Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(12):1600-1615. doi:10.7150/ijms.87430 This issue Cite
Research Paper
Identification of lncRNAs associated with uterine corpus endometrial cancer prognosis based on the competing endogenous RNA network
1. Department of Clinical Laboratory, Yunnan Cancer Hospital, The Third Affiliated Hospital of Kunming Medical University, Kunming, 650106, Yunnan, China.
2. Research and Development Unit, Shenzhen GenDo Medical Technology Co., Ltd., Dapeng, Shenzhen, 518000, China.
3. Qujing Medical College, Qujing, 655000, Yunnan, China.
4. SDIVF R&D Centre, 209,12W, HKSTP, Shatin, Hong Kong, China.
5. Department of Sports Medicine, Qujing First People's Hospital, 650500, Yunnan, China.
6. CUHK-SDU Joint Laboratory on Reproductive Genetics, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China.
7. Department of Clinical Laboratory, Yunnan Molecular Diagnostic Center, The Second Affiliated Hospital of Kunming Medical University, Kunming, 650500, Yunnan, China.
# These authors contribute equally to this work
Received 2023-6-23; Accepted 2023-9-6; Published 2023-9-25
Abstract
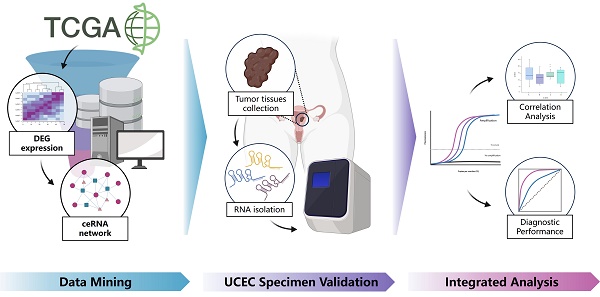
Uterine Corpus Endometrial Carcinoma (UCEC) is one of the major malignant tumors of the female reproductive system. However, there are limitations in the currently available diagnostic approaches for UCEC. Long non-coding RNAs (lncRNAs) play important roles in regulating biological processes as competitive endogenous RNA (ceRNA) in tumors. To study the potential of lncRNAs as non-invasive diagnostic tumor markers, RNA-sequencing dataset of UCEC patients from The Cancer Genome Atlas was used to identify differentially expressed genes. A lncRNA-miRNA-mRNA ceRNA network was constructed by differentially expressed lncRNAs, miRNAs and miRNAs. Pathway enrichment and functional analysis for the mRNAs in the constructed ceRNA network provide the direction of future research for UCEC by demonstrating the most affected processes and pathways. Seven potential lncRNA biomarkers (C20orf56, LOC100144604, LOC100190940, LOC151534, LOC727677, FLJ35390, LOC158572) were validated in UCEC patients by quantitative real-time PCR. Notably, LOC100190940 and LOC158572 were identified as novel RNA molecules with unknown functions. Receiver operating characteristic (ROC) curve analysis demonstrated that the combined 7 lncRNAs had a high diagnostic value for UCEC patients with area under curve (AUC) of 0.941 (95% CI: 0.875-0.947). Our study highlights the potential of the validated 7 lncRNAs panel as diagnostic biomarkers in UCEC, providing new insights into the UCEC pathogenesis.
Keywords: lncRNAs, uterine corpus endometrial cancer, ceRNA network, diagnosis
Introduction
Uterine Corpus Endometrial Carcinoma (UCEC), also referred to as endometrial cancer, is one of the three major malignant tumors of the female reproductive system. It is the second most prevalent malignant tumor (20~30%) of the female reproductive system in China [1]. Among the malignant tumors of the female reproductive system, UCEC has a better prognosis with a 5-year survival rate of 74% to 91% [2, 3]. However, it is important to further improve the survival rate with the development of reliable and accurate tumor markers for UCEC for early diagnosis. The current treatment of UCEC is mainly surgery, supported by radiotherapy, chemotherapy, and targeted therapy [4, 5]. The gold standard for diagnosis is segmental curettage and hysteroscopic endometrial biopsy. However, there are many parameters that may influence the results. For example, the size or location of the tissue sampling may not meet the detection standards. As a result, a single test may not provide a clear diagnosis and it is difficult to achieve the purpose of dynamic monitoring of the disease through multiple sampling [6]. Serum tumor markers were commonly used in clinical diagnosis of UCEC include CA125, CA199, HE4, however their specificity and sensitivity for detecting early-stage tumors remain relatively low [7]. Therefore, effective biomarkers to provide the basis for early diagnosis, treatment, and prognosis of UCEC tumors are urgently needed.
In the past few years, there has been a surge in interest in studying long non-coding RNAs. One popular approach to visualize the complex transcriptional regulatory interactions between microRNAs (miRNAs), messenger RNAs (mRNAs), and lncRNAs is by constructing a competitive endogenous RNA (ceRNA) network [8, 9]. The ceRNA hypothesis, also known as the miRNA sponge, was proposed by Salmena et al. in 2011, which elucidates the intrinsic mechanism by which various RNAs regulate biological processes across transcriptomes [10, 11]. LncRNAs are endogenous RNAs with more than 200 nucleotides without open reading frames and the ability to encode proteins. They provide the binding sites for protein or interact with DNA and RNA through complementary base pairing to regulate multiple biological processes [12]. It has been known that lncRNAs participate in chromatin remodeling, post-translational modifications [13], histone and splicing modifications and other processes in the form of RNAs [14, 15]. They are also able to form nucleic acid-protein complexes or become small RNA molecules to exert different biological functions [16]. Studies have shown that lncRNAs can regulate the expression of oncogenes and tumor suppressor genes through epigenetic transcriptional regulation, post-transcriptional regulation and other mechanisms, leading to gene activation or silencing [17, 18]. It can also act as a regulation of ceRNA and miRNA to mediate the expression level of target genes through the competitive binding of miRNA response elements (MREs) which play an important role in tumorigenesis [19].
The relationship between lncRNAs and UCEC has been suggested in different aspects including the mechanisms by which lncRNA functions. For example, oncogenic lncRNAs promote the occurrence and development of tumors by enhancing cell proliferation and invasion and inhibiting apoptosis [20]. On the other hand, tumor suppressor lncRNAs inhibit tumor cells' proliferation, migration and invasion, and metastasis. They also promote cell apoptosis and inhibit tumorigenesis and development [21]. Studies have suggested that lncRNAs are potential diagnostic biomarkers for UCEC. For instance, Ouyang et al. constructed a model based on seven lncRNAs (AC110491.1, AL451137.1, AC005381.1, AC103563.2, AC007422.2, AC108025.2, and MIR7-3HG) as potential prognostic factors. The patients were categorized into high- and low-risk groups by this model, and survival was significantly improved in the low-risk group [22]. Moreover, lncRNAs can be also used to assess the prognosis of UCEC. For example, the expression level of lncRNA UCA1 in lymph node metastasis tissues is higher than that in proliferative endometrium and primary UCEC tissues. The expression level of UCA1 is closely related to lymph node metastasis, distant metastasis, tumor grade, advanced TNM and vascular invasion and has become a reference indicator for determining the prognosis of patients with endometrial cancer [23]. Furthermore, lncRNAs can predict the response to chemotherapy. Sun found that the expression level of lncRNA HOTAIR in cisplatin-resistant UCEC cells was significantly reduced [24]. Therefore, lncRNAs can be used as molecular markers for predicting the efficacy of treatments and drug resistance in UCEC.
Given there is a lack of comprehensive studies on lncRNAs in UCEC, we used lncRNA, mRNA and miRNA expression data from The Cancer Genome Atlas (TCGA) to construct a ceRNA network. Clinical samples were also retrieved from our hospital for validating the bioinformatics results. The mechanism of action of lncRNAs and their clinical value in early diagnosis and treatment of UCEC is investigated in this study to provide a new insight into the mechanism of UCEC pathogenesis. We mainly focused on lncRNAs of the ceRNAs involved in cancer processes. Finally, a set of lncRNAs (C20orf56, LOC100144604, LOC100190940, LOC151534, LOC727677, FLJ35390, LOC158572) was found to be a potential prognostic indicator in UCEC.
Materials and Methods
Data mining and samples collection
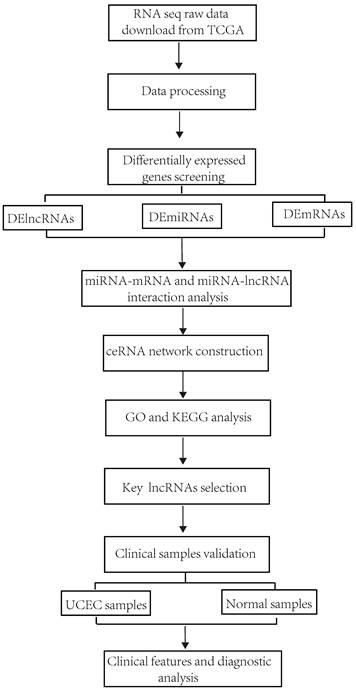
This study's workflow is shown in Figure 1. Clinical information and RNA-sequencing (RNA-seq) data were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/)) by RTCGA Toolbox R package [25]. The RNA-seq dataset is composed of three types of data including lncRNAs, mRNAs and miRNA. According to the exclusion criteria, the samples were excluded if 1) their histological diagnosis was not UCEC; 2) they contained other malignant tumors; 3) incomplete clinical data were provided; 4) they were obtained from patients treated with radiotherapy and chemotherapy before surgery.
Flowchart of the construction of ceRNA network in UCEC.
The tissues of patients with endometrial cancer were diagnosed by the Department of Gynecology of Yunnan Cancer Hospital. The study design was approved by the appropriate ethics review board of Committee of Yunnan Cancer Hospital (No. KYLX202146). All patients were informed consent. A total of 38 patients were collected as the case objects, and 10 relatively normal endometrium of non-UCEC patients were collected as the control group. All cases were from the first onset, without any treatment before surgery, and no history of other malignant tumors. The specimens were confirmed by pathological diagnosis. Intraoperative endometrial tissue specimens were placed in a cryotube containing GeneFresh Tissue ATCG RT Storage solution (GeneDoTech, Shenzhen). All specimens were transferred to the refrigerator at -70°C for storage. Basic information and clinical data of patients with endometrial cancer were collected, including age, menopause status, tumor's location, tumor's size, pathological type, FIGO stage, degree of differentiation, lymph node metastasis and distant metastasis, and follow-up treatment methods after surgery.
Differential expressed genes screening and analysis
The RNASeqV2 system was used to process the raw data of lncRNA and mRNA [26]. The raw data of microRNA sequencing was normalized by Illumina HiSeq 2000 miRNA seq sequencing platforms' high throughput sequencing software. R Studio was used to analyze the difference in expression levels between tumor tissues and normal controls and calculate the False Discovery Rate (FDR), absolute log2 Fold Change (log2FC) and P values. Differentially expressed (DE) miRNAs, lncRNAs and mRNAs were obtained by the thresholds of absolute log2FC > 1.5, FDR < 0.05, and P value < 0.01. We plotted heatmaps and volcano plots using the R package "ggplots" to visualize the results of the differential expression analysis [27].
Construction of ceRNA network of UCEC
To better explore the relationship between DE lncRNAs, miRNAs and mRNAs, the TCGA database was used to construct a lncRNA-mediated ceRNA network in UCECs. First, miRcode (http://www.mircode.org/) was used to predict the miRNAs that interact with differential expressed lncRNAs, and intersect them with the DE miRNAs [28]. Then, the three databases, Targetscan (http://www.targetscan.org/) [29], miRDB (http://www.mirdb.org/) [30] and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) [31] were used for comparative analysis to identify the relationship between the DE miRNAs and the DE mRNAs, while the mRNAs targeted by miRNAs were predicted. After a series of analyses, the lncRNA-miRNA and miRNA-mRNA regulatory pairs were identified. Finally, using Cytoscape (http://www.cytoscape.org/), the corresponding relationships between the three were visualized to construct a ceRNA network diagram [11]. Additionally, mRNAs of tumor suppressor genes (TSGs) and oncogenes were retrieved from the Network of Cancer (NCG) database (v7.1), including 254 TSGs and 256 oncogenes. mRNAs of apoptosis-related genes were retrieved from the Reactome database with the pathway ID “R-HSA-109581) which contains 181 genes for analysis.
Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis
The Database for Annotation Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) was used to perform functional and pathway enrichment analyses for the lncRNA-related DEmRNAs in the ceRNA network [32]. “Human” was selected in the Gene Ontology (GO) database and the GO terms with p-value <0.05 and enrichment scores > 1.5 were considered as significantly enriched. Using the same screening criteria and methods, the significantly enriched pathways of DE genes were identified using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Survival analysis based on differential expressed lncRNAs
The Kaplan‐Meier survival analysis was used to evaluate the association between differentially expressed lncRNAs (C20orf56, LOC100144604, LOC100190940, LOC151534, LOC727677, FLJ35390, LOC158572) and overall survival of the patients [33]. Survival curve was generated using the R package "survival".
RNA extraction and qRT-PCR validation
Total RNA was extracted from tissues using the RNA extraction kit from Shenzhen Genedo Medical Technology Co., Ltd. (Shenzhen, China), which was reverse transcribed into cDNA using a reverse transcription kit (Takara, Dalian, China). Next, qRT-PCR was performed using the FastStart Universal SYBR Green Mastermix (Takara, Dalian, China) on LightCycler® 480 instrument. All the PCR results were calculated using the -ΔCt method, where -ΔCt =- (Ct lncRNA - Ct GAPDH) tumor, and 2-ΔCt represents fold change. The qRT-PCR reactions were repeated in triplicate. Primers for qRT-PCR were synthesized by Shanghai Integrated Biotech Solutions Co.,Ltd (Shanghai, China), and the primers sequences could be found in Table 1.
Construction of prognostic model by validated lncRNAs
To assess the performance of lncRNAs, we used the R package pROC to plot and visualize receiver operating characteristic (ROC) curves and compute the area under the curve (AUC) and its confidence intervals [34]. LASSO (least absolute shrinkage and selection operator) regression model was fitted to the lncRNA-based classifier using the R package glmnet via penalized maximum likelihood [35].
Top 10 upregulated and downregulated miRNAs, lncRNAs, and mRNAs in UCEC
| Differentially expressed lncRNAs | Differentially expressed mRNAs | Differentially expressed miRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene symbol | log2FC | P value | Gene symbol | log2FC | P value | Gene symbol | log2FC | P value | ||
| Upregulation | Upregulation | Upregulation | ||||||||
| LOC642587 | 9.067395 | 1.35E-08 | DEFA5 | 23.45222 | 2.99E-07 | hsa-mir-1269 | 8.30602 | 1.82E-42 | ||
| C20orf56 | 8.891294 | 2.03E-08 | MAGEA9B | 22.23204 | 2.48E-16 | hsa-mir-205 | 5.937647 | 1.91E-63 | ||
| UCA1 | 7.99487 | 2.22E-10 | RPTN | 21.45037 | 2.65E-11 | hsa-mir-516a-2 | 5.424192 | 1.83E-17 | ||
| DSCR8 | 6.476508 | 0.011553 | POU3F3 | 21.31608 | 4.67E-10 | hsa-mir-183 | 4.898356 | 1E-127 | ||
| LOC400794 | 5.662909 | 0.0007 | AMBN | 20.87678 | 1.42E-10 | hsa-mir-522 | 4.739051 | 1.21E-13 | ||
| DLX6AS | 4.997421 | 4.6E-07 | MT4 | 20.8689 | 6.97E-08 | hsa-mir-519a-1 | 4.658677 | 3.55E-20 | ||
| C12orf36 | 4.934214 | 0.005073 | MAGEA3 | 20.83983 | 8.38E-10 | hsa-mir-516a-1 | 4.632546 | 6.43E-15 | ||
| LOC285629 | 4.738387 | 2.61E-05 | SPINK6 | 20.80407 | 1.18E-11 | hsa-mir-138-2 | 4.375384 | 5.4E-17 | ||
| LOC100190940 | 4.674075 | 0.000215 | CST1 | 11.65061 | 6.15E-08 | hsa-mir-96 | 4.318775 | 5.9E-107 | ||
| MGC4473 | 4.474938 | 0.010739 | CST4 | 10.95447 | 5.17E-07 | hsa-mir-891a | 4.224868 | 1.21E-17 | ||
| Downregulation | Downregulation | Downregulation | ||||||||
| LOC572558 | -6.97616 | 1.5E-06 | SLITRK3 | -8.4095 | 6.44E-08 | hsa-mir-1-2 | -3.88584 | 5.31E-25 | ||
| LOC283174 | -5.47984 | 9.36E-08 | BCHE | -7.14543 | 1.54E-07 | hsa-mir-133a-1 | -3.67418 | 5.06E-29 | ||
| C6orf176 | -5.40818 | 0.000527 | TCF23 | -7.00746 | 6.19E-05 | hsa-mir-133a-2 | -3.59272 | 2.53E-16 | ||
| LOC401093 | -4.85392 | 2.96E-20 | C7 | -6.85244 | 2.75E-08 | hsa-mir-133b | -3.5841 | 1.12E-18 | ||
| CCL14-CCL15 | -4.45833 | 1.87E-05 | PTGER3 | -6.81345 | 2.06E-08 | hsa-mir-1247 | -3.34065 | 2.51E-25 | ||
| C20orf200 | -4.31234 | 1.36E-08 | PTGFR | -6.72977 | 2.72E-07 | hsa-mir-1-1 | -3.29388 | 1.58E-08 | ||
| WIT1 | -3.4896 | 0.004975 | MYH11 | -6.65421 | 3.04E-09 | hsa-mir-424 | -3.23424 | 6.98E-37 | ||
| LOC255167 | -3.19814 | 0.004812 | SSTR3 | -6.6446 | 1.1E-06 | hsa-mir-100 | -3.23122 | 3.58E-36 | ||
| LOC100302650 | -3.11601 | 2.04E-06 | DPT | -6.59786 | 9.98E-09 | hsa-mir-3926-1 | -3.22531 | 1.83E-32 | ||
| C9orf110 | -3.11431 | 4.93E-05 | DES | -6.57231 | 1.5E-05 | hsa-mir-143 | -3.2153 | 3.72E-40 | ||
Clinical features analysis of key lncRNAs
We selected the lncRNAs from the subnetwork to study their associations with specific clinical characteristics of the patients, including age, menopause status, tumor's location, tumor's size, pathological type, FIGO stage, degree of differentiation, lymph node metastasis and distant metastasis, and tumor biomarker value. The patients were divided into two groups according to the clinical features cut-off value. The lncRNA expression levels were analyzed for statistical significance of the difference using Student's t test for independent samples by two-group comparisons.
Statistical analysis
R Studio (R version 3.4.1), GraphPad Prism 8.2 and SPSS 19.0 statistical packages were used for statistical analysis. The log-rank test was used in the Kaplan-Meier survival curve analysis, and the Student's t test (two-tailed) was used in qRT-PCR analysis between two groups of data sets. Results with P-value <0.05 were considered statistically significant.
Results
Screening of differentially expressed RNA genes in UCEC
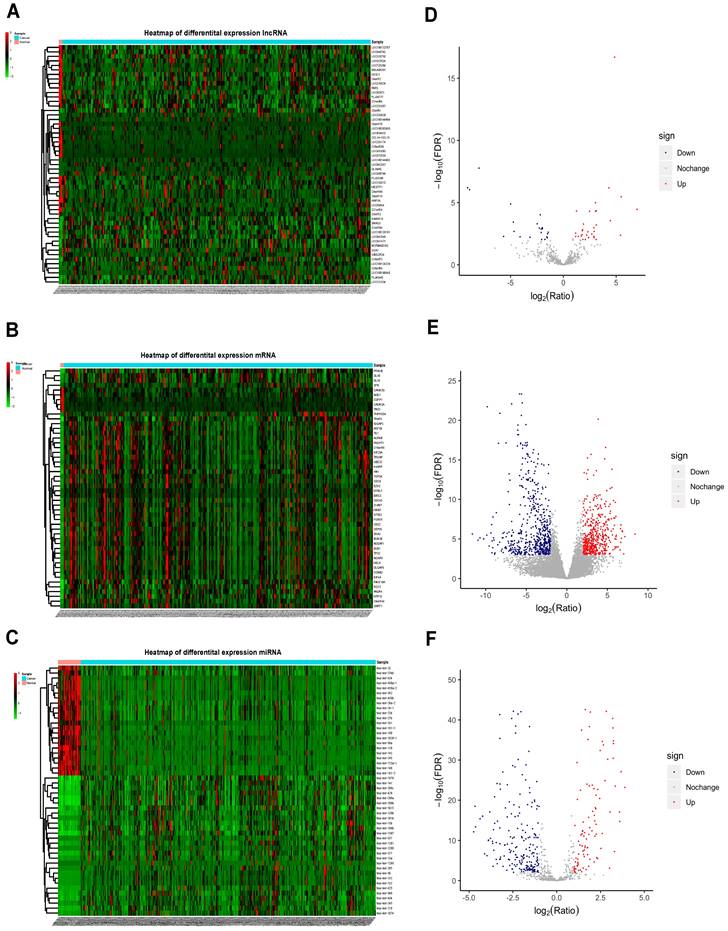
The normalized data obtained from TCGA contained protein-coding RNAs, non-coding RNAs, pseudogenes, immunoglobulins, and other non-coding RNAs. Relevant UCEC data were retrieved from the TCGA database, with a total of 266 UCEC samples and 3 control samples which met the eligibility criteria included for analysis. In total, 53 differentially expressed lncRNAs (DElncRNAs) (31 upregulated and 22 downregulated), 1072 differentially expressed mRNAs (DEmRNAs) (477 up-regulated and 595 down-regulated), and 318 differentially expressed miRNAs (DEmiRNAs) (100 upregulated and 218 downregulated) were identified. The top 50 DElncRNAs, DEmRNAs and DEmiRNAs are shown in Figure 2A, 2B and 2C and the volcano plots are shown in Figure 2D, 2E and 2F. The top 10 mRNAs, lncRNAs, and miRNAs exhibiting significant upregulation and downregulation are listed in Table 1.
Construction of the miRNA-lncRNA-mRNA ceRNA network in UCEC
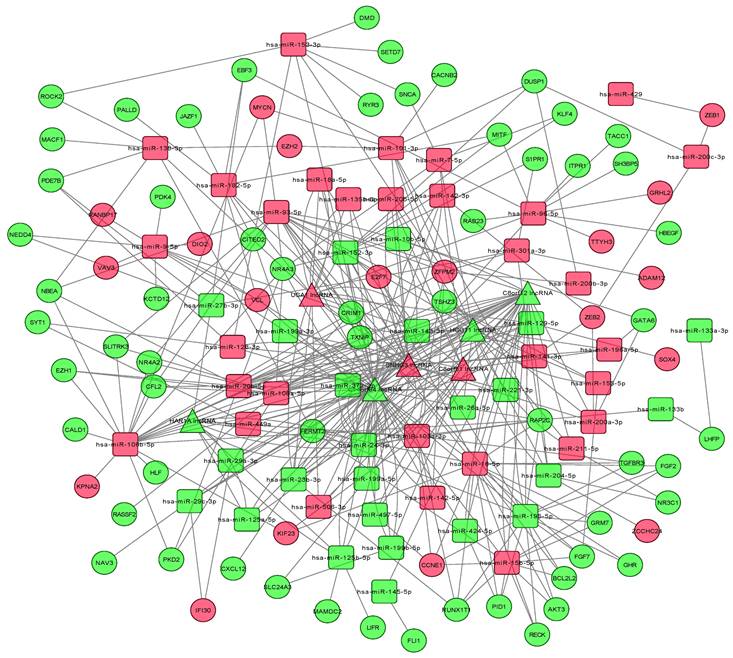
StarBase v2.0 was used to explore potential microRNA response elements (MREs) and predict the relationship between lncRNAs and miRNAs. The miRCode database was used to predict the target lncRNAs of the DEmiRNAs, and the list of relationships between them was obtained. The analyses identified a total of 503 lncRNA-miRNA regulatory pairs consisting of interactions between 97 DEmiRNAs and 15 DElncRNAs while 733 miRNA-mRNA regulatory pairs were composed of interactions between 97 DEmiRNAs and 250 DEmRNAs (Supplementary Tables S1 and S2). Additionally, the relationships between the DEmRNAs of oncogenes, tumor suppressor genes (TSGs) and apoptosis-related genes and DEmiRNAs, DElncRNAs were studied (Supplementary Table S3). A total of 8 mRNAs of TSGs were found to be interacting with 13 DEmiRNAs and 13 DElncRNAs, 16 mRNAs of oncogenes were associated with 40 DEmiRNAs and 14 DElncRNAs, while 5 mRNAs of apoptosis-related genes were related to 9 DEmiRNAs and 11 DElncRNAs. In order to further understand the functions of DElncRNAs, the regulatory ceRNA network of the competitive target relationship between the DElncRNAs, DEmRNAs and DEmiRNAs was constructed that includes 7 lncRNAs, 105 mRNAs and 94 miRNAs (Figure 3).
Functional enrichment analysis the ceRNA network-associated DEmRNAs
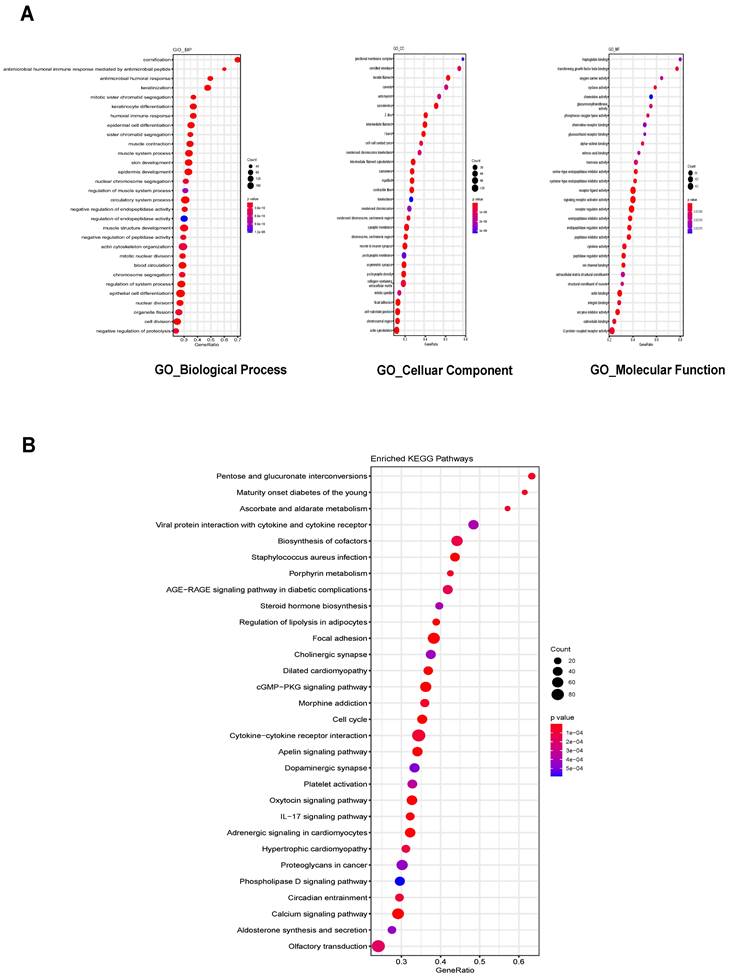
We then predicted the roles of lncRNAs by analyzing ceRNA network-associated DEmRNAs. Gene Ontology (GO) analysis was performed separately for Biological Process (BP), Cellular Component (CC) and Molecular Function (MF). The top 30 enriched GO terms based on the p-value were identified, including the regulation of epidermal cell differentiation for BP, focal adhesion for CC, and signaling receptor activator activity for MF (Figure 4A). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was also performed, with the top 30 enriched pathways identified including biosynthesis of cofactors, cytokine-cytokine receptor interaction and cGMP-PKG signaling pathways (Figure 4B). Our findings provide a valuable resource for discovering additional molecular participants/interactions in UCEC, as these functions have not been studied in UCEC to our knowledge.
Kaplan-Meier survival analysis of DElncRNAs
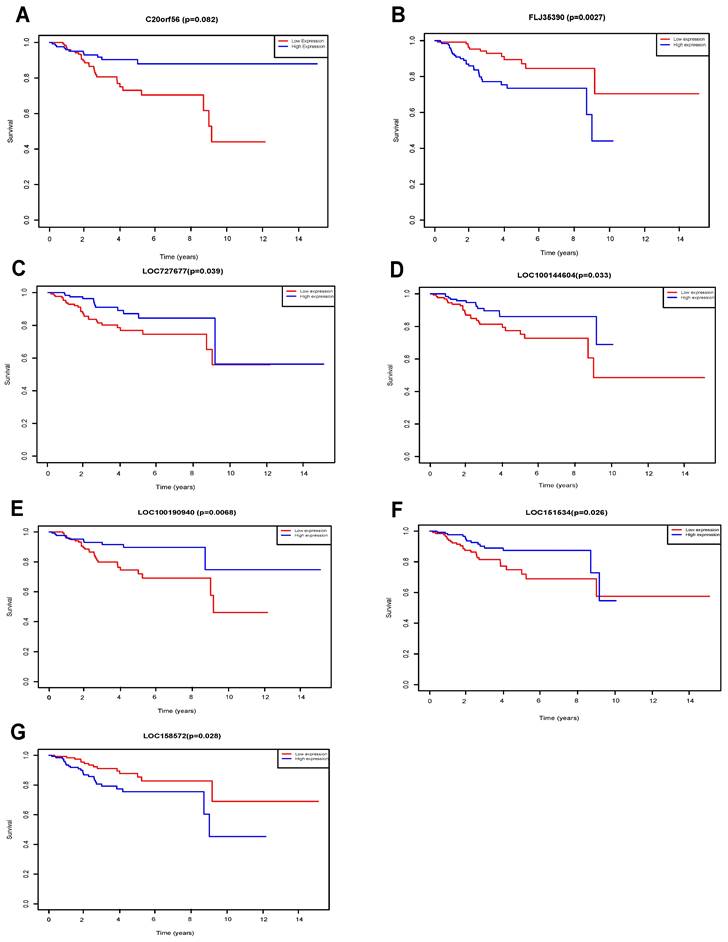
Seven lncRNAs (C20orf56, LOC100144604, LOC100190940, LOC151534, LOC727677, FLJ35390, LOC158572) were identified as being significantly associated with the overall survival of UCEC patients by Kaplan-Meier survival analysis and log-rank test (p<0.05) (Figure 5). We found that high expression of the 4 lncRNAs (FLJ35390, LOC100144604, LOC151534 LOC158572) was associated with poorer prognosis in UCEC patients. In contrast, high expression of 3 lncRNAs (C20orf56, LOC100190940, LOC727677) was associated with better prognosis in UCEC patients.
Heatmap and volcano plots for differentially expressed mRNAs, miRNA and lncRNAs. Left panels, heat maps for all differentially expressed lncRNAs (A), mRNAs (B), and miRNAs (C) in UCEC; Right panels, volcano plots showing lncRNAs (D), mRNAs (E), and miRNAs (F) with |log2FC| ≥ 1.5 (P < .001). Blue, downregulated; red, upregulated; gray, not differentially expressed. lncRNA: long noncoding RNA; miRNA: microRNA
Real-time quantitative PCR primer sequences used in this study
| Gene Symbol | Forward primer | Reverse primer | Product Length (bp) |
|---|---|---|---|
| GAPDH | ATCTCTGCCCCCTCTGCTGA | GATGACCTTGCCCACAGCCT | 303 |
| C20orf56 | CCAAATGGGTGCTGTGTGTG | TACCATGGCAGCGTGATTGT | 195 |
| LOC100190940 | ACTGTGGTCGCTGAGAACTG | GTTTCCGAGACCCACGTCAT | 191 |
| LOC100144604 | ACCCCCAAGGAAGAGTCAGT | ACATGTCAGAAGCCGTCAGG | 134 |
| LOC727677 | TATACACCAGAATGCCCCGC | CCATTGTCAACCGCAACACT | 104 |
| LOC151534 | CGTGGGGAATGGACCCATAG | CGAGCCTTGGTCTTGTCTGT | 118 |
| LOC158572 | TGAATCACGTGTGGAGGGTG | CCAGGTGCATCTACTGCGAA | 173 |
| FLJ35390 | CAATACACGGGTGGGCAGAA | CTGGGCCCCATCATCAACAA | 184 |
The lncRNA-miRNA-mRNA competitive endogenous (ceRNA) network. Squares represent miRNA, circles represent mRNAs, triangles represent lncRNAs.
Validation of DElncRNAs in clinical samples
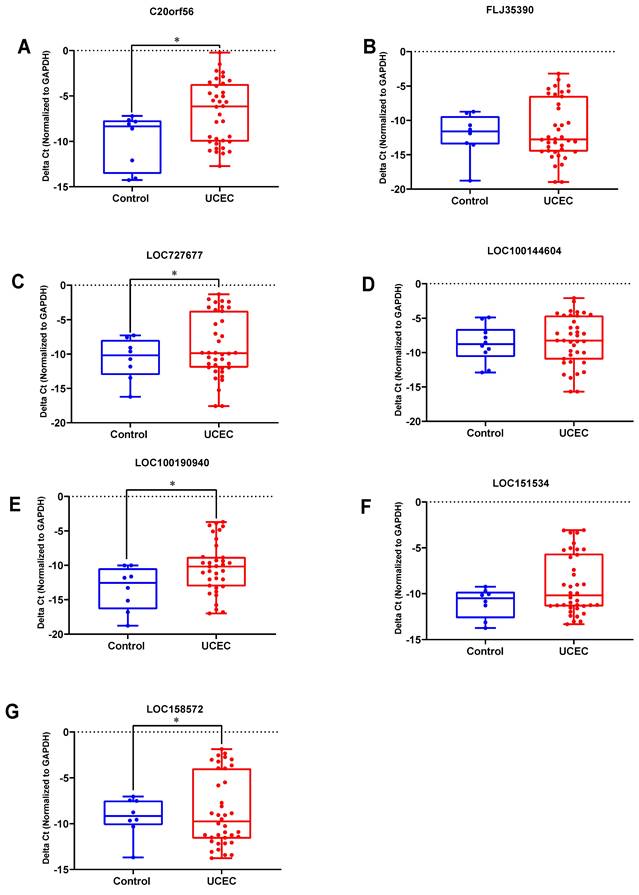
qRT-PCR experiments were used to further validate the expression levels of the 7 key lncRNA genes in 38 UCEC samples and 10 normal endometrial tissues. The expression levels of these lncRNAs are shown in Figure 6. Four lncRNAs (C20orf56, LOC727677, LOC100190940 and LOC158572) were significantly up-regulated in the UCEC samples compared to the control group (Figure 6A, 6C, 6E, 6G). However, there was no significant difference for the three lncRNAs (FLJ35390, LOC100144604 and LOC151534) between the two groups (Figure 6B, 6D, 6F).
The enrichment analysis of GO and KEGG pathway. (A) Top 30 GO biological process terms, Cellular Components terms and Molecular function terms of the DEmRNAs in the ceRNA network (B) Top 30 KEGG pathways of DEmRNAs in the ceRNA network. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
KM survival of DElncRNAs. Survival curves for DElncRNAs that are associated with the overall survival (OS) of UCECpatients. (A) C20orf56, (B) FLJ35390, (C) LOC727677, (D) LOC100144604, (E) LOC100190940, (F) LOC151534, (G) LOC158572.
The expression level of 7 lncRNAs in UCEC and normal tissues. DElncRNAs were detected using qRT-PCR. (A) C20orf56, (B) FLJ35390, (C) LOC727677, (D) LOC100144604, (E) LOC100190940, (F) LOC151534, (G) LOC158572.UCEC represents UCEC patient group, Control represents the normal tissue group. Experiments were performed in triplicate. * p<0.05 by Student's t test.
Correlation analysis of lncRNA expression levels in UCEC patient tissues and clinical data
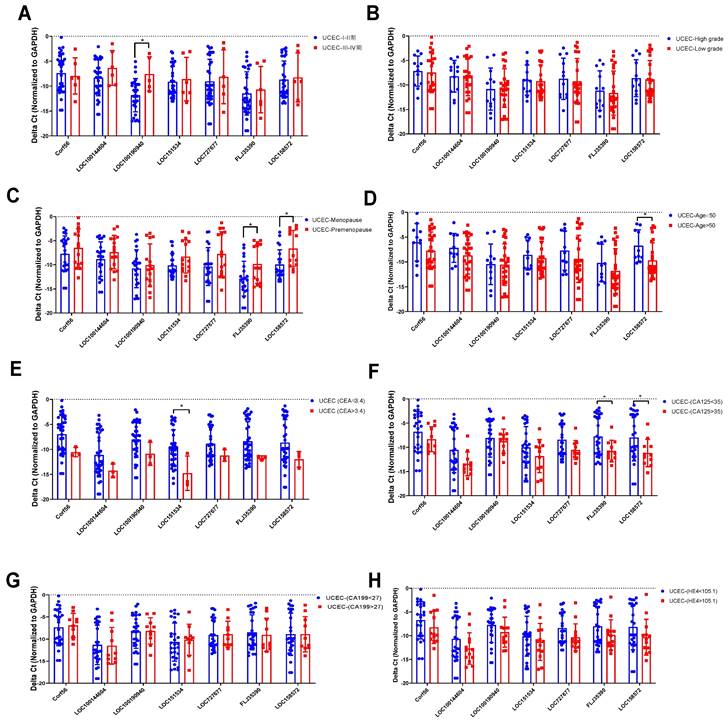
The relationship between the expression levels of 7 lncRNAs in 38 UCEC samples and the clinicopathological characteristics (age, menopause, stage, differentiation, etc.) of endometrial cancer patients was analyzed. The overall correlation between lncRNA expression level and clinical features were listed in Table 3. qRT-PCR was used to assess the correlation between lncRNA expression and clinical features. In clinical staging, the expression of LOC100190940 in endometrial tissue of patients with stage III-IV UCEC was higher than that of patients with stage I-II (Figure 7A). There was no significant difference in the degree of differentiation of different pathological tissues (Figure 7B). The expression levels of FLJ35390 (P=0.0268) and LOC15857 (P=0.0080) in the premenopausal group were higher than those in the postmenopausal group (Figure 7C), while the expression level of LOC158572 in the young (aged ≤50) group was higher than that in the old (aged >50) group (Figure 7D). Moreover, the expression level of LOC151534 was significantly lower in high CEA (>3.4) group than that in low CEA (<3.4) group (Figure 7E), while the expression levels of FLJ35390 and LOC58572 were significantly lower in high CA125(>35) group than that in low CA125 (<35) group (Figure 7F). Among the tumor markers currently used to for the diagnosis of UCEC, the expression of 7 lncRNAs was not significantly associated with the serum levels of CA199 and HE4 (Figure 7G, 7H).
Construction of prognostic model by seven key lncRNAs
Based on the above experimental results, lncRNAs with statistically significant differences in expression between UCEC and control groups (P<0.05) were extracted to analyze their diagnostic value for UCEC. Receiver operating curves (ROC) were drawn based on the expression data of the seven identified lncRNAs, as shown in Figure 8. The ROC analysis showed that the results were statistically significant (P<0.05). Except for FLJ35390, the other six lncRNAs were able to distinguish UCEC and normal endometrial patients. Logistic regression modeling was used to fit the statistical results of the 7 lncRNAs into a combined detection data, and the ROC curve analysis was used to obtain the combined detection results. The area under the curve (AUC) was 0.941 (95% CI: 0.875-0.947). Our result suggests that the combined detection of 7 lncRNAs has a relatively greater diagnostic value for distinguishing endometrial cancer samples from normal endometrial tissues.
Relationship between lncRNA expression levels and clinicopathological characteristics of UCEC patients
| Clinicopathological characteristics | Sample size | Percentage | P-value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| C20orf56 | LOC100144604 | LOC100190940 | LOC151534 | LOC727677 | FLJ35390 | LOC158572 | |||
| Degree of differentiation | |||||||||
| High-mid | 12 | 32.43% | 0.8242 | 0.8861 | 0.8685 | 0.8244 | 0.7584 | 0.7569 | 0.8498 |
| Mid-Low | 25 | 67.57% | |||||||
| Clinical stage | |||||||||
| Stage I-II | 32 | 84.21% | 0.5422 | 0.8252 | 0.0137 | 0.7135 | 0.6551 | 0.6948 | 0.8063 |
| Stage III-IV | 6 | 15.79% | |||||||
| Menopause status | |||||||||
| Menopause | 23 | 60.52% | 0.3433 | 0.2207 | 0.5980 | 0.0725 | 0.0770 | 0.0268 | 0.0080 |
| Pre-menopause | 15 | 39.47% | |||||||
| Age | |||||||||
| ≤50 | 11 | 28.95% | 0.2150 | 0.2325 | 0.9591 | 0.6004 | 0.3140 | 0.3019 | 0.0289 |
| >50 | 27 | 71.05% | |||||||
| CEA | |||||||||
| <3.5 | 35 | 92.11% | 0.1174 | 0.3606 | 0.1892 | 0.0298 | 0.2341 | 0.1542 | 0.2246 |
| >3.5 | 3 | 7.89% | |||||||
| CA125 | |||||||||
| <35 | 27 | 71.05% | 0.2674 | 0.0615 | 0.6035 | 0.2190 | 0.0653 | 0.0282 | 0.0482 |
| >35 | 11 | 28.95% | |||||||
| CA199 | |||||||||
| <27 | 28 | 73.68% | 0.7577 | 0.9037 | 0.9082 | 0.7326 | 0.8458 | 0.6939 | 0.9968 |
| >27 | 10 | 26.32% | |||||||
| HE4 | |||||||||
| <105.1 | 25 | 65.79% | 0.2464 | 0.1712 | 0.2196 | 0.4011 | 0.0739 | 0.1640 | 0.1669 |
| >105.1 | 13 | 34.21% | |||||||
The analysis of 7 lncRNAs expression level and clinical features. DElncRNAs expression levels were compared between the two groups divided according to clinical features cut-off value (A) Tumor stage, (B) Tumor grade, (C) Menopause status, (D) Age, (E) CEA, (F) CA125, (G) CA199, (H) HE4. Experiments were performed in triplicate. * p<0.05 by Student's t test.
Discussion
Endometrial cancer is the second most common malignant tumor of female reproductive system in China. Although many advances have been made in biomedical research on the molecular mechanisms and treatments of UCEC, the overall survival rate of patients remains low since most of the patients are in the advanced stage at the time of diagnosis with poor surgical outcomes and prognosis [36]. UCEC is the leading gynecologic malignancy in developed nations, with about 7% of cases occurring in women under 45. Standard treatments, which involve hysterectomy and salpingo-oophorectomy, often clash with the fertility desires of younger patients. Therefore, a fertility-sparing approach, suitable for early-stage and low-grade endometrial cancer patients, is essential. The efficacy of this approach is studied through the evaluation of various immunohistochemical markers and their response to hormonal therapy [37, 38].
Circulating miRNAs were reported as a promising avenue for early EC diagnosis, staging, and evaluating a woman's receptivity, providing a non-invasive method with reduced error margins. This miR-based approach could be a pivotal tool in fertility-preserving processes. However, the ethical, legal, and regulatory considerations of such innovations need to be addressed alongside their potential benefits [39]. Oocyte vitrification is also a method of fertility preservation for couples diagnosed with UCEC. Upon diagnosis, they undergo ovarian stimulation to retrieve mature oocytes, which are then frozen for future use. Oocytes that have undergone vitrification appear to possess comparable fertilization and implantation potential as fresh oocytes. Collaboration between oncologists and fertility experts ensures optimal cancer treatment while safeguarding reproductive options [40, 41]. Currently, the serum tumor markers used for diagnosis of UCEC include CEA, CA125, CA199, and HE4 [42]. However, the diagnostic efficiency of these methods is hampered by their limited sensitivity and specificity. Therefore, there is an urgent need to identify novel biomarkers of UCEC that are non-invasive, specific, and sensitive for early diagnosis and treatment of the disease. In our study, this study identified specific lncRNAs by constructing the ceRNA network of UCEC using data obtained from the TCGA database, compared and validated the results from bioinformatics analysis using clinical tissue samples. First, 266 UCEC samples and 3 normal control samples were retrieved from the TCGA database and analyzed by RNA sequencing to obtain the raw data of miRNA, mRNA and lncRNA expression. Then, a ceRNA regulatory network was constructed to predict the relationships between them to screen for potential UCEC-related lncRNA biomarkers. Moreover, several interactions between mRNAs, miRNAs and lncRNAs were observed that involve the mRNAs of TSGs, oncogenes, and apoptosis-related genes. These interactions provide an additional direction that they may contribute to tumorigenesis and tumor cell survival which is worth further investigation in future studies. The 250 differentially expressed genes (DEmRNAs) identified were then subjected to GO and KEGG functional enrichment analysis and annotation, and the top 30 enriched biological processes and signalling pathways were selected. The results showed that the most enriched GO terms of DEmRNA include the processes of the muscle system, the cell-cell junction and actin binding. The KEGG enrichment analysis revealed predominantly cancer-related pathways such as cGMP-PKG signaling pathways. In order to validate the potential lncRNA biomarkers of UCEC screened based on the ceRNA network, we selected seven lncRNAs (C20orf56, LOC100190940, LOC100144604, LOC727677, LOC151534, LOC158572, FLJ35390) with significantly different expression in the ceRNA gene network.
Receiver operating characteristic (ROC) curve of 7 lncRNAs.
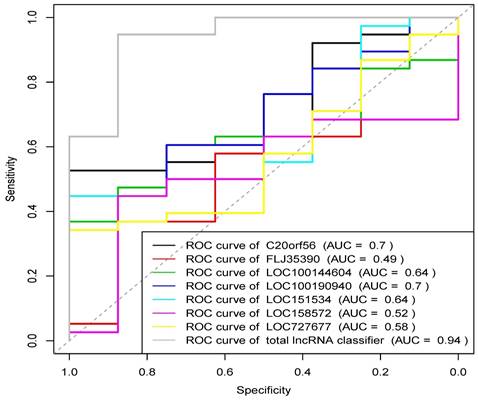
We used qRT-PCR to detect the expression levels of 7 lncRNAs genes in the UCEC samples and normal endometrial tissue, and correlated them with the clinicopathological characteristics of patients with endometrial cancer. Our results showed that these lncRNAs are involved in the differentiation and proliferation of endometrial cancer tumor cells and may play an important role in the malignant transformation process of endometrial cancer, which may aid in the early diagnosis of endometrial cancer. However, further research is needed to investigate the mechanism of expression regulation in the cancer. The results of ROC analysis also showed that the combined detection of the 7 lncRNAs has greater diagnostic value in distinguishing endometrial cancer samples from normal endometrial tissues. The up-regulated expression of the five lncRNAs was consistent with the results of the bioinformatics analysis. It was found that the expression level of LOC100190940 was higher in stage III and IV UCEC patients than in stage I and II patients. Moreover, the expression level of LOC158572 was negatively correlated with patient's age. The expression levels of FLJ35390 and LOC15857 in premenopausal patients were higher than those in postmenopausal patients. As a well-studied lncRNA, C20orf56 (LINC00261) has been widely studied in pancreatic cancer, prostate cancer, non-small cell lung cancer, bile duct cancer, colon cancer, endometrial cancer and other tumors. It may be used as early diagnosis, prognosis and target indicators of treatment. The results of this study also showed that LINC00261 was significantly up-regulated in UCEC samples which is in line with the literature's findings. LINC00261 can also be over-expressed in pancreatic cancer tissues by binding miR-222-3p to activate the HIPK2/ERK/c-myc pathway [43], which unmasked a new epigenetic and post-transcriptional regulatory mechanism that contributes to targeted therapy for pancreatic cancer. Another study showed that the down-regulation of LINC00261 expression was considered to be an independent risk factor affecting the postoperative recurrence-free survival rate of colon cancer patients (P<0.05) [44]. Being significantly correlated with clinical stage, LINC00261 may serve as a novel molecular biomarker for predicting colon cancer metastasis and survival. The expression level of LOC100190940 is positively correlated to the clinical stage of endometrial cancer and its high expression is closely related to the metastasis of UCEC. As a result, it may be used as an early diagnosis indicator of endometrial cancer and provides a direction for future research. Currently, LOC100190940 has not been reported in UCEC while other literatures have found that LOC100190940 can promote the occurrence and development of colorectal cancer and lung cancer. In colorectal cancer, LINC02418 negatively regulates apoptosis through the LINC02418/miR-34b-5p/BCL2 axis, acts as a tumor driver and can be used as an indicator for predicting prognosis [45]; The expression levels of LOC151534 (LBX2-AS1) and FLJ35390 (LINC00957) were down-regulated in UCEC based on the data obtained from TCGA, and their expression levels were higher in premenopausal patients than in postmenopausal patients, which also suggests their effects as tumor suppressor. In this study, the expression level of LOC151534 (LBX2-AS1) was also significantly correlated with the age of patients with UCEC. However, due to the small sample size of the experiment, the difference was not obvious and further confirmation was needed through increasing the sample size. In this study, it was found that LncRNA LBX2-AS1 was identified as an oncogene in some tumors. It is also abnormally expressed in liver cancer [46], non-small cell lung cancer [47] and associated with tumor cell proliferation, migration and invasion. It may also act as a novel prognostic biomarker and therapeutic target. Tumor biomarkers have extremely important clinical application value in disease screening, monitoring recurrence and metastasis as well as evaluating treatment outcome and prognosis of patients.
Conclusion
Based on our results, 7 potential lncRNA biomarkers (C20orf56, LOC100144604, LOC100190940, LOC151534, LOC727677, FLJ35390, LOC158572) were validated in UCEC patients by quantitative real-time PCR. We further analyzed the correlation between these lncRNAs and tumor biomarkers and found that the expression levels of FLJ35390 and LOC15857 in UCEC were correlated with that of CA125 while LOC151534 was correlated with CEA. Therefore, they can reflect the onset, differentiation, and disease progression of UCEC and are of great value in diagnosis, treatment outcome monitoring and prognosis evaluation of UCEC. However, the number of clinical samples collected in this experiment is small and the results need to be further validated by increasing the number of samples to improve the reliability of the results. In this study, LOC100190940 and LOC158572 are considered as novel RNA molecules with unknown functions reported. Future studies will employ larger sample size for confirming our findings.
Abbreviations
UCEC: Uterine Corpus Endometrial Carcinoma; lncRNAs: Long non-coding RNAs; ceRNA: competitive endogenous RNA; MREs: miRNA response elements; miRNAs: microRNAs; mRNAs: messenger RNAs; KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene Ontology; FDR: False Discovery Rate; AUC: Area under curve; ROC: Receiver operating characteristics; LASSO: Least absolute shrinkage and selection operator.
Supplementary Material
Supplementary tables.
Acknowledgements
This work was supported by the Scientific Research Fund of Department of Yunnan Education (Grant No. 2021J0266), Special Funds for Science Technology Innovation and Industrial Development of Shenzhen Dapeng New District (KJYF202001-21), Shenzhen Science and Technology Program (Grant No. SGDX20211123141201002).
Author contributions
LXW and XWS performed conceptualization, data curation, and writing-original draft. LYW, JBL and ZQX contributed to investigation, material preparation, and validation, GHDL, QL, JYZ and LZ contributed to investigation and data collection. XZ and GY interpreted the patient data and provide the valuable comments on the material preparation. GL and YMW participated in supervision and writing-review and editing. All authors contributed to the article and approved the submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
2. Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387:1094-108
3. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300
4. Cisek P, Kieszko D, Kordzinska-Cisek I, Kutarska E, Grzybowska-Szatkowska L. Retrospective Analysis of Intravaginal Brachytherapy in Adjuvant Treatment of Early Endometrial Cancer. Biomed Res Int. 2018;2018:7924153
5. Chen F, Fan Y, Cao P, Liu B, Hou J, Zhang B. et al. Pan-Cancer Analysis of the Prognostic and Immunological Role of HSF1: A Potential Target for Survival and Immunotherapy. Oxid Med Cell Longev. 2021;2021:5551036
6. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-92
7. Bian J, Sun X, Li B, Ming L. Clinical Significance of Serum HE4, CA125, CA724, and CA19-9 in Patients With Endometrial Cancer. Technol Cancer Res Treat. 2017;16:435-9
8. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393-407
9. Sun Y, Zhou Y, Bai Y, Wang Q, Bao J, Luo Y. et al. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer. 2017;16:162
10. Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858-61
11. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
12. Guallar D, Wang J. RNA-binding proteins in pluripotency, differentiation, and reprogramming. Front Biol (Beijing). 2014;9:389-409
13. Fernandes JCR, Acuña SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA. 2019 5
14. Uroda T, Anastasakou E, Rossi A, Teulon JM, Pellequer JL, Annibale P. et al. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol Cell. 2019;75:982-95 e9
15. Lu Z, Guo JK, Wei Y, Dou DR, Zarnegar B, Ma Q. et al. Structural modularity of the XIST ribonucleoprotein complex. Nat Commun. 2020;11:6163
16. Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093-100
17. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62
18. Wang Q, He R, Tan T, Li J, Hu Z, Luo W. et al. A novel long non-coding RNA-KAT7 is low expressed in colorectal cancer and acts as a tumor suppressor. Cancer Cell Int. 2019;19:40
19. Wang J, Zhang C. Identification and validation of potential mRNA- microRNA- long-noncoding RNA (mRNA-miRNA-lncRNA) prognostic signature for cervical cancer. Bioengineered. 2021;12:898-913
20. Liu L, Chen X, Zhang Y, Hu Y, Shen X, Zhu W. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget. 2017;8:31386-94
21. Fang Q, Sang L, Du S. Long noncoding RNA LINC00261 regulates endometrial carcinoma progression by modulating miRNA/FOXO1 expression. Cell Biochem Funct. 2018;36:323-30
22. Ouyang D, Li R, Li Y, Zhu X. A 7-lncRNA signature predict prognosis of Uterine corpus endometrial carcinoma. J Cell Biochem. 2019;120:18465-77
23. Liu T, Wang X, Zhai J, Wang Q, Zhang B. Long Noncoding RNA UCA1 Facilitates Endometrial Cancer Development by Regulating KLF5 and RXFP1 Gene Expressions. Cancer Biother Radiopharm. 2021;36:521-33
24. Sun MY, Zhu JY, Zhang CY, Zhang M, Song YN, Rahman K. et al. Autophagy regulated by lncRNA HOTAIR contributes to the cisplatin-induced resistance in endometrial cancer cells. Biotechnol Lett. 2017;39:1477-84
25. Samur MK. RTCGAToolbox: a new tool for exporting TCGA Firehose data. PLoS One. 2014;9:e106397
26. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323
27. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G. et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24-6
28. Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062-3
29. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 4
30. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146-52
31. Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC. et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48:D148-D54
32. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57
33. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
34. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77
35. Zhang A, Yang J, Ma C, Li F, Luo H. Development and Validation of a Robust Ferroptosis-Related Prognostic Signature in Lung Adenocarcinoma. Front Cell Dev Biol. 2021;9:616271
36. Makker V, Taylor MH, Aghajanian C, Oaknin A, Mier J, Cohn AL. et al. Lenvatinib Plus Pembrolizumab in Patients With Advanced Endometrial Cancer. J Clin Oncol. 2020;38:2981-92
37. Giampaolino P, Cafasso V, Boccia D, Ascione M, Mercorio A, Viciglione F. et al. Fertility-Sparing Approach in Patients with Endometrioid Endometrial Cancer Grade 2 Stage IA (FIGO): A Qualitative Systematic Review. Biomed Res Int. 2022;2022:4070368
38. Mutlu L, Manavella DD, Gullo G, McNamara B, Santin AD, Patrizio P. Endometrial Cancer in Reproductive Age: Fertility-Sparing Approach and Reproductive Outcomes. Cancers (Basel). 2022 14
39. Piergentili R, Gullo G, Basile G, Gulia C, Porrello A, Cucinella G. et al. Circulating miRNAs as a Tool for Early Diagnosis of Endometrial Cancer-Implications for the Fertility-Sparing Process: Clinical, Biological, and Legal Aspects. Int J Mol Sci. 2023 24
40. Gullo G, Perino A, Cucinella G. Open vs. closed vitrification system: which one is safer? Eur Rev Med Pharmacol Sci. 2022;26:1065-7
41. Zaami S, Stark M, Signore F, Gullo G, Marinelli E. Fertility preservation in female cancer sufferers: (only) a moral obligation? Eur J Contracept Reprod Health Care. 2022;27:335-40
42. Xia L, Wang Y, Meng Q, Su X, Shen J, Wang J. et al. Integrated Bioinformatic Analysis of a Competing Endogenous RNA Network Reveals a Prognostic Signature in Endometrial Cancer. Front Oncol. 2019;9:448
43. Zhou Z, Ma J. Expression and Clinical Significance of Long-chain Noncoding RNA LINC00261 in Colon Cancer. Clin Lab. 2019 65
44. Zhai S, Xu Z, Xie J, Zhang J, Wang X, Peng C. et al. Epigenetic silencing of LncRNA LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene. 2021;40:277-91
45. Han B. LncRNA LINC02418 regulates proliferation and apoptosis of non-small cell lung cancer cells by regulating miR-4677-3p/SEC61G. Eur Rev Med Pharmacol Sci. 2019;23:10354-62
46. Wang Y, Zhao Y, Zhang X, Zhang A, Ma J. Long noncoding RNA LBX2-AS1 drives the progression of hepatocellular carcinoma by sponging microRNA-384 and thereby positively regulating IRS1 expression. Pathol Res Pract. 2020;216:152903
47. Tang LX, Su SF, Wan Q, He P, Xhang Y, Cheng XM. Novel long non-coding RNA LBX2-AS1 indicates poor prognosis and promotes cell proliferation and metastasis through Notch signaling in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23:7419-29
Author contact
![]() Corresponding authors: Prof. Gang Lu, CUHK-SDU Joint Laboratory on Reproductive Genetics, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China Phone: 852-39435739, Email: lugangedu.hk. Prof. Yuming Wang, Department of Clinical Laboratory, Yunnan Molecular Diagnostic Center, The Second Affiliated Hospital of Kunming Medical University, Kunming650500, Yunnan, P.R. China. Email: wangym992011com.
Corresponding authors: Prof. Gang Lu, CUHK-SDU Joint Laboratory on Reproductive Genetics, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China Phone: 852-39435739, Email: lugangedu.hk. Prof. Yuming Wang, Department of Clinical Laboratory, Yunnan Molecular Diagnostic Center, The Second Affiliated Hospital of Kunming Medical University, Kunming650500, Yunnan, P.R. China. Email: wangym992011com.

 Global reach, higher impact
Global reach, higher impact