ISSN: 1449-1907
Int J Med Sci 2023; 20(13):1698-1704. doi:10.7150/ijms.89242 This issue Cite
Research Paper
Use of metagenomic next-generation sequencing for diagnosis of peritonitis in end-stage liver disease
Department of Infectious Diseases, Key Laboratory of Molecular Biology for Infectious Diseases (Ministry of Education), Institute for Viral Hepatitis, the Second Affiliated Hospital, Chongqing Medical University, Chongqing, China.
*These authors contributed equally to this work
Abstract
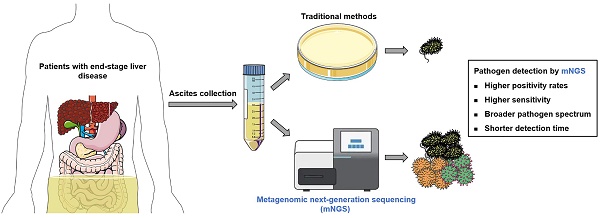
Background: Conventional methods are low in positive rates and time-consuming for ascites pathogen detection in patients with end-stage liver disease (ESLD). With many advantages, metagenomic next-generation sequencing (mNGS) may be a good alternative method. However, the related studies are still lacking.
Methods: Ascites from 50 ESLD patients were sampled for pathogen detection using mNGS and conventional methods (culture and polymorphonuclear neutrophils detection) in this prospective observational study.
Results: Forty-two samples were detected positive using mNGS. 29 strains of bacteria, 11 strains of fungi, and 9 strains of viruses were detected. 46% of patients were detected to be co-infected with 2 or more pathogens by mNGS. Moreover, mNGS showed similar and high positive rates in ESLD patients with different clinical characteristics. Compared to conventional methods, mNGS had higher positivity rates (84% vs. 20%, P<0.001), sensitivity (45.2% vs. 23.8%, P=0.039), broader pathogen spectrum, shorter detection time (24 hours vs. 3-7 days), but lower specificity (25% vs 100%, P = 0.010). Furthermore, compared to conventional methods, mNGS showed similar consistence with final diagnosis (42% vs. 36%, P=0.539).
Conclusions: mNGS may be a good supplement for conventional methods and helpful to early etiological diagnosis of peritonitis, and thus improve ESLD patients' survival.
Keywords: Metagenomic next-generation sequencing, conventional methods, peritonitis, end-stage liver disease, diagnosis

 Global reach, higher impact
Global reach, higher impact