Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(3):464-473. doi:10.7150/ijms.90829 This issue Cite
Research Paper
Text Mining and Drug Discovery Analysis: A Comprehensive Approach to Investigate Diabetes-Induced Osteoporosis
Department of Orthopedic Surgery, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang 310030, China.
#These authors contributed equally to this work.
Received 2023-10-6; Accepted 2023-12-13; Published 2024-1-1
Abstract
Purpose: Osteoporosis (OP) and diabetes are prevalent diseases in orthopedic and endocrinology departments, with OP potentially arising as a complication of diabetes. However, the mechanism underlying diabetes-induced osteoporosis (DOP) remains enigmatic, and drug discovery in this domain is restricted, hindering research into the DOP's etiology and treatment. With the ultimate goal of preventing OP in diabetic patients, the objective of this study is to mine the genes and pathways linked to DOP using bioinformatics and databases.
Method: The present study employed text mining as the initial approach to retrieve genes commonly associated with diabetes and OP. Subsequently, functional annotation was conducted to investigate the roles and functionalities. In order to explore the interactions between proteins relevant to DOP, we constructed protein-protein interaction (PPI) networks. Furthermore, to obtain key genes and candidate drugs for DOP treatment, we conducted drug-gene interaction (DGI) analysis, complemented by a thorough examination of transcriptional factors (TFs)-miRNA co-regulation.
Results: The results through text mining revealed 110 genes that are commonly associated with both diabetes and OP. Subsequent enrichment analysis narrowed down the list to 95 symbols that were involved in PPI analysis. After DGI analysis, we identified 7 genes targeted by 11 drugs, which represent candidates for treating DOP.
Conclusion: This study unveils ANDECALIXIMAB, SILTUXIMAB, OLOKIZUMAB, SECUKINUMAB, and IXEKIZUMAB as promising potential drugs for DOP treatment, demonstrating the significance of utilizing text mining and pathway analysis to investigate disease mechanisms and explore existing therapeutic options.
Keywords: Mining, Diabetes, Osteoporosis, Drug
Introduction
Osteoporosis (OP) is a debilitating condition characterized by weakened bones and increased vulnerability to fractures. A complication arising from diabetes mellitus, OP manifests through reduced bone mass and microstructural changes in bone tissue [1]. Despite a focus on disease regression, the underlying pathogenesis of diabetes-induced osteoporosis (DOP) remains a slow area of exploration. Additionally, addressing inefficient clinical treatment guidelines and limited drug discovery for DOP is of utmost importance.
The multifaceted realm of diabetes mellitus can be neatly divided into two categories, namely type 1 (T1DM) and type 2 diabetes mellitus (T2DM), each brandishing their distinct pathophysiological implications for the aggravated risk of fractures. It is fascinating to observe how both types display a conspicuous commonality in the form of decreased bone formation, osteoblast dysfunction, and low bone turnover [2]. There is a possible association between diabetes and serum osteocalcin—notably, patients with either T1DM or T2DM have lower serum osteocalcin levels compared to non-diabetic individuals [3]. Furthermore, osteocalcin levels demonstrate a negative correlation with glycosylated hemoglobin levels in T1DM patients [4]. Only with a good understanding of the pathophysiology and pathogenesis of DOP can doctors be more effective in the treatment of these patients, such as developing targeted therapies and improving patient outcomes.
At present, there are very few available therapies for the prevention and treatment of DOP. Recent research suggests that physical activity can improve glucose intolerance and bone loss caused by diabetes, as well as promote skeletal development, possibly due to the effects of skeletal irisin [5]. In addition, the interplay of choices takes center stage, with the selection of medication for patients ensnared in the clutches of both OP and diabetes evolving into a matter of paramount importance. The alluring first-line drug recommendation of bisphosphonates for OP therapy in diabetes-stricken patients whispers the promise of a brighter future [6]. Scientific research verified that metformin has a positive effect on bone mineral density (BMD) in diabetics [7,8]. However, the frowns return with the advisory not to embrace sulfonylurea drugs for individuals entangled in the web of bone mineral disorders and diabetes, for fear of the lurking menace of hypoglycemia [9], while thiazolidinediones are also not recommended due to their mechanism of action[10]. Therefore, it is crucial to prioritize research efforts in exploring potential pharmacological interventions given the increased demand from DOP patients.
With the rapid development of bioinformatics and drug discovery databases, new research ideas and strategies are given to the treatment of various diseases, which may be a potential way to explore the mechanism and therapeutic for the DOP. A therapeutic target database has been updated, which is facilitating the progress of drug discovery [11]. Meanwhile, a simplified integrated database called TCMSID is exploited for mining candidate medicines from traditional Chinese medicine (TCM) [12]. Additionally, a web-based knowledge graph database on synthetic lethality has also been created by researchers for the purpose of discovering new anticancer drugs [13]. Therefore, making full use of existing bioinformatics technologies and drug databases may provide a shortcut to finding potential drugs for DOP.
The study aims to explore the pathomechanism and potential therapeutic options for DOP using text mining, functional annotation, protein network analysis, drug-gene interaction analysis, and miRNA-transcription factor (TF) coregulatory network construction, with the ultimate goal of informing clinical prevention and treatment for DOP.
Method
Text mining
Diabetes and OP-related gene sets were obtained through the GenCLiP3 website. In the GenCLiP3 database, using "All human genes" as the gene dataset, selecting "Search in MEDLINE" and using the default values in the journal filter, we searched for the keywords "diabetes" and "osteoporosis" respectively. Once the lists of associated genes are obtained, the intersection of the two sets of associated genes is utilized for functional enrichment analysis.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
GO and signaling pathway annotations for the genes corresponding to the intersection of OP and diabetes were carried out using the DAVID website. In addition, GO, which included biological processes, cellular components, and molecular functions, was a crucial method for achieving functional annotation. The key symbols that are closely related to the pathology of DOP were further mined using KEGG analysis. The false discovery rate (FDR) of the whole analysis should be less than 0.05.
Protein-protein interaction (PPI) network
The common symbols were inputted into the STRING website among the high confidence (score 0.900) and the Cytoscape software to screen out hub genes by apps of Molecular Complex Detection (MCODE) and cytoHubba. The cutoff parameters were “degree cutoff=2”, “node score cutoff=0.2”, “k-core=2”, and “max depth=100”. And the cutoff parameters of MCODE were defaults, and that of cytoHubba were the top 10 genes.
Drug-gene interaction (DGI)
Interacting genes between cytoHubba and MCODE apps were utilized for DGI analysis. The strict criteria are a DGI score transcending at a height of 3 and an unequivocal classification, the final genes were selected for the subsequent microRNA-transcription factor (TF) coregulatory network analysis.
MiRNA-TFs regulatory network
The miRNAs and TFs associated with the final genes were investigated using the NetworkAnalyst platform.
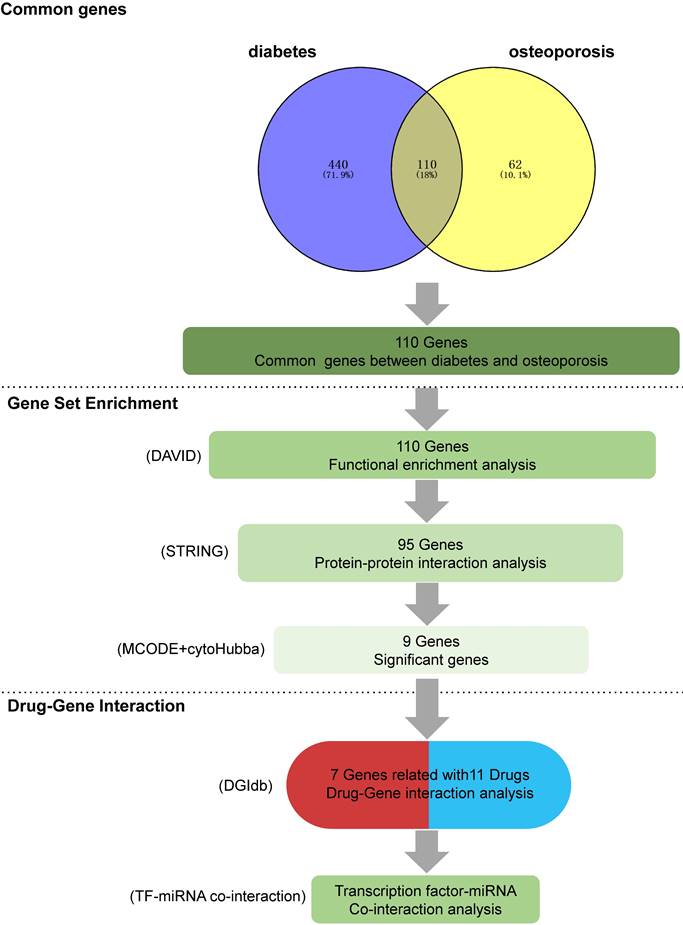
A comprehensive summary of the overall results obtained from data mining. (1) Intersecting genes: the process of obtaining intersecting genes involved the utilization of specific search terms, 'diabetes' and 'osteoporosis' in GENCLIP3. This yielded 550 genes for diabetes and 172 genes for OP, ultimately leading to the identification of 110 common genes shared between the two conditions (2) Functional annotations: gene functional enrichment analysis was performed using the DAVID tool. Subsequently, 9 hub symbols were identified using STRING and Cytoscape software. (3) Drug-gene interaction analysis: 11 potential medicines were acquired by DGIdb. For the final TF-miRNA co-interaction analysis, seven genes were chosen.
Result
Attainment of common symbols
Following text mining, we identified 550 unique genes associated with diabetes and 172 unique genes related to OP. Among these, there were 110 common genes shared between diabetes and OP, forming the basis for further investigation.
Functional annotations
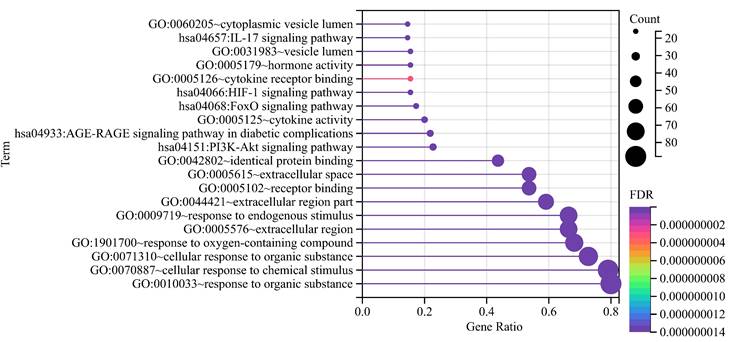
The top 5 biological processes were (1) response to oxygen-containing compound (False discovery rate = 1.16E-48), (2) response to endogenous stimulus (False discovery rate = 8.68E-46), (3) response to organic substance (False discovery rate = 8.96E-45), (4) cellular response to chemical stimulus (False discovery rate = 2.68E-44), and (5) cellular response to organic substance (False discovery rate = 6.05E-43). Regarding cellular components, the top 5 were (1) extracellular space (False discovery rate = 1.01E-23), (2) extracellular region (False discovery rate = 1.03E-15), (3) extracellular region part (False discovery rate = 1.11E-14), (4) vesicle lumen (False discovery rate = 1.99E-08), and (5) cytoplasmic vesicle lumen (False discovery rate = 1.40E-07). As for the molecular function, (1) receptor binding (False discovery rate = 2.07E-26), (2) cytokine activity (False discovery rate = 6.54E-16), (3) hormone activity (False discovery rate = 4.46E-14), (4) identical protein binding (False discovery rate = 4.74E-13), and (5) cytokine receptor binding (False discovery rate = 3.00E-09) were the top 5.
KEGG pathway analyses revealed the involvement of (1) AGE-RAGE signaling pathway in diabetic complications (False discovery rate = 6.33E-22), (2) FoxO signaling pathway (False discovery rate = 1.55E-13), (3) HIF-1 signaling pathway (False discovery rate = 1.61E-12), (4) IL-17 signaling pathway (False discovery rate = 2.50E-12), and (5) PI3K-Akt signaling pathway (False discovery rate = 1.93E-12), hinting at potential mechanisms underlying DOP (Figure 2).
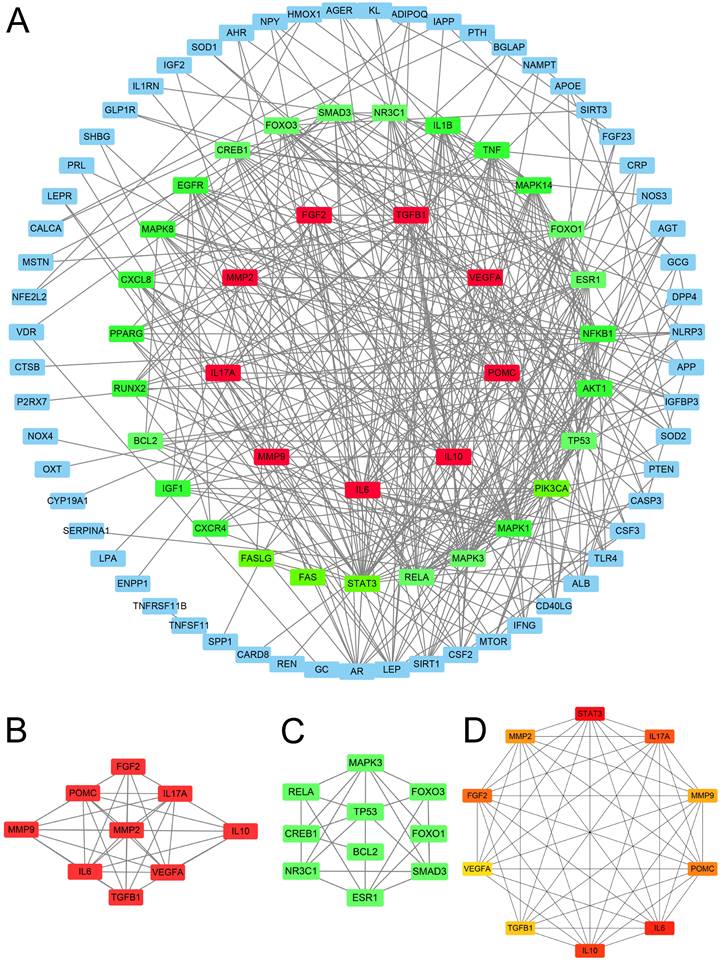
PPI network and hub genes obtainment
The PPI network analysis yielded 67 genes with medium to high confidence interactions (Figure 3A). Subsequently, cluster analysis using MCODE identified 9 genes in the first module and 10 genes in the second module as hub genes (Figure 3B-C), while cytoHubba apps obtained the top 10 symbols (Figure 3D).
Potential therapeutics
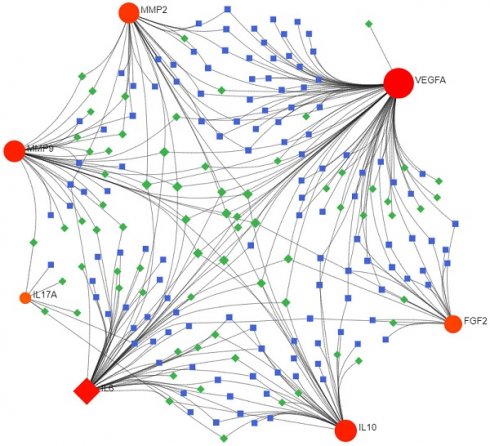
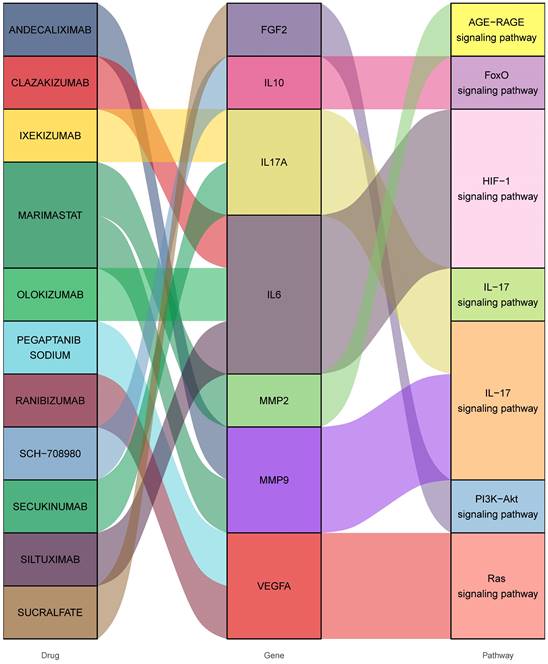
Consequently, 11 drugs (RANIBIZUMAB, PEGAPTANIB SODIUM, SUCRALFATE, SECUKINUMAB, IXEKIZUMAB, MARIMASTAT, SCH-708980, SILTUXIMAB, OLOKIZUMAB, CLAZAKIZUMAB, ANDECALIXIMAB) corresponding to 7 core genes (IL6-interleukin 6, VEGFA-vascular endothelial growth factor A, IL17A- interleukin 17 alpha, MMP2- matrix metallopeptidase 2, IL10- interleukin 10, FGF2- fibroblast growth factor 2, MMP9- matrix metallopeptidase 9) were found to affect DOP (Figure 4 and Table 1).
Functional annotations. With dots of varying size and color denoted the number of genes involved and their reliability, respectively.
Protein network analysis. (A) The protein-protein interaction network was constructed using STRING. (B) Cluster 1: There are 35 edges and 9 nodes. (C) Cluster 2: There are 10 nodes and 21 edges. (D) CytoHubba analysis: The top 10 genes were analyzed.
Interactions between drug-gene-pathway. The diagram provides insights into the interplay between 7 genes targeting 11 drugs and 6 pathways, providing an insightful visualization of the intricate relationships involved.
MiRNA-TF coregulatory network
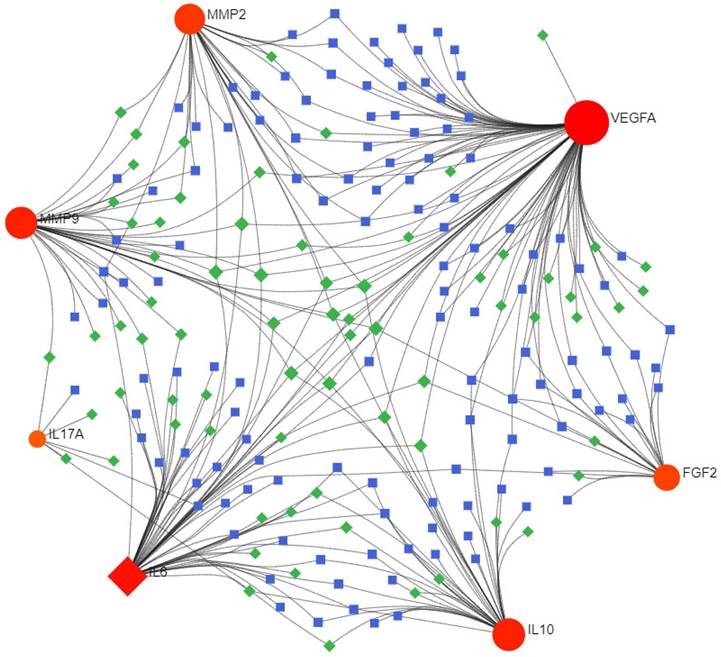
The miRNA-TF coregulatory network analysis revealed five significant TFs, namely DPF2, ZBTB7A, ARID1B, IRF1, and CREM, and the top five miRNAs, including hsa-miR-107, hsa-miR-203a, hsa-miR-335, hsa-miR-15a, and hsa-miR-16, which might play important roles in the regulation of hub genes associated with DOP (Figure 5 and Table 2).
Discussion
Diabetes can cause bone loss, which can lead to OP. The clinical prevention and treatment of DOP depend greatly on the definition of its mechanisms and therapeutic targets. The purpose of this study was to screen out key genes, identify targetable therapeutic agents and explore the underlying pathogenesis through multiple bioinformatics methods.
Consequently, 7 hub genes correlated to 11 drugs and 6 pathways were filtrated. FGF2, produced by epithelial and mesenchymal cells, plays a crucial role in skeletal development and the formation of osteoblasts [14,15]. However, its expression decreases in a high-glucose environment, leading to osteoblast proliferation disorder [16]. IL10, renowned for its potent immune and inflammatory suppression properties, is thought to inhibit bone resorption and reduce bone loss [17], but some studies have proved that IL10 is significantly increasing in DM patients [18,19], which also just goes to show that DOP development and its regulatory mechanisms are quite complex. Moving on to IL17A, a proinflammatory cytokine, it can indirectly stimulate bone resorption by RANKL (receptor activator of NF-κB ligand)[20,21]. Besides, diabetes induces cells to secrete IL17A, which in turn leads to a variety of complications [22].
Candidate drugs for diabetes-induced osteoporosis
| Number | Name | Symbol | Type | Score | PMID |
|---|---|---|---|---|---|
| 1 | RANIBIZUMAB | VEGFA | Inhibitor | 8.54 | 18046235 |
| 2 | PEGAPTANIB SODIUM | VEGFA | Antibody, Agonist | 3.25 | 23953100 |
| 3 | SUCRALFATE | FGF2 | Agonist | 5.89 | 7948825 |
| 4 | SECUKINUMAB | IL17A | Antibody, Agonist | 7.73 | 25354738 |
| 5 | IXEKIZUMAB | IL17A | Antibody, Agonist | 7.73 | None |
| 6 | MARIMASTAT | MMP2 | Inhibitor | 3.31 | 15748894 |
| 7 | SCH-708980 | IL10 | Inhibitor | 3.64 | None |
| 8 | SILTUXIMAB | IL6 | Inhibitor | 9.89 | 8823310 |
| 9 | OLOKIZUMAB | IL6 | Inhibitor | 9.89 | 24641941 |
| 10 | CLAZAKIZUMAB | IL6 | Inhibitor | 7.42 | None |
| 11 | ANDECALIXIMAB | MMP9 | Inhibitor | 10.3 | None |
| 12 | MARIMASTAT | MMP9 | Inhibitor | 3.68 | 17234180 |
Each drug-gene interaction was rigorously assessed to ensure that the putative drug exerted the anticipated effect on the specific medical condition, whereby the screening criterion necessitated an interacting score exceeding 3. Subsequently, the provenance of the information was diligently monitored to validate the report's veracity and appraise associated metadata. Drugs that effectively targeted the candidate genes via suitable interactions were subsequently aggregated in the conclusive inventory.
* The total number of Pubmed and database references determines the score.
Summary of TFs and miRNA
| TFs | Symbols | Count |
|---|---|---|
| DPF2 | IL6, IL17A | 2 |
| ZBTB7A | IL6, IL17A | 2 |
| ARID1B | IL6, FGF2 | 2 |
| IRF1 | IL6, FGF2 | 2 |
| CREM | IL6, FGF2 | 2 |
| hsa-miR-107 | IL6, IL17A, FGF2 | 3 |
| hsa-miR-203a | IL6, IL17A, FGF2 | 3 |
| hsa-miR-335 | IL6, FGF2, VEGFA | 3 |
| hsa-miR-15a | IL6, IL17A | 2 |
| hsa-miR-16 | IL6, IL17A | 2 |
Additionally, this investigation delved into IL6, an inflammatory factor known not only to encourage osteoclastogenesis, resulting in excessive osteoclastic activity and osteolysis [23], but also involves in the pathophysiology of diabetes [24]. HIF-1 controls VEGFA, an efficient therapeutic target for OP, to encourage bone formation. Reduced bone formation could result from OP if its expression is reduced [25]. When it comes to MMP2 and MMP9, they are playing a key role in bone physiology. On the one hand, the expression of MMP-2 and MMP-9 are negatively correlated with bone mineral density and considered as the essential biomarkers of OP [26]. On the other hand, they can hold potential as biomarkers useful in predicting the risk and progression of T2DM [27].
As mentioned above, the expressions of IL17A, MMP2, IL10 and IL6 are up-regulated with diabetes that promote the progression of OP, whereas the expressions of FGF2 and VEGFA are down-regulated. Therefore, this study only addresses inhibitors of up-regulated genes. Based on the DGI scores, the top 5 potential drugs are ANDECALIXIMAB (MMP9 inhibitor, 10.3), SILTUXIMAB (IL6 inhibitor, 9.89), OLOKIZUMAB (IL6 inhibitor, 9.89), SECUKINUMAB (IL17A inhibitor, 7.73) and IXEKIZUMAB (IL17A inhibitor, 7.73). ANDECALIXIMAB is mainly applied for gastric cancer and ulcerative colitis treatment [28,29], and it is currently in the clinical trial stage. SILTUXIMAB and OLOKIZUMAB, both as IL6 inhibitors, are now respectively being studied for Castleman disease [30] and rheumatoid arthritis [31]. SECUKINUMAB and IXEKIZUMAB are used in clinical trials for the therapy of plaque psoriasis and psoriatic arthritis with a remarkable therapeutic effect [32-34]. Despite not being currently used for DOP treatment, targeting these key genes may hold the potential to prevent the onset and progression of the disease.
It is worth noting that miRNA and TF may play a big part in the appearance and regulation of DOP. For example, a study shows that circulating miR-107 facilitates osteogenesis and serves as a potential diagnostic biomarker and therapeutic target for OP [35], but miR-107 is dysregulated in diabetes [36,37]. IRF1, as a transcription factor (TF), promotes osteogenic differentiation [38] and is also involved in the progress of diabetes [39]. Therefore, miRNA and TF may also be an important breakthrough in the fight against DOP.
DOP, characterized by metabolic bone loss, involves the endocrine system as a crucial mediator. Advanced glycation end products (AGEs) interact with the receptor for AGEs (RAGE) and may significantly impact bone cell metabolism and function [40]. The AGE-RAGE pathway emerged as a critical player in diabetic complications, including diabetic osteopathy, potentially contributing to reduced bone density, suppressed bone turnover markers, and impaired bone quality in diabetic patients [41,42]. AGEs play a crucial role in both OP and diabetes. They not only promote osteoclastogenesis by increasing RANKL expression but also negatively impact osteoblasts [43,44]. Another important pathway involved in bone and immune cell regulation is the IL-17 signaling pathway, where elevated plasma IL-17 levels are closely linked to BMD [45]. The PI3K-Akt signaling pathway is essential for inhibiting OP through promoting bone formation [46], whereas in the context of diabetes, the pathway becomes impaired, exacerbating the pathogenic cascade. The aforementioned key genes are intricately involved in the pathogenesis of both diabetes and OP, suggesting that post-diabetic up- or down-regulation of genes significantly contributes to the occurrence and progression of OP via complex pathways, including the PI3K-Akt signaling pathway and AGE-RAGE pathway. Consequently, perturbations in the delicate balance between osteogenesis and osteoclasts lead to the manifestation of OP.
Of particular concern is the potential for these interacting genes to engender a vicious cycle, thereby exacerbating the severity of both OP and diabetes. As such, early interventions in diabetes patients through targeted medical therapy present an advantageous approach to inhibit the development of OP. This approach encompasses the control of the internal environmental disturbances inherent in diabetes to promote physiological stability, as well as the timely implementation of interventions to postpone the progression of OP and diabetes, ultimately disrupting the deleterious cycle.
Unfortunately, this study is not without limitations. The dynamic nature of the utilized database necessitates periodic updates to maintain congruence with ongoing developments and to ensure replicability. Moreover, the subjectivity involved in the criteria for core genes, such as the parameter choices for application analysis (MCODE analysis and PPI construction) and DGI score selection, warrants consideration and further refinement. To bolster the credibility of this investigation, experimental verification remains imperative.
TF-miRNA coregulatory network construction. The TF-miRNA coregulatory network of the 7 hub genes is depicted, with red nodes signifying the hub genes, blue squares representing TFs, and green squares symbolizing miRNAs. This depiction offers a comprehensive understanding of the regulatory mechanisms at play.
Conclusion
In conclusion, the present study successfully identified a total of 110 intersecting genes that are implicated in both diabetes and OP. The implementation of functional annotation has revealed a potentially innovative avenue for understanding the mechanisms underlying DOP. Consequently, a refined selection comprising seven genes (MMP2, MMP9, IL17A, IL10, FGF2, IL6 and VEGFA) and eleven pharmacological agents has emerged, exhibiting promise in delivering therapeutic effects aimed at mitigating the occurrence of OP in individuals afflicted with diabetes.
Abbreviations
OP: osteoporosis; DOP: diabetes-induced osteoporosis; PPI: protein-protein interaction; DGI: drug-gene interaction; TF: transcriptional factors; T1DM: type 1 diabetes mellitus; T2DM: type 2 diabetes mellitus; BMD: bone mineral density; GO: Gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; FDR: false discovery rate; RANKL: receptor activator of NF-κB ligand; AGEs: advanced glycation end products.
Supplementary Material
Supplementary information.
Acknowledgements
Funding
This study was supported by the National Natural Science Foundation of China (NSFC), NO.82272452.
Date availability statement
The datasets presented in this study can be found in online repositories.
Author contributions
Chenfeng Wang designed the experiments, analyzed the data, and wrote the manuscript. Yihe Hu and Feng Liang devised the concept, designed the research, supervised the research, and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sheu A, Greenfield JR, White CP, Center JR. Assessment and treatment of osteoporosis and fractures in type 2 diabetes. Trends in endocrinology and metabolism: TEM. 2022;33:333-44
2. Schacter GI, Leslie WD. Diabetes and Osteoporosis: Part I, Epidemiology and Pathophysiology. Endocrinol Metab Clin North Am. 2021;50:275-85
3. McCabe LR. Understanding the pathology and mechanisms of type I diabetic bone loss. J Cell Biochem. 2007;102:1343-57
4. Kanazawa I, Yamaguchi T, Yamauchi M, Yamamoto M, Kurioka S, Yano S. et al. Serum undercarboxylated osteocalcin was inversely associated with plasma glucose level and fat mass in type 2 diabetes mellitus. Osteoporos Int. 2011;22:187-94
5. Behera J, Ison J, Voor MJ, Tyagi N. Exercise-Linked Skeletal Irisin Ameliorates Diabetes-Associated Osteoporosis by Inhibiting the Oxidative Damage-Dependent miR-150-FNDC5/Pyroptosis Axis. Diabetes. 2022;71:2777-92
6. Zawada A, Ratajczak AE, Rychter AM, Szymczak-Tomczak A, Dobrowolska A, Krela-Kaźmierczak I. Treatment of Diabetes and Osteoporosis-A Reciprocal Risk? Biomedicines. 2022 10
7. Hidayat K, Du X, Wu MJ, Shi BM. The use of metformin, insulin, sulphonylureas, and thiazolidinediones and the risk of fracture: Systematic review and meta-analysis of observational studies. Obes Rev. 2019;20:1494-503
8. Salari-Moghaddam A, Sadeghi O, Keshteli AH, Larijani B, Esmaillzadeh A. Metformin use and risk of fracture: a systematic review and meta-analysis of observational studies. Osteoporos Int. 2019;30:1167-73
9. Starup-Linde J, Gregersen S, Frost M, Vestergaard P. Use of glucose-lowering drugs and risk of fracture in patients with type 2 diabetes. Bone. 2017;95:136-42
10. Sedlinsky C, Molinuevo MS, Cortizo AM, Tolosa MJ, Felice JI, Sbaraglini ML. et al. Metformin prevents anti-osteogenic in vivo and ex vivo effects of rosiglitazone in rats. Eur J Pharmacol. 2011;668:477-85
11. Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F. et al. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022;50:D1398-d407
12. Zhang LX, Dong J, Wei H, Shi SH, Lu AP, Deng GM. et al. TCMSID: a simplified integrated database for drug discovery from traditional chinese medicine. J Cheminform. 2022;14:89
13. Wang J, Wu M, Huang X, Wang L, Zhang S, Liu H, et al. SynLethDB 2.0: a web-based knowledge graph database on synthetic lethality for novel anticancer drug discovery. Database (Oxford). 2022; 2022
14. Vincent T, Hermansson M, Bolton M, Wait R, Saklatvala J. Basic FGF mediates an immediate response of articular cartilage to mechanical injury. Proc Natl Acad Sci U S A. 2002;99:8259-64
15. Naganawa T, Xiao L, Abogunde E, Sobue T, Kalajzic I, Sabbieti M. et al. In vivo and in vitro comparison of the effects of FGF-2 null and haplo-insufficiency on bone formation in mice. Biochem Biophys Res Commun. 2006;339:490-8
16. Huang H, Luo L, Liu Z, Li Y, Tong Z, Liu Z. Role of TNF-α and FGF-2 in the Fracture Healing Disorder of Type 2 Diabetes Model Induced by High Fat Diet Followed by Streptozotocin. Diabetes Metab Syndr Obes. 2020;13:2279-88
17. Rong K, Lang Y, Zhou Y, Ni L, Wang L, Wang L. et al. Risk Genetic Variants (IL-10) for Osteoporosis in Han Population from Northwest China. J Inflamm Res. 2023;16:1091-102
18. Dos Santos Haber JF, Barbalho SM, Sgarbi JA, de Argollo Haber RS, de Labio RW, Laurindo LF. et al. The Relationship between Type 1 Diabetes Mellitus, TNF-α, and IL-10 Gene Expression. Biomedicines. 2023 11
19. Saxena M, Srivastava N, Banerjee M. Association of IL-6, TNF-α and IL-10 gene polymorphisms with type 2 diabetes mellitus. Mol Biol Rep. 2013;40:6271-9
20. Kotake S, Udagawa N, Takahashi N, Matsuzaki K, Itoh K, Ishiyama S. et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest. 1999;103:1345-52
21. Horiuchi K, Kimura T, Miyamoto T, Miyamoto K, Akiyama H, Takaishi H. et al. Conditional inactivation of TACE by a Sox9 promoter leads to osteoporosis and increased granulopoiesis via dysregulation of IL-17 and G-CSF. J Immunol. 2009;182:2093-101
22. Sigurdardottir S, Zapadka TE, Lindstrom SI, Liu H, Taylor BE, Lee CA. et al. Diabetes-mediated IL-17A enhances retinal inflammation, oxidative stress, and vascular permeability. Cell Immunol. 2019;341:103921
23. Harmer D, Falank C, Reagan MR. Interleukin-6 Interweaves the Bone Marrow Microenvironment, Bone Loss, and Multiple Myeloma. Frontiers in endocrinology. 2018;9:788
24. Akbari M, Hassan-Zadeh V. IL-6 signalling pathways and the development of type 2 diabetes. Inflammopharmacology. 2018;26:685-98
25. Singh P, Singh M, Khinda R, Valecha S, Kumar N, Singh S. et al. Genetic Scores of eNOS, ACE and VEGFA Genes Are Predictive of Endothelial Dysfunction Associated Osteoporosis in Postmenopausal Women. Int J Environ Res Public Health. 2021 18
26. Wang F, Rong P, Wang J, Yu X, Wang N, Wang S. et al. Anti-osteoporosis effects and regulatory mechanism of Lindera aggregata based on network pharmacology and experimental validation. Food Funct. 2022;13:6419-32
27. Gajewska B, Śliwińska-Mossoń M. Association of MMP-2 and MMP-9 Polymorphisms with Diabetes and Pathogenesis of Diabetic Complications. Int J Mol Sci. 2022 23
28. Sandborn WJ, Bhandari BR, Randall C, Younes ZH, Romanczyk T, Xin Y. et al. Andecaliximab [Anti-matrix Metalloproteinase-9] Induction Therapy for Ulcerative Colitis: A Randomised, Double-Blind, Placebo-Controlled, Phase 2/3 Study in Patients With Moderate to Severe Disease. J Crohns Colitis. 2018;12:1021-9
29. Shah MA, Cunningham D, Metges JP, Van Cutsem E, Wainberg Z, Elboudwarej E. et al. Randomized, open-label, phase 2 study of andecaliximab plus nivolumab versus nivolumab alone in advanced gastric cancer identifies biomarkers associated with survival. J Immunother Cancer. 2021 9
30. van Rhee F, Rosenthal A, Kanhai K, Martin R, Nishimura K, Hoering A. et al. Siltuximab is associated with improved progression-free survival in idiopathic multicentric Castleman disease. Blood Adv. 2022;6:4773-81
31. Nasonov E, Fatenejad S, Feist E, Ivanova M, Korneva E, Krechikova DG. et al. Olokizumab, a monoclonal antibody against interleukin 6, in combination with methotrexate in patients with rheumatoid arthritis inadequately controlled by methotrexate: efficacy and safety results of a randomised controlled phase III study. Annals of the rheumatic diseases. 2022;81:469-79
32. Yiu ZZN, Becher G, Kirby B, Laws P, Reynolds NJ, Smith CH. et al. Drug Survival Associated With Effectiveness and Safety of Treatment With Guselkumab, Ixekizumab, Secukinumab, Ustekinumab, and Adalimumab in Patients With Psoriasis. JAMA Dermatol. 2022;158:1131-41
33. Gottlieb AB, Deodhar A, McInnes IB, Baraliakos X, Reich K, Schreiber S. et al. Long-term Safety of Secukinumab Over Five Years in Patients with Moderate-to-severe Plaque Psoriasis, Psoriatic Arthritis and Ankylosing Spondylitis: Update on Integrated Pooled Clinical Trial and Post-marketing Surveillance Data. Acta Derm Venereol. 2022;102:adv00698
34. Paller AS, Seyger MMB, Magariños GA, Pinter A, Cather JC, Rodriguez-Capriles C. et al. Long-term Efficacy and Safety of Up to 108 Weeks of Ixekizumab in Pediatric Patients With Moderate to Severe Plaque Psoriasis: The IXORA-PEDS Randomized Clinical Trial. JAMA Dermatol. 2022;158:533-41
35. Zheng ZZ, Xu JH, Dai Y, Jiang B, Tu ZM, Li L. et al. Circulating miR-107 as a diagnostic biomarker of osteoporotic vertebral compression fracture increases bone formation in vitro and in vivo. Life Sci. 2023;323:121693
36. Šimonienė D, Stukas D, Daukša A, Veličkienė D. Clinical Role of Serum miR107 in Type 2 Diabetes and Related Risk Factors. Biomolecules. 2022 12
37. Foley NH, O'Neill LA. miR-107: a toll-like receptor-regulated miRNA dysregulated in obesity and type II diabetes. J Leukoc Biol. 2012;92:521-7
38. Song C, Guo Y, Chen F, Liu W. IRF-1-inhibited lncRNA XIST regulated the osteogenic differentiation via miR-450b/FBXW7 axis. Apoptosis. 2023;28:669-80
39. Shin J, Toyoda S, Nishitani S, Onodera T, Fukuda S, Kita S. et al. SARS-CoV-2 infection impairs the insulin/IGF signaling pathway in the lung, liver, adipose tissue, and pancreatic cells via IRF1. Metabolism. 2022;133:155236
40. Asadipooya K, Uy EM. Advanced Glycation End Products (AGEs), Receptor for AGEs, Diabetes, and Bone: Review of the Literature. J Endocr Soc. 2019;3:1799-818
41. Tamaki J, Kouda K, Fujita Y, Iki M, Yura A, Miura M. et al. Ratio of Endogenous Secretory Receptor for Advanced Glycation End Products to Pentosidine Predicts Fractures in Men. J Clin Endocrinol Metab. 2018;103:85-94
42. Lamb LS, Alfonso H, Norman PE, Davis TME, Forbes J, Müench G. et al. Advanced Glycation End Products and esRAGE Are Associated With Bone Turnover and Incidence of Hip Fracture in Older Men. J Clin Endocrinol Metab. 2018;103:4224-31
43. Franke S, Siggelkow H, Wolf G, Hein G. Advanced glycation endproducts influence the mRNA expression of RAGE, RANKL and various osteoblastic genes in human osteoblasts. Arch Physiol Biochem. 2007;113:154-61
44. Notsu M, Yamaguchi T, Okazaki K, Tanaka K, Ogawa N, Kanazawa I. et al. Advanced glycation end product 3 (AGE3) suppresses the mineralization of mouse stromal ST2 cells and human mesenchymal stem cells by increasing TGF-β expression and secretion. Endocrinology. 2014;155:2402-10
45. Bhadricha H, Patel V, Singh AK, Savardekar L, Patil A, Surve S. et al. Increased frequency of Th17 cells and IL-17 levels are associated with low bone mineral density in postmenopausal women. Sci Rep. 2021;11:16155
46. Ma Y, Ran D, Zhao H, Song R, Zou H, Gu J. et al. Cadmium exposure triggers osteoporosis in duck via P2X7/PI3K/AKT-mediated osteoblast and osteoclast differentiation. Sci Total Environ. 2021;750:141638
Author contact
![]() Corresponding author: Feng Liang, email: frankfengedu.cn. Yihe Hu, email: xy_huyhcom.
Corresponding author: Feng Liang, email: frankfengedu.cn. Yihe Hu, email: xy_huyhcom.

 Global reach, higher impact
Global reach, higher impact