and their paired metastatic lymph nodes
The result documented that the differential expression between penile cancer and the paired metastatic lymph nodes (Figure 3). Figure 3A showed the experimental procedure of bulk RNA sequencing. Figure 3B and 3C displayed that the different expression level between penile cancer and their metastatic lymph nodes. There was a noticeable difference in the expression of JAK3, STAT4 and STAT5A between primary penile cancer and their metastatic lymph nodes.
Comprehensive analysis of STAT4 and its 26 accessory protein molecules in immune infiltration and immune checkpoint inhibitor response
As we discovered in our current study, STAT4 appeared to be a protagonist in mediating tumorigenesis, malignant progression and lymphatic metastasis of penile cancer. Therefore, our subsequent research focused on STAT4. In order to make a comprehensive investigation to reveal the mechanism of STAT4 in penile cancer, molecular interaction analysis of STAT4 were applied to reveal its important accessory molecules in our study (Figure 4). Figure 4A showed the molecular interaction network of STAT4. Figure 4B displayed that the immune infiltration analysis of STAT4 and its 26 accessory protein molecules (POLDIP3, SEC24A, PWP1, TFAM, LAMTOR5, SMG7, DIS3L, KMT2D, SEC24B, NUP62, NMI, WDCP, PCGF1, ECH1, STAT1, FLOT2, SH2D1B, SEC23B, SEC16A, NUP58, IL12RB2, BCL7A, ALMS1, FLOT1, KLF11, CRK) in homo sapiens, which were contributed to wake up the suppressor T cells and silence antitumor cells in pan cancers, thereby forming an immunosuppressive tumor microenvironment. Figure 4C found that STAT4 was associated with the response to anti PD-1 therapy (pembrolizumab) and anti CTLA4 therapy (ipilimumab) respectively.
Differential expression between penile cancer and normal penile tissue based on GSE196978 data. (A and B: Differences in gene expression via volcano plot and mean-difference plot; C: Venn diagram for 44687 significantly different genes; D and E: UMAP plot and box plot of GSE196978; F The diagram of JAK-STAT-SOCS1 axis expression; G: The up-regulation expression of JAK2, STAT1, STAT2, STAT3, STAT4 and the down-expression of JAK3 TYK2, SOCS1 in penile cancer comparing with normal penile tissue.)
Comprehensive analysis of T cell subpopulation in tumor microenvironment
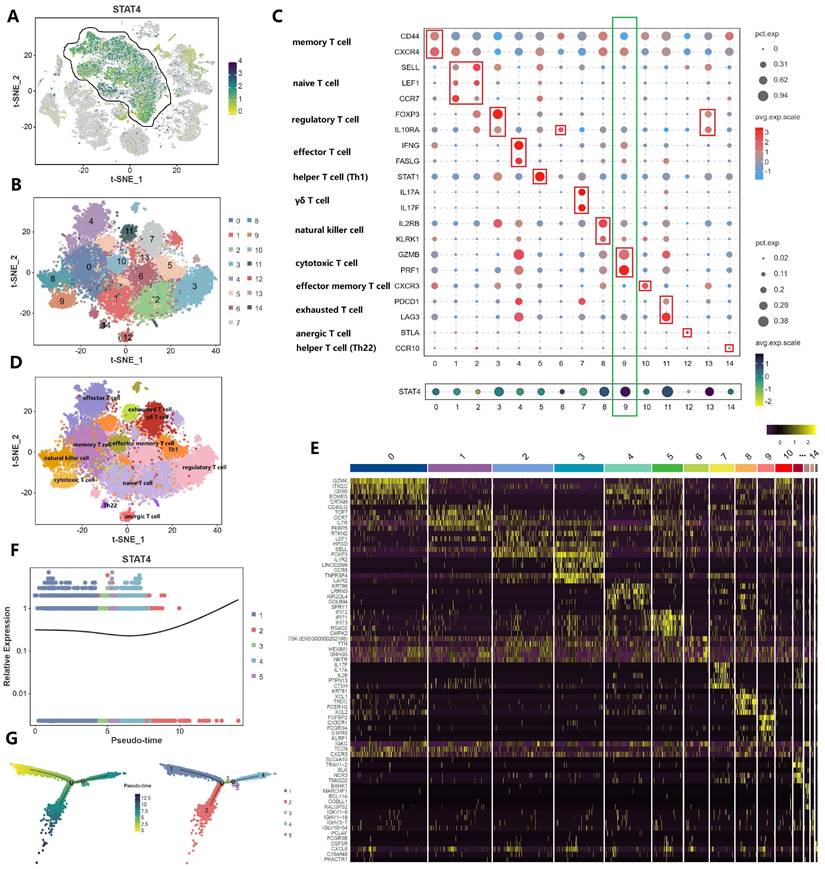
From the previously mentioned results, STAT4 was mainly expressed in T cell subpopulation in the immune microenvironment. Therefore, we make a comprehensive analysis of T cell subpopulation (Figure 5). Figure 5A and 5B regrouped T cell subpopulation into 14 clusters. Figure 5C and 5D made the annotation of T cell subpopulation by marker genes (memory T cell: CD44,CXCR4; naïve T cell: SELL, LEF1, CCR7; regulatory T cell: FOXP3, IL10RA; effector T cell: IFNG, FASLG; helper T cell [Th1]: STAT1; yδ T cell:IL17A, IL17F; natural killer cell: IL2RB, KLRK1; cytotoxic T cell: GZMB, PRF1; effector memory T cell: CXCR3; exhausted T cell: PDCD1, LAG3; anergic T cell: BTLA; helper T cell [Th22]:CCR10). Notedly, STAT4 expression was highest in cluster 9, suggesting that it was associated with cytotoxic T cell. Figure 5E demonstrated that the top 5 upregulated genes in each cluster. Figure 5F applied the single cell trajectory analysis for STAT4, which was suggested that STAT4 expression in penile cancer gradually increased with time. Figure 5G revealed the expression abundance and expression status of T cell subpopulation based on pseudo-time variation.
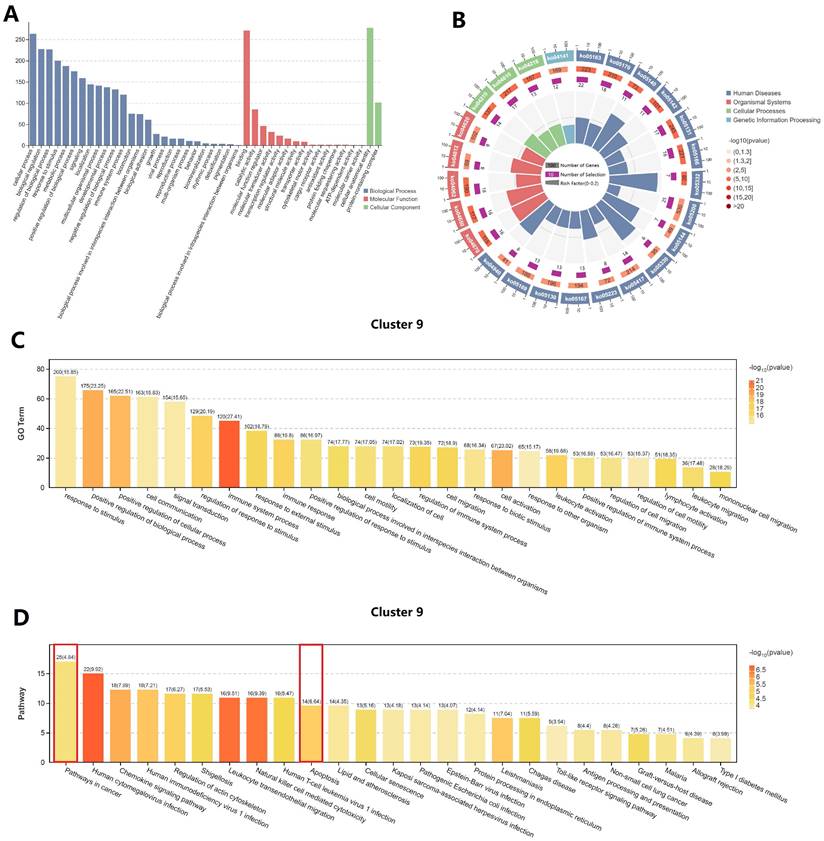
Function enrichment analysis of cluster 9 (cytotoxic T cell)
GO and KEGG enrichment analysis of the upregulated genes in cluster 9 was indicated two important terms, “pathway in cancer” and “apoptosis” (Figure 6). Therefore, we inferred that cytotoxic T cells in penile cancer were in an active state of apoptosis and their ability to destroy cancer cells was diminished. At the same time, the previous results of our study suggested the existence of regulatory T cell and exhausted T cell, which further proved that the immunosuppressive tumor microenvironment in penile cancer, thereby promoting the malignant progression and escape of cancer cells.
Difference between advanced (IV stage) and early (I stage) penile cancer based on single-cell RNA sequencing. (A: The experimental procedure of single-cell RNA sequencing; B: The up-regulation expression of JAK1, JAK3, TYK2, STAT4, STAT5A, STAT6 and the down-expression of SOCS1 in advanced penile cancer comparing with early penile cancer; C: Tumor microenvironment landscape of six penile cancer specimens; D: The expression degree of JAK1, JAK3, TYK2, STAT4, STAT5A and STAT6 in tumor microenvironment; E: The diagram of JAK-STAT-SOCS1 axis expression.)
Difference between penile cancer and the paired metastatic lymph nodes based on bulk RNA sequencing. (A: The experimental procedure of Bulk RNA sequencing; B: The diagram of JAK-STAT-SOCS1 axis expression; C: The up-expression of JAK3, STAT4 and STAT5A in metastatic lymph nodes comparing with primary penile cancer.)
Discussion
Due to the rare characteristics of penile cancer, the progression landscape of malignancy remains largely unknown, which limits our approach to accurately evaluate the progression, prognosis, and treatment options of this disease. Penile cancer, as a malignant tumor that seriously affects men's physical and mental health, family harmony and social stability, its prognosis is closely related to clinical and histological characteristics [21]. The most essential prognostic factor affecting patient survival is the status of lymph node metastasis, with the 5-year cancer-specific survival rate for patients with N0 to N3 dropped from 95% to 35% [22]. In addition, the prognosis of distant metastasis in penile cancer is even worse [23]. Thus, in order to have a better understanding, the potential mechanisms of occurrence, progression and tumor metastasis of penile cancer were urgently needed to be investigated and explored. Currently, research on biomarkers helped to gain a deeper understanding of individuals with penile cancer who are at high risk of metastatic progression [24]. At the same time, designing targeted therapy strategies aimed at inhibiting metastatic biomarkers could hold promise for improving patient prognosis. Our study provided important supplementary contributions to this field.
Comprehensive analysis of STAT4 and its 26 accessory protein molecules in immune infiltration and immune checkpoint inhibitor response. (A: The molecular interaction network of STAT4; B: The immune infiltration analysis of STAT4 and its 26 accessory protein molecules [POLDIP3, SEC24A, PWP1, TFAM, LAMTOR5, SMG7, DIS3L, KMT2D, SEC24B, NUP62, NMI, WDCP, PCGF1, ECH1, STAT1, FLOT2, SH2D1B, SEC23B, SEC16A, NUP58, IL12RB2, BCL7A, ALMS1, FLOT1, KLF11, CRK] in homo sapiens; C: The association between STAT4 expression and immune checkpoint inhibitors response.)
Comprehensive analysis of T cell subpopulation in tumor microenvironment. (A and B: Regrouping of T cell subpopulation; C and D: Annotation of T cell subpopulation by marker genes; E: The top 5 upregulated genes in each cluster; F: The single cell trajectory of STAT4; G: The expression abundance and expression status of T cell subpopulation based on pseudo-time variation.)
The JAK-STAT signaling was first discovered in 1957, and after approximately 30 years of exploration, there have been landmark discoveries in the field of cancer [14]. The putative biological process is that cytokines bind to their receptors and can activate the JAKs with associated-receptor. Subsequently these JAKs can activate STATs by phosphorylating. The dimers, formed by tyrosine-phosphorylated STATs, are allowed to translocate to the nucleus and further combine with target DNA sequences, thereby regulating gene expression [25,26]. In term of tumorigenesis, Huang et al. revealed that the activation of JAK-STAT axis can promote bladder cancer formation, and JAK-STAT inhibitors can significantly inhibit tumor activation [27]. In term of metastases, Wang et al. found that JAK2 and STAT3 can contribute to cancer cell proliferation and migration in colorectal cancer [28]. In term of treatment, JAK-STAT inflammatory signaling was enhanced in prostate cancer tumor cells which can lead to drug resistance [13, 29]. JAK-STAT axis is a core signaling axis in a wide variety of t cancer [30]. A previous pan cancer study showed that although all STATs follow similar molecular activation pathways, different STATs differed in their involvement in carcinogenesis [31]. In current study, we observed an altered expression of JAK-STAT-SOCS1 axis during three different stages of penile cancer, from tumorigenesis to malignant progression to lymphatic metastasis. Notably, STAT4 was an important dominant player in the three-phase process. Therefore, it was reasonable to infer that the STAT4 was an essential biomarker of penile cancer.
Function enrichment analysis of cluster 9 (cytotoxic T cell). (A: GO classification bar chart; B: KEGG enrichment circle diagram; C: GO enrichment analysis of cluster 9; D: KEGG enrichment analysis of cluster 9.)
During tumorigenesis phase of penile cancer, the expression evolution of JAK-STAT-SOCS1 axis was characterized by the upregulation of JAK2, STAT1, STAT2, STAT3 and STAT4. Previous evidence suggested that the combined activation of the JAK2-STAT3 signal was frequently found in various solid cancer. Persistently activated JAK2-STAT3 signal promotes cancer cells proliferation and survival, and also induces angiogenesis and immunosuppression in the tumor microenvironment [31,32]. The deficiency of STAT2 was proved to inhibit tumorigenesis due to low levels of inflammatory factors [33]. During malignant progression phase of penile cancer, the expression evolution of JAK-STAT-SOCS1 axis was characterized by the upregulation of JAK1, JAK3, STAT4, STAT5A and STAT6. Malignant signaling JAK1 can catalyze glycosylation and maintain stability of PD-L1 to suppress the antitumor effect of immune microenvironment [34]. Yuan et al. found that JAK3 had an ability to promote the cancer malignant progression by cooperating to T cells [35]. By contrast, STAT6 was documented to drive the M2 macrophage polarization to attenuate antitumor response [36]. Additionally, the overexpression of STAT5A was significant related to cancer proliferation and invasion [37]. During lymph node metastasis phase of penile cancer, the expression evolution of JAK-STAT-SOCS1 axis was characterized by the upregulation of JAK3, STAT4 and STAT5B. We noted that JAK3 has been reported to participate in cancer metastasis through phospholipase D [38]. Similarly, STAT5B has been found to have an ability to promote tumor growth and metastasis [39]. These findings firstly provide valuable implications to halt the progress of penile cancer by targeting the JAK-STAT-SOCS1 axis.
A growing number of studies have identified that STAT4 was closely related to the poor prognosis of various solid cancers [40]. Study have found a robust correlation between immune checkpoint and STAT pathways, especially STAT4, in glioblastomas [41]. Other scholars have also demonstrated that STAT4 can increased the expression of PD-1, enhancing the anti-cancer efficacy of immune checkpoint inhibitor therapy in prostate cancer [42]. STAT4 and its 26 partner genes were mainly related to the activation of suppressor T cells and the inhibition of anti-tumor immune cells, such as CD4+ T cell, CD8+ T cell, NK and NKT, thereby forming an immunosuppressive tumor microenvironment, leading to cancer occurrence, progression and metastasis. This is consistent with what we observed in tumor microenvironment of penile cancer. We speculated that the overexpression of STAT4 contributed to make the formation of an immunosuppressive microenvironment in penile cancer and further promote the malignant progression. Importantly, we also found that STAT4 was associated with the response to two immune checkpoint inhibitors, including anti PD-1 therapy (pembrolizumab) and anti CTLA4 therapy (ipilimumab). We further hypothesized that STAT4 overexpression may enhance the expression of PD-1 or CTLA4, which suggested its potential as an important predictive biomarker for immunotherapy response in penile cancer patients.
Conclusion
In summary, our findings revealed that the complexity of JAK-STAT-SOCS1 axis and the predominant role of STAT4 in penile cancer, which can mediate tumorigenesis, malignant progression, and lymphatic metastasis. This insight provided valuable information for developing precise treatment strategies for patients with penile cancer.
Acknowledgements
Funding
This study was supported by grants from National Natural Science Foundation of China (82103639) and Natural Science Foundation of Guangdong Province (2021A1515220182, 2022A1515012321).
Ethics approval
The Institutional Review Board of Sun Yat-sen University Cancer Center approved this research (B2023-390-01) and confirmed the study design and conduct complied with all relevant regulations regarding the use of human study participants and was conducted in accordance with the criteria set by the Declaration of Helsinki.
Patient consent statement
Written informed consent was received from all participants.
Author contributions
All authors made a significant contribution to the work reported. Hui Han and Miao Mo designed the study. Da-Ming Xu, Ling-Xiao Chen and Xiao-Yu Zhuang analyzed and interpreted the data. Da-Ming Xu and Ling-Xiao Chen collected clinical samples. Da-Ming Xu and Xiao-Yu Zhuang conducted the experiments. Da-Ming Xu and Xiao-Yu Zhuang wrote the manuscript. Hui Han, Miao Mo and Da-Ming Xu edited the manuscript. Hui Han and Miao Mo final reviewed the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Brouwer OR, Albersen M, Parnham A. et al. European Association of Urology-American Society of Clinical Oncology Collaborative Guideline on Penile Cancer: 2023 Update. Eur Urol. 2023;83(6):548-560 doi:10.1016/j.eururo.2023.02.027
2. Joshi VB, Spiess PE, Necchi A, Pettaway CA, Chahoud J. Immune-based therapies in penile cancer. Nat Rev Urol. 2022;19(8):457-474 doi:10.1038/s41585-022-00617-x
3. Pagliaro LC, Crook J. Multimodality therapy in penile cancer: when and which treatments? World J Urol. 2009;27(2):221-225 doi:10.1007/s00345-008-0310-z
4. Douglawi A, Masterson TA. Updates on the epidemiology and risk factors for penile cancer. Transl Androl Urol. 2017;6(5):785-790 doi:10.21037/tau.2017.05.1
5. Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol. 2016;70(1):93-105 doi:10.1016/j.eururo.2016.02.029
6. Olesen TB, Sand FL, Rasmussen CL. et al. Prevalence of human papillomavirus DNA and p16INK4a in penile cancer and penile intraepithelial neoplasia: a systematic review and meta-analysis. Lancet Oncol. 2019;20(1):145-158 doi:10.1016/S1470-2045(18)30682-X
7. Emmanuel A, Nettleton J, Watkin N, Berney DM. The molecular pathogenesis of penile carcinoma-current developments and understanding. Virchows Arch. 2019;475(4):397-405 doi:10.1007/s00428-019-02607-8
8. De Paula AA, Motta ED, Alencar Rde C. et al. The impact of cyclooxygenase-2 and vascular endothelial growth factor C immunoexpression on the prognosis of penile carcinoma. J Urol. 2012;187(1):134-140 doi:10.1016/j.juro.2011.09.027
9. Kayes O, Ahmed HU, Arya M, Minhas S. Molecular and genetic pathways in penile cancer. Lancet Oncol. 2007;8(5):420-429 doi:10.1016/S1470-2045(07)70137-7
10. Chaux A, Munari E, Cubilla AL. et al. Immunohistochemical expression of the mammalian target of rapamycin pathway in penile squamous cell carcinomas: a tissue microarray study of 112 cases. Histopathology. 2014;64(6):863-871 doi:10.1111/his.12338
11. Zhou QH, Deng CZ, Li ZS. et al. Molecular characterization and integrative genomic analysis of a panel of newly established penile cancer cell lines. Cell Death Dis. 2018;9(6):684 doi:10.1038/s41419-018-0736-1
12. Huang KB, Guo SJ, Li YH. et al. Genome-Wide Profiling Reveals HPV Integration Pattern and Activated Carcinogenic Pathways in Penile Squamous Cell Carcinoma. Cancers (Basel). 2021;13(23):6104 doi:10.3390/cancers13236104
13. Chan JM, Zaidi S, Love JR. et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science. 2022;377(6611):1180-1191 doi:10.1126/science.abn0478
14. Philips RL, Wang Y, Cheon H. et al. The JAK-STAT pathway at 30: Much learned, much more to do. Cell. 2022;185(21):3857-3876 doi:10.1016/j.cell.2022.09.023
15. Liu CJ, Hu FF, Xie GY. et al. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief Bioinform. 2023;24(1):bbac558 doi:10.1093/bib/bbac558
16. Fekete JT, Győrffy B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int J Cancer. 2019;145(11):3140-3151 doi:10.1002/ijc.32369
17. Trapnell C, Cacchiarelli D, Grimsby J. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381-386 doi:10.1038/nbt.2859
18. Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14(3):309-315 doi:10.1038/nmeth.4150
19. Ashburner M, Ball CA, Blake JA. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25-29 doi:10.1038/75556
20. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27-30
21. Thomas A, Necchi A, Muneer A. et al. Penile cancer. Nat Rev Dis Primers. 2021;7(1):11 doi:10.1038/s41572-021-00246-5
22. Horenblas S, van Tinteren H. Squamous cell carcinoma of the penis. IV. Prognostic factors of survival: analysis of tumor, nodes and metastasis classification system. J Urol. 1994;151(5):1239-1243 doi:10.1016/s0022-5347(17)35221-7
23. Deng X, Liu Y, Zhan X. et al. Trends in Incidence, Mortality, and Survival of Penile Cancer in the United States: A Population-Based Study. Front Oncol. 2022;12:891623 doi:10.3389/fonc.2022.891623
24. Pekarek L, Ortega MA, Fraile-Martinez O. et al. Clinical and Novel Biomarkers in Penile Carcinoma: A Prospective Review. J Pers Med. 2022;12(9):1364 doi:10.3390/jpm12091364
25. Xin P, Xu X, Deng C. et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int Immunopharmacol. 2020;80:106210 doi:10.1016/j.intimp.2020.106210
26. Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther. 2021;6(1):402 doi:10.1038/s41392-021-00791-1
27. Huang W, Li Y, Zhang C. et al. IGF2BP3 facilitates cell proliferation and tumorigenesis via modulation of JAK/STAT signalling pathway in human bladder cancer. J Cell Mol Med. 2020;24(23):13949-13960 doi:10.1111/jcmm.16003
28. Wang J, Zhang Y, Song H. et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol Cancer. 2021;20(1):81 doi:10.1186/s12943-021-01375-x
29. Lo UG, Chen YA, Cen J. et al. The driver role of JAK-STAT signalling in cancer stemness capabilities leading to new therapeutic strategies for therapy- and castration-resistant prostate cancer. Clin Transl Med. 2022;12(8):e978 doi:10.1002/ctm2.978
30. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798-809 doi:10.1038/nrc2734
31. Mengie Ayele T, Tilahun Muche Z, Behaile Teklemariam A, Bogale Kassie A, Chekol Abebe E. Role of JAK2/STAT3 Signaling Pathway in the Tumorigenesis, Chemotherapy Resistance, and Treatment of Solid Tumors: A Systemic Review. J Inflamm Res. 2022;15:1349-1364 doi:10.2147/JIR.S353489
32. Liang L, Hui K, Hu C. et al. Autophagy inhibition potentiates the anti-angiogenic property of multikinase inhibitor anlotinib through JAK2/STAT3/VEGFA signaling in non-small cell lung cancer cells. J Exp Clin Cancer Res. 2019;38(1):71 doi:10.1186/s13046-019-1093-3
33. Nan J, Wang Y, Yang J, Stark GR. IRF9 and unphosphorylated STAT2 cooperate with NF-κB to drive IL6 expression. Proc Natl Acad Sci U S A. 2018;115(15):3906-3911 doi:10.1073/pnas.1714102115
34. Chan LC, Li CW, Xia W. et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019;129(8):3324-3338 doi:10.1172/JCI126022
35. Yuan S, Wang X, Hou S. et al. PHF6 and JAK3 mutations cooperate to drive T-cell acute lymphoblastic leukemia progression. Leukemia. 2022;36(2):370-382 doi:10.1038/s41375-021-01392-1
36. Yu T, Gan S, Zhu Q. et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat Commun. 2019;10(1):4353 doi:10.1038/s41467-019-12384-2
37. Wang S, Fan R, Gao H. et al. STAT5A modulates gastric cancer progression via upregulation of CD44. Genomics. 2023;115(3):110638 doi:10.1016/j.ygeno.2023.110638
38. Gomez-Cambronero J. Phospholipase D in cell signaling: from a myriad of cell functions to cancer growth and metastasis. J Biol Chem. 2014;289(33):22557-22566 doi:10.1074/jbc.R114.574152
39. Moser C, Ruemmele P, Gehmert S. et al. STAT5b as molecular target in pancreatic cancer-inhibition of tumor growth, angiogenesis, and metastases. Neoplasia. 2012;14(10):915-925 doi:10.1593/neo.12878
40. Erdogan F, Radu TB, Orlova A. et al. JAK-STAT core cancer pathway: An integrative cancer interactome analysis. J Cell Mol Med. 2022;26(7):2049-2062 doi:10.1111/jcmm.17228
41. Wang QW, Sun LH, Zhang Y. et al. MET overexpression contributes to STAT4-PD-L1 signaling activation associated with tumor-associated, macrophages-mediated immunosuppression in primary glioblastomas. J Immunother Cancer. 2021;9(10):e002451 doi:10.1136/jitc-2021-002451
42. Xiao J, Sun F, Wang YN. et al. UBC9 deficiency enhances immunostimulatory macrophage activation and subsequent antitumor T cell response in prostate cancer. J Clin Invest. 2023;133(4):e158352 doi:10.1172/JCI158352
Author contact
![]() Corresponding authors: Hui Han, State Key Laboratory of Oncology in South China, Guangdong Provincial Clinical Research Center for Cancer, Sun Yat-sen University Cancer Center, Guangzhou 510060, P. R. China. Department of Urology, Sun Yat-sen University Cancer Center, Guangzhou 510060, P.R. China. E-mail: hanhuiorg.cn. Miao Mo, Department of Urology, Xiangya Hospital, Central South University, Changsha 410008, P.R. China. E-mail: momiaocom.
Corresponding authors: Hui Han, State Key Laboratory of Oncology in South China, Guangdong Provincial Clinical Research Center for Cancer, Sun Yat-sen University Cancer Center, Guangzhou 510060, P. R. China. Department of Urology, Sun Yat-sen University Cancer Center, Guangzhou 510060, P.R. China. E-mail: hanhuiorg.cn. Miao Mo, Department of Urology, Xiangya Hospital, Central South University, Changsha 410008, P.R. China. E-mail: momiaocom.

 Global reach, higher impact
Global reach, higher impact