Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(13):1662-1670. doi:10.7150/ijms.87836 This issue Cite
Review
The Structural Proteins of Membrane Rafts, Caveolins and Flotillins, in Lung Cancer: More Than Just Scaffold Elements
Department of Molecular Biomedicine and Translational Research, Instituto Nacional de Enfermedades Respiratorias “Ismael Cosío Villegas”. Mexico City, Mexico.
Received 2023-7-6; Accepted 2023-8-25; Published 2023-10-2
Abstract
Lung cancer is one of the most frequently diagnosed cancers worldwide. Due to its late diagnosis, it remains the leading cause of cancer-related deaths. Despite it is mostly associated to tobacco smoking, recent data suggested that genetic factors are of the highest importance. In this context, different processes meaningful for the development and progression of lung cancer such endocytosis, protein secretion and signal transduction, are controlled by membrane rafts. These highly ordered membrane domains contain proteins such as caveolins and flotillins, which were traditionally considered scaffold proteins but have currently been given a preponderant role in lung cancer. Here, we summarize current knowledge regarding the involvement of caveolins and flotillins in lung cancer from a molecular point of view.
Keywords: Lung cancer, membrane rafts, caveolins, flotillins, carcinogenesis.
Introduction
Primary lung cancer (LC) is the leading cause of cancer death worldwide. In 2020, it was the most diagnosed cancer after breast cancer [1,2]. Non-small cell lung cancer (NSCLC) is the most frequent type of LC, accounting for approximately 80% to 85% of the cases, while small cell lung cancer (SCLC) accounts for approximately 15% of all the patients [3]. LC is associated with tobacco smoking, but recent data indicate that 14% of LC deaths can be attributed to air pollution exposure; these values vary between 5% to more than 20% in different countries [4]. These data imply that LC currently affects populations with low smoking prevalence.
LC is influenced by endogenous and external factors that impact different molecular mechanisms involved in cell division, differentiation, and death, eventually leading to cellular transformation, cancer, and metastasis. LC have significant differences in cellular morphology, and despite progress in treatment approaches, the overall survival rate within five years is still less than 20% [5,6]. Therefore, it is crucial to understand better the molecular mechanisms underlying LC development, progression, and metastasis in order to improve prevention, treatment efficacy, and survival.
Recent advances in cancer research have revealed the complex scenario, and dynamic nature of cancer pathogenesis. In particular, research in epigenetics has evidenced that cancer involves profound changes in cellular functions due to alterations in genome structure, stability, and gene expression [7-9]. Growth receptors, transducing mediators, and effectors play a crucial role in regulating cell proliferation, and significant advances have been reported regarding the contribution of these molecules in carcinogenesis, tumor progression, and drug targeting [10-12].
Lipid rafts (LR) are membrane subdomains involved in the transport and localization of different signaling transduction elements regulating cell proliferation. For example, it is a well-documented fact, that LR participates in the translocation of the tumor necrosis factor receptor, which is essential for TNF-α-mediated NF-κB activation. Disturbing LR organization switches the outcome of TNF-α signaling from NF-κB activation to apoptosis [13]. However, the spatiotemporal organization of this signaling network is incompletely understood. Current knowledge states that the endoplasmic reticulum delivers secreted and membrane proteins to the Golgi apparatus through transport vesicles. After they are properly folded and modified, mature proteins are transported into the plasma membrane (PM) or different organelles through endosomal, regulated, or constitutive secretory pathways. Distinct sorting proteins regulate cellular trafficking, and malfunctioning of this process causes diseases, including cancer [14-16]. In this review, we will focus on recent data about the role of LR-structural proteins, caveolins, and flotillins in LC. These proteins are important because the appropriate assembly of membrane subdomains partly determines the functionality of several proteins implied in cancer. Based on the experimental work performed in LC and other types of cancer, we propose a rationale to explore the role of LR-associated proteins in cancer development.
The role of MR proteins in LC
LR are small (ranging from 10 to 200 μm), specialized dynamic cellular membrane subdomains. Currently, the term membrane rafts (MR) is preferred [17], and two distinct types are recognized based on the density of their packing and chemical nature: planar and flask-shaped or caveolae. MR are composed of sphingolipids, mainly sphingomyelin and gangliosides [18], cholesterol [19], and particular proteins (proteolipids). MR are resistant to solubilization by non-ionic detergents (DRMs) [20]; this allows its isolation from cellular membranes at low temperature [21-23]. However, DRM are MR aggregates and do not actually represent the native state of MR in living cells [24]. The harmonious interactions between lipids and lipid-associated proteins determine MR structure and function. Therefore, the traditional view of MR proteins as “scaffold supports” has been replaced by the recognition that these proteins organize and control several cell membrane attributes [25,26]. Ordered aggregation of homologous MR through protein-protein or lipid-protein interactions can stabilize larger structures functioning as platforms, which could include or exclude particular proteins for specific biochemical processes. MR have diverse cellular functions and are associated with a growing number of specialized functions at the PM level. However, the functions of these structures in subcellular membrane compartments, such as the Golgi, endocytic endosomes or exosomes, are not fully understood. Moreover, since cholesterol is synthesized in the endoplasmic reticulum (ER) but sphingolipids are synthesized in the Golgi, the first MR assemblage largely occurs in Golgi membranes [27,28], and MR participates in the subsequent delivery of transport vesicles and recycling pathways [29].
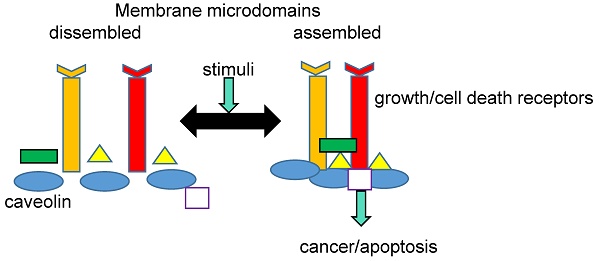
The PM plays a key role in signal transduction, and a close association between cell signaling processes and MR has been well documented. Many proteins involved in cell signaling, such as receptors, GTP-binding proteins, kinases, and phosphatases, can be selectively incorporated into the membrane in response to endogenous or exogenous stimuli. This is possible because of the asymmetric structure of both PM leaflets and the physicochemical properties conferred by the liquid-ordered phases of the MR. Currently, there is a general agreement that individual MR domains can harbor only a few proteins; thus, MRs must cluster together [29]. Raft clustering on the extracellular face of PM can be triggered by different molecules like antigens, antibodies, or other MR-binding proteins, including cholera toxin B. [30]. Cytoplasmic leaflet proteins such as flotillins and annexins could serve as clustering agents [31,32]. In addition, the recruitment and concentration of death receptors could also aggregate MR, forming clusters of apoptotic signaling molecule-enriched rafts (CASMERs). CASMERs favor the activation of procaspase 8 and apoptosis through downstream signaling molecules [33]. Finally, it is important to emphasize that the dynamic changes in MR composition could guarantee the proper chemical modification of signal transducers and protect MR resident proteins from degradation. Therefore, the dynamic MR composition would ensure the functionality of PM transducers in signaling pathways [34,35].
Several proteins are structurally and functionally related to MR in both physiological and pathological states. In fact, a comprehensive MR-proteomic database providing information about this topic is available (Mohamed A. et al. [36]). MR-associated proteins include caveolins, which are found in caveolae, and flotillins and raft-linking proteins (raftlins), which are found in flat rafts [37-39]. These proteins have important roles in cancer development and the regulation of tumor microenvironment and metastasis.
Caveolins
Caveolae are 50-100 nm flask-shaped plasma membrane domains enriched in lipids such as ceramide, phosphatidic acid, diacylglycerides, and glycosphingolipids [40]. Caveolae are involved in different physiological processes, including transport, cell survival, proliferation, migration, and apoptosis. Since caveolae were first described [41], several reports have depicted roles for caveolae in human pathologies [42,43]. The main structural proteins of caveolae are a family of proteins known as caveolins. These proteins play an important role in the pathogenesis of several diseases, including different types of carcinomas.
So far, the role of caveolin 1, (cav 1) is the best characterized in LC. Cav 1 is a 22 kDa MR-associated protein involved in different normal cell functions, including lipid transportation, cell growth, and death regulation. Different studies support that cav 1 may have opposite roles: it may act as a tumor suppressor and a cancer promoter. The role of cav1 in cancer depends on the cell type and cancer stage [42-44]. In spite of CAV1 expression is mostly reduced in LC cell lines [45,46], it was expressed in 76% of NSCLC cell lines compared with normal human lung epithelial cells. Moreover, silencing CAV1 in NSCLC cells inhibited cell proliferation and colony formation. However, CAV1 was downregulated in 95% of SCLC cell lines, and re-expression of cav1 in SCLC resulted in a decreased capacity to form cell colonies [47]. These findings account for a differentiated role of this protein: as an oncoprotein in NSCLC and as a tumor suppressor in SCLC. Other lines of evidence from in vivo, in vitro, and clinical studies have shown that CAV1 expression may account for anti-tumor properties or the aggressiveness of carcinomas [48-54]. Caveolae are important for regulating cancer signaling because they hold different signaling molecules [55]. Cav 1 and other MR-associated proteins act as oncoproteins or tumor suppressors by activating cell proliferation and survival pathways or by blocking specific components of transduction cascades. For example, residues 82-101 in cav 1-N-terminal domain (caveolin scaffolding domain, CSD) interact with many signaling proteins, such as the nonfunctional forms of endothelial nitric oxide synthase (soluble form of cav 1), heterotrimeric G proteins, and MAP kinase, to negatively regulate their activation [36,56-58].
The role of cav 1-tyrosine 14
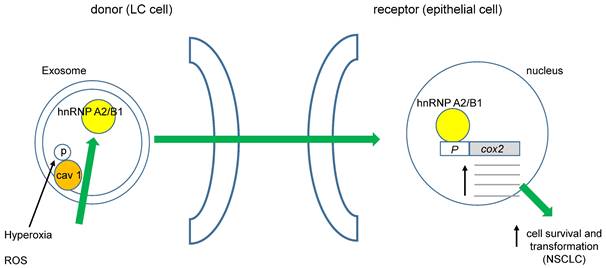
It was recently demonstrated that cav 1 phosphorylated at tyrosine 14 (pY14) becomes exacerbated in exosome membranes (microvesicles, MVs) during cell-hyperoxia and the production of reactive oxygen species (ROS). This increase in pY14 cav1 resulted in a higher release of the heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNPA2/B1) into MVs [59]. Furthermore, hnRNPA2/B1 specifically binds to the COX-2 core promoter, favoring NSCLC [60], (figure 1). Interestingly, ROS also could directly affect cav 1's role in cell migration of human LC cells by activating Akt (protein kinase B) through phosphorylation of Akt-T308 or S473. However, opposite effects of superoxide anion and hydrogen peroxide with respect to the hydroxyl radical were observed [61]. Several studies have demonstrated that oxidative stress induces the phosphorylation of cav 1-Y14, increasing cell migration. Recently, Jiang and coworkers found that cav 1- pY14 restricts the mitochondrial recruitment of mitofusin 2 in breast cancer cells. Since mitofusin 2 is critical for mitophagy, this process results in ROS accumulation and damaged mitochondria [62]. Moreover, hydrogen peroxide favors the phosphorylation of cav 1-Y14. These data point to a reciprocal interaction between oxidative stress and cav 1 function (figure 2).
ROS could induce cell transformation in NSCLC. Phosphorylation of cav 1-Y14 increases load of hnRNAPA2/B1 into exosomes, they move towards non-transformed epithelial cells favoring its transformation through enhanced cox 2 transcription.
Phosphorylation of cav 1-Y14 and its oncogenic activities in LC is differentially regulated by ROS. Superoxide anion and hydrogen peroxide promote Y14 dephosphorylation and cav 1 proteasome degradation, whereas hydroxyl radical provoke enhanced cell migration, invasion and metastasis.
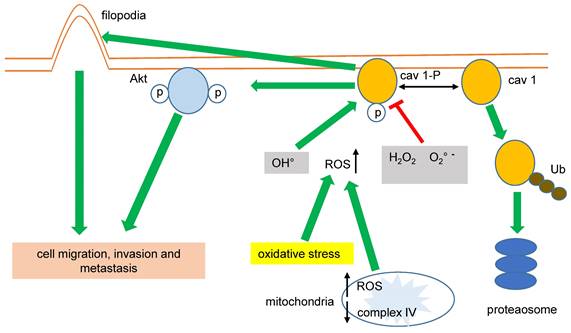
Impaired mitochondrial function and increased glycolysis (Warburg effect) are hallmarks of cancer cells. In this sense, the expression of CAV1 was greatly correlated with higher glycolysis rates and the inhibition of the mitochondrial complex IV in MDA-MB-231 breast cancer cells. Conversely, the inhibition of the mitochondrial complex IV increased the mitochondrial levels of ROS associated with cav 1-pY14, favoring cell migration and invasion [61,63], (figure 2).
Cav 1 improves the migration of cancer cells by other mechanisms; in lung adenocarcinoma (LUAD) cells, cav 1 increases invasion favoring both filopodia and lamellipodia formation [64,65]. Downregulating CAV1 gene expression by shRNA-cav 1-transduction into NSCLC H460 and H292 significantly decreased cell migration and the expression of integrins β1 and β3 [66]. Again, these data suggest that cav1-pY14 promotes cancer cell migration, invasion, and metastasis, (figure 2). Interestingly, cav 1-Y14 could also link cell mobility, migration, invasion, and oxidative stress in LC because overexpression of cav 1-Y14F displayed a dominant negative phenotype, similar to the cav 1 RNAi-mediated knockdown [67].
Another recent report based on bioinformatics identified a novel connection between cav 1 and cell motility. Cav 1 modulated the activity of cellular calcium fluxes, and both cav 1 and c-Myc displayed a favorable effect on the O-GlcNAcylation. Since O-GlcNAcylation coordinates calcium signaling via the TRPM7 channel, this process increased cell migration and invasion in LC cell lines [68]. Finally, as mentioned previously, cav 1 regulates several signal transduction pathways. Activating MR-resident proteins such as epidermal growth factor receptor (EGFR) increased the proliferation, migration, invasion, and volume of xenograft tumors, both in human lung tumor tissues and the human LUAD cell line GLC-82 [69]. siRNA-mediated downregulation of cav 1 caused a stable proliferation arrest in human LUAD and SCLC cell lines, showing a substantial decrease of pAkt and its downstream effectors, phosphorylated ERK (pERK) and STAT3 (pSTAT3). These findings highlight that the tumorigenic effects displayed by the overexpression of cav 1 are partly mediated by the EGFR/ERK signaling pathway [70].
Genetic and epigenetic regulation of cav 1 and LC
Promoter hypermethylation
Cancer development is associated with hypermethylation of specific cytosine residues of the CpG dinucleotide located in or near gene promoters (CpG-islands), global hypomethylation, and selective demethylation of the regulatory regions of some genes [71,72]. Promoter methylation and acetylation/deacetylation of histones are common epigenetic mechanisms that lead to activation/repression of transcription of several genes, mainly tumor suppressor genes. In some types of cancer, including LC, DNA-methylation represses the CAV1 gene at cancer onset, but demethylation and the concomitant re-expression of this gene occur before metastasis [73-75]. Human cav 1 is encoded on chromosome 7q31.1, and there are two isoforms, product of different mRNAs, α and β. Cav 1 α has 31 more amino acids in the N-terminus than cav 1 β [76,77]. Analyses of the CAV1 gene showed 30 CpG sites distributed in two promoter regions containing 7 and 28 CpG sites, respectively. Hypermethylation of the first three sites was strongly associated with decreased expression of cav 1 in SCLC, breast cancer and human T-leukemia cell lines, and neuronal cells [47,58,78,79]. Furthermore, the CAV1 promoter has two ETS binding sites and single binding sites for GATA6, p53, SP1, E cadherin, 3-hydroxy- 3-methylglutaryl coenzyme-A-synthase-1 (HMGCS-1), and CD44. Interestingly, the CAV1 promoter also has sterol regulatory elements (SREs) [80,81]. In fact, cav 1 directly binds cholesterol, a fundamental component of the MR; this may indicate that the role of cav 1 in cholesterol trafficking into the PM and mitochondria is linked with its function in MR signal transduction and metabolic regulation [81-84]. Other regulatory mechanisms like loss of heterozygosity have been described within chromosome 7q31.1 [85], but there is no evidence for deletions of the CAV1 gene in human tumors [86].
Regulation of cav 1 expression by miRNAs and LncRNAs. (A) miRNA-204 downregulates cav 1-mRNA, decreasing cav 1 protein, pSTAT3, and cisplatin resistance. Lnc-BMP1-1 decreases cav1 transcription, favoring apoptosis. (B) Reducing miRNA-204 levels, increases cav 1 and pSTAT3, and consequently, cell proliferation and invasion are favored. On the other hand, pSTAT3 blocks the expression of the miRNA, generating a positive feedback loop. Diminished or increased levels of miR-1827 and LncRNA-TUG1 respectively, favoring cell proliferation and metastasis. Red arrows indicate negative effects, green ones, activating effects.
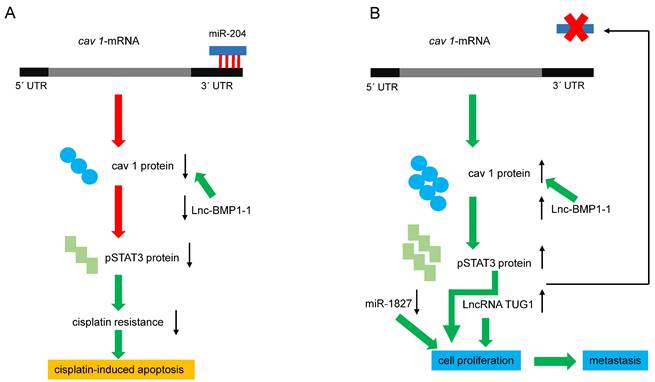
Micro RNA and Long Non-coding RNA regulating cav 1
Recent reports have clarified the mechanisms underlying the positive effect of reducing CAV 1 expression on cisplatin cell damage in human LC cells. Silencing CAV1 in A549 cells, enhanced cisplatin‐mediated mitochondrial apoptotic signaling [87]. Conversely, a higher expression of microRNA-204 (miR‐204) in A549 and PC9 cells reduces cisplatin resistance, which was related to decreased CAV1 expression [88]. The 3´ UTR of cav 1-mRNA contains a conserved 5´ AAGGGAA 3´ site targeted by miR-204 [89], and CAV1 expression is negatively modulated by miR-204 (figure 3A). The phosphorylation of AKT and Bad can be further suppressed by the miR-204/cav 1 pathway, promoting cisplatin-induced apoptosis by silencing Bcl-2 and Bcl-xl [88]. Moreover, the human gene MIR204, encodes a 110 bp pre-miR-204 stem-loop located in intron 6 of transient receptor potential nonselective cation channel, subfamily M, member 3 (TRPM3) in the locus 9q21.12-q21.13 [89-91]. Both genes are co-transcribed in clear renal carcinoma cells, and miR-204 negatively regulates the translation of TRPM3 by binding to its 3´UTR. Interestingly, the expression of TRPM3 also requires another direct target of miR-204, cav 1: in fact, knocking down cav 1 reduced the expression of TRPM3 [91]. Since TRPM3 regulates autophagy, the previous results imply that cav 1 modulates this cellular process through another mechanism. The expression of miR-204 is regulated by DNA methylation, long non-coding RNAs (LncRNAs), and transcription factors. LncRNA taurine upregulated 1 (LncRNA TUG1) like DNA methylation blocks the expression of miR-204, whereas the transcription factors Pax6 and STAT3 activate and inhibit its expression, respectively [90]. As mentioned previously, silencing CAV1 gene in human lung carcinoma cells decreases activated pSTAT3, suggesting that high levels of cav 1 could increase pSTAT3 and thereby block the expression of miR-204, as described in renal cancer cells (figure 3B). Using functional and bioinformatics analyses, Guo and coworkers recently identified that microRNA-1827 (miR-1827) directly targets cav 1. In this context, miR-1827 level was decreased in NSCLC tissues and A549 cells, and decreased miR-1827 levels in a manner directly correlated with metastasis. Restoring miR-1827 prevented anoikis resistance, suppressed cell viability, and induced apoptosis in A549 cells, whereas over-expression of cav 1 attenuated these effects [91].
In addition to TUG1, other LncRNAs, such as LET [93] and TARID [94], also play crucial roles in LC tumorigenesis [92]. TUG1 is an LncRNA with 6.7-kb nucleotides located at chromosome 22q12 [95]; this gene is overexpressed in several other types of cancer [96-100]. TUG1 shows higher expression in lung tumors than in adjacent tissue, and its expression in LC cell lines favors proliferation and migration and inhibits apoptosis [101] (figure 3B). A recent report found that TUG1 is among the most expressed LncRNAs in LC and may be a biomarker of lung neoplasms. Other LncRNAs, such as PTENP1 and UCA1, could be biomarkers of NSCLC and LUAD, respectively [102]. Serum levels of TUG1 provide a high diagnostic value for NSCLC and LUAD [103,104], so it has been proposed as a therapeutic target in SCLC [105].
Lnc-BMP1-1 is another LncRNA related to cav 1 in LC. This gene is transcribed from the intron area of surfactant protein C; thus, highly expressed in the lung. Recent studies show that Lnc-BMP1-1 might promote the transcription of CAV1 by downregulating the expression of histone deacetylase 2 [106] (figure 3A). Lnc-BMP1-1 expression was decreased in LC tissues, especially in patients with a history of cigarette smoking; therefore, it is logical to expect a lower expression of CAV1 in those patients. Interestingly, it is known that oxidative stress induced by cigarette smoke causes lung tumorigenesis, and smokers are more prone to have lower expression of Lnc-BMP1-1 and cav 1 [106]. Hence, the following question arises: Does Lnc-BMP1-1 promote the tumor-suppressing effects of cav 1? Considering that cav 1 prevents hydrogen peroxide-induced oxidative damage to lung carcinoma cells [107], and higher cav 1 expression enhances the sensitivity of A549 cells to the anti-cancer drug doxorubicin [106], it is evident that downregulating CAV1 could be unfavorable for NSCLC. In this context, it was reported that the anti-cancer drug cordycepin is involved in the JNK/Foxo3a axis, which in turn, activates the Bax/caspase-3-mediated pathway. Since cav1 signaling directly targets the Bax/caspase-3-apoptosis mediated pathway, this may be the mechanism whereby cav 1 causes cell death of A549 cells. [108]. Therefore, a second question arises: How does NSCLC avoid the negative effects exerted by Lnc-BMP1-1 on CAV1 expression? The answer still is beyond the currently available experimental data, but clarifying this paradox will be important for LC therapy.
Caveolin 2 (cav 2) in LC
The importance of cav 2 in cancer pathogenesis was recognized almost at the same time as that of cav 1. Increased CAV2 expression is related to poor prognosis of pancreatic cancer both in vivo and in vitro. The mechanism involves a specific genetic variant that impairs miR-548s binding to the 3´UTR, affecting the expression of genes related to focal adhesion and extracellular matrix organization [109]. Regarding LC, only a few reports have implicated cav 2 in cancer progression. Murine lung carcinoma cell lines implanted into cav 2-KO mice were unable to grow, and tumor size was dramatically reduced two weeks after implantation [110,111]. In these murine models, cav 2 deficiency triggered an anti-tumor immune response and impaired tumor angiogenesis. Host cav 2 deficiency reduced tumor cell proliferation in the late stages of the implanted tumor's development, when defective neovascularization is essential for supporting tumor growth. In fact, cav 2 knockout mice showed more necrotic than apoptotic cell death within the implanted tumors [111]. Interestingly, cav 2 deficiency significantly increased anti-angiogenic thrombospondin 1 (TSP1) mRNA levels, with the concomitant decrease in endothelial nitric oxide synthase (eNOS) phosphorylation and a decrease in pro-angiogenic factors such as vascular endothelial growth factor-A (AVEGF-A).
Flotillins in LC
Flotillin 1 and flotillin 2 (flot 1 and flot 2) are MR-associated proteins involved in several cellular functions, such as signaling, endocytosis, and other cytoskeleton-related roles. The expression of both proteins is interdependent, i.e., the downregulation of flot 2 severely reduces flot 1 expression [112]. Both flot 1 and 2 are associated with LC [112-115]. Data from knockdown models and flot overexpression in cell culture indicate that overexpression of flot 1 inhibits apoptosis and enhances the malignant behavior of LUAD by promoting cell growth, invasion, and migration. The expression of several cell cycle regulatory markers, such as cyclin D kinase 2 (CDK2), cyclin E, and cyclin D1, were significantly increased, whereas p16 expression was reduced [114]. Epithelial-mesenchymal transition is also favored by overexpression flot 1, and it is mediated by higher Akt phosphorylation of and FOXO3a downregulation [114]. Interestingly, it has been reported that, in NSCLC, overexpression of cytoskeleton protein 4.1N significantly reduces the expression of flot 1, avoiding its carcinogenic effects through lower activation of the Wnt/β-catenin/c-Myc pathway and higher E-cadherin expression [115]. Nevertheless, increased flot 2 expression was significantly associated with higher EGFR expression in NSCLC compared to the non-cancerous lung control tissues [116]. EGFR overexpression and EGFR mutations play a key role in NSCLC, mainly by resistance to its degradation. Clathrin-independent EGFR endocytosis is mediated by flot 1 and MR, and induces the lysosomal degradation of EGFR [117]. Taken together, these data indicate that flot 1 is essential for tumor suppressor in NSCLC. Therefore, flotillins seem to have oncogenic and tumor suppressor effects similar to those of cav 1.
Final remarks
Cancer cells can evade apoptosis through well-known mechanisms involving defects in cell signaling or apoptosis mediators. Since MRs participate in cell signaling and the activation of apoptosis, the proteins that determine the assemblage and proper functioning of MR, such as caveolins and flotillins, are targets for regulating these physiological processes. However, the fact that these proteins act both as tumor suppressors and oncoproteins is intriguing and still unclarified. Beyond caveolins and flotillins, this phenomenon is extensive to other MR “scaffolding” proteins, for example, several MR proteins with the MARVEL (MAL and related proteins for vesicle trafficking and membrane link) domain. However, MARVEL proteins differ from caveolins and flotillins because they cross the membrane four times, with extracellular, transmembrane, and intracellular segments. Myelin and lymphocyte protein (MAL), MAL2, MAL like protein (MALL, formerly BENE) are involved in several human cancers [23,118,119], either promoting or suppressing tumor growth and metastasis [120-122]. The effect of these proteins on cancer, whether beneficial or detrimental, depends on epigenetic mechanisms. Therefore, considering the initial proposal by doctors Weinberg and Hanahan [123], we must continue searching for common ground between the processes and mediators driving cell transformation.
Acknowledgements
This publication was supported by the financial assignment by the Instituto Nacional de Enfermedades Respiratorias “Ismael Cosío Villegas” to the research project B31-18.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
2. Fitzmaurice C, Abate D, Abate D. et al. Global Burden of Disease Cancer Collaboration. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5:1749-68 doi:10.1001/jamaoncol.2019.2996
3. Ferlay J, Colombet M, Soerjomataram I. et al. Cancer Statistics for the Year 2020: An Overview. Int J Cancer. 2021 doi: 10.1002/ijc.33588
4. Turner MC, Andersen ZJ, Baccarelli A. et al. Outdoor air pollution and cancer: an overview of the current evidence and public health recommendations. CA Cancer J Clin. 2020;70:460-479
5. Lv Y, Huang Z, Lin Y. et al. miRNA expression patterns are associated with tumor mutational burden in lung adenocarcinoma. Onco immunology. 2019;8:e1629260. doi:10.1080/2162402X.2019.1629260
6. Luo W, Wang Z, Zhang T. et al. Immunotherapy in non-small cell lung cancer: rationale, recent advances and future perspectives. Precis Clin Med. 2021;4:258-270
7. Belinsky SA. Unmasking the lung cancer epigenome. Annu Rev Physiol. 2015;77:453-74
8. Cooper WA, Lam CDL, O'Toole SA. et al. Molecular biology of lung cancer. J Thorac Dis. 2013;5(Suppl 5):S479-S490
9. Alexandre JM, Diaz-Lagares A, Villalba M. et al. Translating cancer epigenomics into the clinic: focus on lung cancer. Transl Res. 2017;189:76-92
10. Jiang Y, Xie W-J, Chen R-W. et al. The Hippo signaling core components YAP and TAZ as new prognostic factors in lung cancer. Front Surg. 2022;9:813123
11. Sanaei M-J, Razi S, Pourbagheri-Sigaroodi A. et al. The PI3K/Akt/mTOR pathway in lung cancer; oncogenic alterations, therapeutic opportunities, challenges, and a glance at the application of nanoparticles. Transl Oncol. 2022;18:101364
12. Corral de la Fuente E, Olmedo-Garcia ME, Gomez Rueda A. et al. Targeting KRAS in non-small cell lung cancer. Front Oncol. 2022;11:792635. doi.org/10.3389/fonc.2021.792635
13. Legler DF, Micheau O, Doucey MA. et al. Recruitment of TNF receptor 1 to lipid rafts is essential for TNFα-mediated NF-κB activation. Immunity. 2003;18:655-64
14. Howell GJ, 1 Holloway ZG, Cobbold C. et al. Cell biology of membrane trafficking in human disease. Int Rev Cytol. 2006;252:doi 10.1016/S0074-7696(06)52005-4
15. Bajaj R, Warner AN, Fradette JF. et al. Dance of the Golgi: understanding Golgi dynamics in cancer metastasis. Cells. 2022;11:1484. doi.org/10.3390/cells11091484
16. Del Giudice S, De Luca V, Parizadeh S. et al. Endogenous and exogenous regulatory signaling in the secretory pathway: role of Golgi signaling molecules in cancer. Front Cell Dev Biol. 2022;10:833663
17. Pike LJ. Rafts defined: a report on the Keystone Symposium on lipid rafts and cell function. J Lipid Res. 2006;47:1597-8
18. Gabandé-Rodríguez E, Boya P, Labrador V. et al. High sphingomyelin levels induce lysosomal damage and autophagy dysfunction in Niemann Pick disease type A. Cell Death Differ. 2014;21:864-875
19. Simons K, Ehehalt R. Cholesterol, lipid rafts, and disease. J Clin Invest. 2002;110:597-603
20. Gajate C, Mollinedo F. Lipid raft isolation by sucrose gradient centrifugation and visualization of raft-located proteins by fluorescence microscopy: the use of combined techniques to assess Fas/CD95 location in rafts during apoptosis triggering. Methods Mol Biol. 2021;2187:147-186
21. Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46-50
22. Schroeder RJ, Ahmed SN, Zhu Y. et al. Cholesterol and sphingolipid enhance the triton X-100 insolubility of glycosylphosphatidylinositol anchored proteins by promoting the formation of detergent-insoluble ordered membrane domains. J Biol Chem. 1998;273:1150-57
23. Rubio-Ramos A, Labat-de-Hoz L, Correas I. et al. The MAL protein, an integral component of specialized membranes, in normal cells and cancer. Cells. 2021;10:1065-1101 doi.org/10.3390/cells10051065
24. Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377-88
25. Magal LG, Yaffe Y, Shepshelovich J. et al. Clustering and lateral concentration of raft lipids by the MAL protein. Mol Biol Cell. 2009;20:3751-62
26. van Deventer S, Arp AB, van Spriel AB, Dynamic plasma membrane organization. a complex symphony. Trends Cell Biol. 2021;31:119-129 doi: 10.1016/j.tcb.2020.11.004
27. Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111-136
28. Ikonen E, Simons K. Protein and lipid sorting from the trans-Golgi network to the plasma membrane in polarized cells. Semin Cell Dev Biol. 1998;9:503-09
29. Rajendran L, Simons K. Lipid rafts and membrane dynamics. J Cell Sci. 2005;11:1099-1102
30. Harder T, Engelhardt KR. Membrane domains in lymphocytes-from lipid rafts to protein scaffolds. Traffic. 2004;5:265-75
31. Gauthier-Rouvière C, Bodin S, Comunale F. et al. Flotillin membrane domains in cancer. Cancer Metastasis Rev. 2020;39:361-74
32. Draeger A, Wray S, Babiychuk EB. Domain architecture of the smooth-muscle plasma membrane: regulation by annexins. Biochem J. 2005;387:309-14
33. Mollinedo F, Gajate C. Lipid rafts, death receptors and CASMERs: new insights for cancer therapy. Future Oncol. 2010;6:491-94
34. Mollinedo F, Gajate C. Lipid rafts as signaling hubs in cancer cell survival/death and invasion: implications in tumor progression and therapy: thematic review series: biology of lipid rafts. J Lipid Res. 2020;61:611-35
35. Li B, Qin Y, Yu X. et al. Lipid raft involvement in signal transduction in cancer cell survival, cell death and metastasis. Cell Prolif. 2022;55:e13167 doi: 10.1111/cpr.13167
36. Mohamed A, Shah AD, Chen D. et al. Raft prot V2: understanding membrane microdomain function through lipid raft proteomes. Nucleic Acids Res. 2018 gky948
37. Lajoie P, Nabi IR. Lipid rafts, caveolae, and their endocytosis. In: Jeon KW, ed. International review of cell and molecular biology. 1st ed. Academic Press-Elsevier; San Diego. 2010:135-153
38. Banning A, Tomasovic A, Tikkanen R. Functional aspects of membrane association of reggie/flotillin proteins. Curr Protein Pept Sci. 2011;12:725-35 doi: 10.2174/138920311798841708
39. Saeki K, Miura Y, Aki D. et al. The B cell-specific major raft protein, Raftlin, is necessary for the integrity of lipid raft and BCR signal transduction. EMBO J. 2003;22:3015-26 doi: 10.1093/emboj/cdg293
40. Simons K, Vaz WL. Model systems, lipid rafts and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269-95
41. Palade G E. Fine structure of blood capillaries. J Appl Physics. 1953;24:1424
42. Schwencke C, Braun-Dullaeus RC, Wunderlich C. et al. Caveolae and caveolin in transmembrane signaling: Implications for human disease. Cardiovasc Res. 2006;70:42-49
43. Wichera SA, Prakasha YS, Pabelick CM. Caveolae, caveolin-1 and lung diseases of aging. Expert Rev Resp Med. 2019;13:291-300
44. Yang W, Geng Ch, Yang Z. et al. Deciphering the roles of caveolin in neurodegenerative diseases. The good, the bad and the importance of context. Ageing Res Rev. 2020;62:101116
45. Racine C, Bélanger M, Hirabayashi H. et al. Reduction of caveolin 1 gene expression in lung carcinoma cell lines. Biochem Biophys Res Commun. 1999;255:580-86
46. Shatz M, Liscovitch M. Caveolin-1: A tumor-promoting role in human cancer. Int J Radiat Biol. 2008;84:177-189
47. Sunaga N, Miyajima K, Suzuki M. et al. Different roles for caveolin-1 in the development of non-small cell lung cancer versus small cell lung cancer. Cancer Res. 2004;64:4277-85
48. Campos A, Burgos-Ravanal R, González MF. et al. Cell intrinsic and extrinsic mechanisms of caveolin-1-enhanced metastasis. Biomolecules. 2019;9:314. doi.org/10.3390/biom9080314
49. Sun M-Z, Guan Z, Liu S. et al. Caveolin-1 interferes cell growth of lung cancer NCI-H446 cell through the interactions with phospho-ERK1/2, estrogen receptor and progestin receptor. Biomed Pharmacother. 2012;66:242-48
50. Ho CC, Huang PH, Huang HY. et al. Up-regulated caveolin-1 accentuates the metastasis capability of lung adenocarcinoma by inducing filopodia formation. Am J Pathol. 2002;161:1647-56
51. Kim Y-J, Kim J-H, Kim O. et al. Caveolin-1 enhances brain metastasis of non-small cell lung cancer, potentially in association with the epithelial-mesenchymal transition marker SNAIL. Cancer Cell Int. 2019;19:171. doi.org/10.1186/s12935-019-0892-0
52. Pancotti F, Roncuzzi L, Maggiolin M. et al. Caveolin-1 silencing arrests the proliferation of metastatic lung cancer cells through the inhibition of STAT3 signaling. Cell Signal. 2012;24:1390-97
53. Volonte D, Vyas AR, Chen C. et al. Caveolin-1 promotes the tumor suppressor properties of oncogene-induced cellular senescence. J Biol Chem. 2018;293:1794-809
54. Duregon E, Senetta R, Bertero L. et al. Caveolin 1 expression favors tumor growth and is associated with poor survival in primary lung adenocarcinomas. Tumor Biol. 2017;39:1010428317694311
55. Wang Y, Song Y, Che X. et al. Caveolin-1 enhances RANKL-induced gastric cancer cell migration. Oncol Rep. 2018;40:1287-96
56. Okamoto T, Schlegel A, Scherer PE. et al. Caveolins, a family of scaffolding proteins for organizing 'preassembled signaling complexes' at the plasma membrane. J Biol Chem. 1998;273:5419-422
57. Williams TM, Lisanti PM. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol. 2005;288:C494-C506
58. Tsuji Y, Nakagawa T, Hatanaka M. et al. Quantification of caveolin isoforms using quantitative real-time RT-PCR, and analysis of promoter CpG methylation of caveolin-1 in human T cell leukemia cell lines. Int J Mol Med. 2006;18:489-95
59. Lee H, Li C, Zhang Y. et al. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J Exp Med. 2019;216:2202-20
60. Xuana Y, Wanga J, Ban L. et al. hnRNPA2/B1 activates cyclooxygenase-2 and promotes tumor growth in human lung cancers. Mol Oncol. 2016;10:610-24
61. Luanpitpong S, Talbott SJ, Rojanasakul Y. et al. Regulation of lung cancer cell migration and invasion by reactive oxygen species and caveolin-1. J Biol Chem. 2010;285:38832-40
62. Jiang Y, Krantz S, Qin X. et al. Caveolin-1 controls mitochondrial damage and ROS production by regulating fission-fusion dynamics and mitophagy. Redox Biol. 2022;52:102304
63. Díaz-Valdivia N, Simón L, Díaz J. et al. Mitochondrial Dysfunction and the Glycolytic Switch Induced by Caveolin-1 Phosphorylation Promote Cancer Cell Migration, Invasion, and Metastasis. Cancers. 2022;14:2862. doi.org/10.3390/cancers14122862
64. Ho C-C, Huang P-H, Huang H-Y. et al. Up-Regulated caveolin-1 accentuates the metastasis capability of lung adenocarcinoma by inducing filopodia formation. Am J Pathol. 2002;161:1647-56
65. Chanvorachote P, Chunhacha P, Pongrakhananon V. Caveolin-1 induces lamellipodia formation via an Akt-dependent pathway. Cancer Cell Int. 2014;14:52
66. Petpiroon N, Bhummaphan N, Tungsukrutha S. et al. Chrysotobibenzyl inhibition of lung cancer cell migration through Caveolin-1-dependent mediation of the integrin switch and the sensitization of lung cancer cells to cisplatin-mediated apoptosis. Phytomedicine. 2019;58:152888
67. Shatza M, Lustiga G, Reich R. et al. Caveolin-1 mutants P132L and Y14F are dominant negative regulators of invasion, migration and aggregation in H1299 lung cancer cells. Exp Cell Res. 2010;316:1748-62
68. Luanpitpong S, Rodboon N, Samart P. et al. A novel TRPM7/O-GlcNAc axis mediates tumour cell motility and metastasis by stabilising c-Myc and caveolin-1 in lung carcinoma. Br J Cancer. 2020;123:1289-1301 doi.org/10.1038/s41416-020-0991-7
69. Luan T-Y, Zhu T-N, Cui Y-J. et al. Expression of caveolin-1 is correlated with lung adenocarcinoma proliferation, migration, and invasion. Med Oncol. 2015;32:207
70. Pancotti F, Roncuzzi L, Maggiolini M. et al. Caveolin-1 silencing arrests the proliferation of metastatic lung cancer cells through the inhibition of STAT3 signaling. Cell Signal. 2012;24:1390-97
71. Pantazi P, Acha-Sagredo A, Papaioannou A. et al. DNA methylation: its role in transcriptional regulation and association with lung cancer. Res Rep Biochem. 2015;5:11-30
72. Patra S, Bettuzzi S. Epigenetic DNA-methylation regulation of genes coding for lipid raft-associated components: A role for raft proteins in cell transformation and cancer progression. Oncol Rep. 2007;17:1279-90
73. Wiechen K, Diatchenko L, Agoulink A. et al. Caveolin-1 is down-regulated in human ovarian carcinoma and acts as candidate tumour suppressor gene. Am J Pathol. 2001;159:1635-43
74. Cui J, Rohr LR, Swanson G. et al. Hypermethylation of the caveolin-1 gene promoter in prostate cancer. Prostate. 2001;46:249-56
75. Ho C-C, Huang P-H, Huang H-Y. et al. Up-regulated caveolin-1 accentuates the metastasis capability of lung adenocarcinoma by inducing filopodia formation. Am J Pathol. 2002;161:1647-56
76. Kogo H, Aiba T, Fujimoto T, Cell type-specific occurrence of caveolin-1alpha, -1beta in the lung caused by expression of distinct mRNAs. J Biol Chem. 2004; 279:25574-81.
77. Engelman JA, Zhang XL, Lisanti MP. Sequence and detailed organization of the human caveolin-1 and -2 genes located near the D7S522 locus (7q31.1). Methylation of a CpG island in the 5' promoter region of the caveolin-1 gene in human breast cancer cell lines. FEBS Lett. 1999;448:221-30
78. Zschocke J, Manthey D, Bayatti N. et al. Estrogen receptor-mediated silencing of caveolin gene expression in neuronal cells. J Biol Chem. 2002;277:38772-38780
79. Boopathi E, Mendes-Gomes C, Goldfarb R. et al. Transcriptional Repression of Caveolin-1 (CAV1) gene expression by GATA-6 in bladder smooth muscle hypertrophy in mice and human beings. Am J Pathol. 2011;178:2236-50
80. Roy S, Luetterforst R, Harding A. et al. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat Cell Biol. 1999;1:98-105
81. Nwosu ZC, Ebert MP, Dooley S. et al. Caveolin-1 in the regulation of cell metabolism: a cancer perspective. Mol Cancer. 2016;15:71. doi 10.1186/s12943-016-0558-7
82. Bosch M, Mari M, Herms A. et al. Caveolin-1 deficiency causes cholesterol-dependent mitochondrial dysfunction and apoptotic susceptibility. Curr Biol. 2011;21:681-686
83. Rothberg KG, Heuser JE, Donzell WC. et al. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673-82
84. Fernandez-Rojo MA, Gongora M, Fitzsimmons RL. et al. Caveolin-1 is necessary for hepatic oxidative lipid metabolism: evidence for crosstalk between caveolin-1 and bile acid signaling. Cell Rep. 2013;4:238-47
85. Engelman JA, Zhang XL, Lisanti MP. Genes encoding human caveolin-1 and -2 are colocalized to the D7S522 locus (7q31.1), a known fragile site (FRA7G) that is frequently deleted in human cancers. FEBS Lett. 1998;436:403-10
86. Hurlstone AF, Reid G, Reeves JR. et al. Analysis of the CAVEOLIN-1 gene at human chromosome 7q31.1 in primary tumours and tumour-derived cell lines. Oncogene. 1999;18:1881-90
87. Liu Y, Fu Y, Hu X. et al. Caveolin-1 knockdown increases the therapeutic sensitivity of lung cancer to cisplatin-induced apoptosis by repressing Parkin-related mitophagy and activating the ROCK1pathway. J Cell Physiol. 2020;235:1197-08
88. Huang G, Lou T, Pan J. et al. MiR-204 reduces cisplatin resistance in non-small cell lung cancer through suppression of the caveolin-1/AKT/Bad pathway. Aging (Albany N. Y.). 2019;11:2138-50
89. Deo M, Yu J-Y, Chung K-H. et al. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538-48
90. Liu J, Liu Y, Wang F. et al. miR-204: molecular regulation and role in cardiovascular and renal diseases. Hypertension. 2021;78:270-81
91. Guo X, Wang Z, Sun Q. et al. The inhibitory effect of microRNA-1827 on anoikis resistance in lung adenocarcinoma A549 cells via targeting caveolin-1. Acta Biochim Biophys Sin. 2020;52:1148-55
92. Niu Y, Ma F, Huang W. et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16:5. doi 10.1186/s12943-016-0575-6
93. Yang F, Huo XS, Yuan SX. et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083-96
94. Arab K, Park YJ, Lindroth AM. et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604-14
95. Rapicavoli NA, Blackshaw S. New meaning in the message: noncoding RNAs and their role in retinal development. Dev Dyn. 2009;238:2103-14
96. Zhang Q, Geng PL, Yin P. et al. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311-15
97. Han Y, Liu Y, Gui Y. et al. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:555-59
98. Esfandi F, Taheri M, Omrani MD. et al. Expression of long non-coding RNAs (lncRNAs) has been dysregulated in non-small cell lung cancer tissues. BMC Cancer. 2019;19:222. doi.org/10.1186/s12885-019-5435-5
99. Hall DP, Cost NG, Hegde S. et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell. 2014;26:738-753
100. Ghaforui-Fard S, Vafaee R, Taheri M. Taurine-upregulated gene 1: A functional long noncoding RNA in tumorigenesis. J Cell Physiol. 2019;234:17100-12
101. Liu H, Wang S-S, Guan J-Z. et al. Regulatory effect and mechanism of the long non-coding RNA TUG1 on the proliferation, apoptosis, and migration of lung cancer cells. Asian J Sur. 2022;45:1338e1340
102. Guo Z, Hui Y, Kong F. et al. Finding lung-cancer-related lncRNAs based on Laplacian regularized least squares with unbalanced bi-random walk. Front Genet. 2022;13:933009
103. Yuan S, Xiang Y, Guo X. et al. Circulating long noncoding RNAs act as diagnostic biomarkers in non-small cell lung cancer. Front Oncol. 2020;10:537120
104. Liu H, Zhou G, Fu X. et al. Long noncoding RNA TUG1 is a diagnostic factor in lung adenocarcinoma and suppresses apoptosis via epigenetic silencing of BAX. Oncotarget. 2017;8:101899-910
105. Ou C, Li G. Long non-coding RNA TUG1: a novel therapeutic target in small cell lung cancer. J Thorac Dis. 2017;9:E644-E645
106. Ling X, Li Y, Qiu F. et al. Down expression of lnc-BMP1-1 decreases that of Caveolin-1 is associated with the lung cancer susceptibility and cigarette smoking history. Aging. 2020;12:462-80
107. Suchaoin W, Chanvorachote P. Caveolin-1 attenuates hydrogen peroxide-induced oxidative damage to lung carcinoma cells. Anticancer Res. 2012;32:483-90
108. Joo JC, Hwang JH, Jo E. et al. Cordycepin induces apoptosis by caveolin-1-mediated JNK regulation of Foxo3a in human lung adenocarcinoma. Oncotarget. 2017;8:12211-24
109. Zhu Y, Tian J, Peng X. et al. A genetic variant conferred high expression of CAV2 promotes pancreatic cancer progression and associates with poor prognosis. Eur J Cancer. 2021;151:94-105
110. Liu Y, Qi X, Li G. et al. Caveolin-2 deficiency induces a rapid anti-tumor immune response prior to regression of implanted murine lung carcinoma tumors. Sci Rep. 2019;9:18970
111. Liu Y, Jang S, Xie L. et al. Host deficiency in caveolin-2 inhibits lung carcinoma tumor growth by impairing tumor angiogenesis. Cancer Res. 2014;74:6452-62
112. Zhang L, Mao Y, Mao Q. et al. FLOT1 promotes tumor development, induces epithelial-mesenchymal transition, and modulates the cell cycle by regulating the Erk/Akt signaling pathway in lung adenocarcinoma. Thorac Cancer. 2019;10:909-917 doi: 10.1111/1759-7714.13027
113. Wang Y-L, Yao E-J, Guo L. et al. Expression of flotillin-2 in human non-small cell lung cancer and its correlation with tumor progression and patient survival. Int J Clin Exp Pathol. 2015;8:601-07
114. Zhang PF, Zeng GQ, Hu R. et al. Identification of Flotillin-1 as a novel biomarker for lymph node metastasis and prognosis of lung adenocarcinoma by quantitative plasma membrane proteome analysis. J Proteomics. 2012;77:202-14
115. Yang Q, Zhu M, Wang Z. et al. 4.1N is involved in a flotillin-1/β-catenin/Wnt pathway and suppresses cell proliferation and migration in non-small cell lung cancer cell lines. Tumor Biol. 2016;37:12713-23
116. Wen Q, Wang W, Chu S. et al. Flot-2 expression correlates with EGFR levels and poor prognosis in surgically resected Non-Small Cell Lung Cancer. PLoS One. 2015 doi:10.1371/journal.pone.0132190
117. Yao N, Wang C-R, Liu M-Q. et al. Discovery of a novel EGFR ligand DPBA that degrades EGFR and suppresses EGFR-positive NSCLC growth. Signal Transduct Target Ther. 2020;5:214
118. Bhandari A, Shen Y, Sindan N. et al. MAL2 promotes proliferation, migration, and invasion through regulating epithelial-mesenchymal transition in breast cancer cell lines. Biochem Biophys Res Commun. 2018;504:434-39
119. Li J, Li Y, Liu H. et al. The four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are highly expressed in colorectal cancer and correlated with poor prognosis. PLoS One. 2017;12:e0178515
120. Wang K, Yang Y, Zheng S. et al. Association mining identifies MAL2 as a novel tumor suppressor in colorectal cancer. Onco Targets Ther. 2022;15:761-69
121. Hatta M, Nagai H, Okino K. et al. Down-regulation of members of glycolipid-enriched membrane raft gene family, MAL and BENE, in cervical squamous cell cancers. J Obstet Gynaecol Res. 2004;30:53-58
122. Lara-Lemus R. On the role of myelin and lymphocyte protein (MAL) in cancer: a puzzle with two faces. J Cancer. 2019;10:2312-18 doi: 10.7150/jca.30376
123. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation”. Cell. 2011;144:646-74 doi: 10.1016/j.cell.2011.02.013
Author contact
![]() Corresponding author: betony44com; 52 55 54871705.
Corresponding author: betony44com; 52 55 54871705.

 Global reach, higher impact
Global reach, higher impact