Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(12):1562-1569. doi:10.7150/ijms.87174 This issue Cite
Research Paper
Decreased miR-329-3p upregulates Adamts4 and Dnajb1 in mouse hepatic I/R injury in an age-independent manner
1. Department of Pediatrics, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
2. Division of Endocrinology, Department of Internal Medicine, Qilu Hospital of Shandong University, Jinan 250000, China.
3. Department of Endocrinology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
4. Institute of Organ Transplantation, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
5. Key Laboratory of Organ Transplantation, Ministry of Education, Ministry of Public Health, Chinese Academy of Medical Sciences, Wuhan 430030, China.
6. Reproductive Medicine Center, Tongji Hospital, Tongji Medicine College, Huazhong University of Science and Technology, Wuhan 430030, China.
Received 2023-6-15; Accepted 2023-9-1; Published 2023-9-18
Abstract
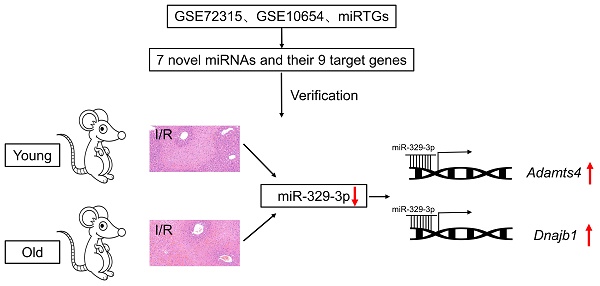
Introduction: Hepatic ischemia/reperfusion (I/R) injury is common after liver surgery, particularly in patients of older age. However, an understanding of the mechanism of injury remains incomplete. In this study, we explored the molecular mechanisms underlying hepatic I/R injury and associations with age in a murine model.
Methods: Gene expression profiling datasets (GSE72315 and GSE10654) and a microRNA (miRNA) expression profiling dataset (GSE72315) were downloaded from Gene Expression Omnibus. Differentially expressed genes (DEGs) and miRNAs (DEMiRs) were identified using online GEO2R or R before and after hepatic I/R injury in mice. Significant Gene Ontology (GO) terms were analyzed with the DAVID functional annotation tool. The DEMiR-miRNA target gene (miRTG) networks were constructed with miRTarBase, and the differentially expressed miRNAs and genes were analyzed with real-time quantitative polymerase chain reaction and immunofluorescence staining.
Results: Through bioinformatic analysis, seven novel candidate miRNAs were identified that may regulate the expression of nine genes in hepatic I/R injury. Before and after hepatic I/R injury, mmu-miR-9-5p, mmu-miR-329-3p, and mmu-miR-290a-5p showed significant differential expression both in young (1 month old) and old (1 year old) mice. miR-329-3p had the most significant differential expression, and its predicted target genes Adamts4 and Dnajb1 were also significantly upregulated.
Conclusions: The decrease in miR-329-3p expression upregulated Adamts4 and Dnajb1 expression in mouse hepatic I/R injury in an age-independent manner. This finding contributes to our understanding of hepatic I/R injury, and highlights novel molecular targets for future therapeutic development.
Keywords: Ischemia/reperfusion injury, liver surgery, microRNA, bioinformatics analysis, age-independent manner.
Introduction
Hepatic ischemia/reperfusion (I/R) injury is a common pathophysiological process after liver surgery. It significantly affects both recovery of liver function and perioperative outcomes after liver resection and transplantation [1, 2]. Hepatic I/R injury increases the risk of organ failure, acute and chronic tissue rejection, and satellite organ damage after liver transplantation; this limits the indications for liver resection and transplantation of marginal donor livers [3, 4]. An aging global population has led to a corresponding increase in the age of both patients undergoing liver surgery and transplant donors. The compensatory capacity of the liver after I/R injury is significantly reduced with age, as such the rate of graft failure, postoperative complications, and mortality is increasing despite continuous improvements in perioperative and critical care [5, 6]. Multiple alterations at the cellular and molecular levels contributed to the increased liver injury post‐IR in the aged mice. But to date, the relationship between donor age and sensitivity to I/R injury after liver transplantation and the effect of the former on the pathophysiological mechanism of the latter has not been fully elucidated.
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules with a length of 22 nucleotides. It is considered a post-transcriptional regulator of gene expression and can bind to the 3 '-untranslated region (3' -UTR) of target messenger RNA (mRNA)[7]. More and more studies have shown that miRNAs are often implicated in the pathophysiology of liver disease and is thought to be involved in the development of toxic liver damage, viral hepatitis, and liver tumors [8-10]. A number of miRNAs have been found to play a crucial role in liver I/R injury, such as miR-125b, miR-155, miR-182-5p, and so on[11, 12]. In this study, we amid to explore the differential expression of miRNAs and target genes between older and younger livers in a murine model of hepatic I/R injury using bioinformatic analysis. This sought to improve the understanding of the association between age and I/R injury, its underlying mechanism, and inform potential future therapeutic targets.
Materials and Methods
Gene Expression Omnibus (GEO) database analysis
The gene expression profiling datasets (GSE72315 and GSE10654) and a miRNA expression profiling dataset (GSE72315) were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information (NCBI). Data from each microarray were separately analyzed with GEO2R or R (version 3.6.1) to identify DEGs and DEMiRs. Firstly, we explored the differential expression of miRNA and target genes in mice with and without I/R injury. Six mice with I/R and six sham-operated were adopted in GSE72315 (GPL13112, Illumina HiSeq 2000 platform) to profile miRNA expression. For the construction of the gene expression microarray, six mice with I/R and six sham-operated mice were included in an analysis using GSE10654 (GPL5759, Affymetrix GeneChip Mouse Genome 430 2.0 Array) and GSE72315 (GPL13112, Illumina HiSeq 2000 platform). The GSE10654 dataset was divided into two groups based on the mouse's age (1-month-old mice ('younger mice') versus 1-year-old mice ('older mice')). For miRNA expression profiling, the threshold was determined according to the following values: P < 0.05 and fold change ≥ 3.0. For each gene expression profiling, the threshold was determined according to the following values: P < 0.05 and fold change ≥ 1.5. We identified differentially expressed genes (DEGs) between the GSE72315 and GSE10654 datasets, and any differentially expressed miRNAs (DEMiRs) were grouped according to whether they were seen to be upregulated or downregulated[13].
Identification of miRNA target genes (miRTGs) and potential binding sites
We used miRTarBase to identify experimentally validated miRTGs. miRNA target DEGs (miRT-DEGs) were subsequently obtained from overlapping DEGs and miRTGs. Finally, high DEMiR-low miRT-DEGs and low DEMiR-high miRT-DEGs were derived from overlapping DEMiRs and miRT-DEGs. To outline the potential functions of these 31 upregulated and 58 downregulated DEGs, a functional enrichment analysis was performed. Significant GO terms were identified with the DAVID functional annotation tool.
Animals
C57BL/6 male mice (1 month or 1 year) were purchased from the Beijing HFK Bio-Technology Company (Beijing, PR China). All experimental procedures involving animals were approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. The mice received humane care according to the Guide for the Care and Use of Laboratory Animals of the Chinese Academy of Sciences. All animals were housed under standard environmental conditions with controlled temperature (22 ± 2°C), humidity (50 ± 10%), and light (12 h light/dark cycle) with free access to standard diet and water.
Hepatic I/R injury mode
A hepatic I/R injury murine model was generated using a 70% hepatic I/R model [14]. Induction of anesthesia was performed with an intraperitoneal injection of sodium pentobarbital (60 mg/kg), and all efforts were made to minimize suffering. The artery and portal vein were interrupted with an atraumatic clip for 60 min to block blood flow to the left and middle lobes of the liver. As the right liver lobe was excluded, mesenteric venous congestion was prevented by allowing portal decompression through the right and caudate liver lobes. During the operation, normothermia was maintained (37°C) using a warming pad and incubator. Maintenance of anesthesia was performed with intraperitoneally administered ketamine (100 mg/kg) and xylazine (20 mg/kg). Care was taken to avoid exsanguination from the abdominal aorta, and every effort was made to minimize any suffering of the animals throughout the study. Plasma samples and liver tissues were collected 6 hours after the liver I/R injury. Sham operation group (sham group): The remaining steps were the same as those I/R group except for blocking hepatic blood flow.
Serum aminotransferase activities
To estimate the degree of hepatocyte damage after I/R injury, serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were measured using a Hitachi 7600 automatic analyzer (Hitachi, Ltd., Tokyo, Japan).
Histopathological examination
Liver tissue samples were fixed with 10% formalin for 24 hours before being embedded in paraffin. The sections were stained with hematoxylin and eosin (H&E). Suzuki's histological grading was performed to evaluate for damage. This assessment has three components including sinusoidal congestion, hepatocyte necrosis, and ballooning degeneration. Each criterion was scored on a scale of 0 (no damage) to 4 (severe damage) depending on the degree of liver damage. In the absence of any congestion, vacuolization, or necrosis, the best score of 0 was assigned. Severe congestion accompanied by severe vacuolization and > 60% necrosis area was assigned a score up to 12.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
Cell death in liver paraffin sections was detected using the TUNEL assay. The tissue samples were fixed in 10% neutral-buffered formalin before being embedded in paraffin. Hepatocyte death assays were performed with an in-situ cell death detection kit (Dead End Fluorometric TUNEL System; Promega, Madison, WI, USA) according to the manufacturer's instructions. The number of TUNEL-positive nuclei was counted in six randomly chosen images from non-overlapping areas of each group. Data were presented as the percentage of TUNEL-positive cells.
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA, including miRNAs, was extracted from the liver tissue using TRIzol reagent (Invitrogen, USA). RNA concentration was assessed with NanoDrop 2000 (Wilmington, DE) and the levels of miRNAs were measured with qRT-PCR using Bulge-Loop™ miRNA qRT-PCR Starter Kit (RiboBio, Guangzhou, China) on ABI 7500 System (Applied Biosystems). The primers for miR-9-5p, miR-592-5p, miR-124-3p, miR-329-3p, miR-290a-5p, miR-709, miR-760-3p, and U6 small nuclear RNA were obtained from RiboBio Company (Guangzhou, China). The sequences are covered by a patent. miRNA expression levels were normalized to the expression of the internal control U6 using the 2-ΔΔCT method [15].
Immunofluorescence staining
Prepared paraffin sections from liver tissues were de-waxed in xylene twice for 15 minutes, dehydrated in graded concentrations of ethanol (5 minutes each), and then rinsed 3 times with 0.01 M phosphate-buffered saline (PBS; pH 7.4) (5 minutes each). The tissue sections on glass slides were placed in a humid box. Non-specific antibody binding was blocked by incubation with 10% bovine serum albumin for 30 min at 37°C. Primary antibodies to Adamts4 (diluted at 1:80) and Dnajb1 (diluted at 1:100) were added to the tissue sections and the sections were incubated in the humid box at 4°C overnight. After rinsing with PBS (pH 7.4) (3 times, 5 minutes each), the samples were incubated with fluorescent-tagged secondary antibodies (diluted at 1:100) in the dark, in the humid box at 37°C for 1 hour. Tissue sections were mounted in buffered glycerol, viewed under a fluorescence microscope, and photographed.
Statistical analysis
All data are represented as mean ± standard error of the mean (SEM). Individual group statistical comparisons were analyzed by unpaired Student t-test, and the difference in mRNA expression between mice with hepatic I/R injury and controls was statistically analyzed by one-way analysis of variance using SPSS (Version 22.0). A value of P < 0.05 was considered statistically significant.
Results
Identification of aberrantly expressed miRNA-gene network in I/R
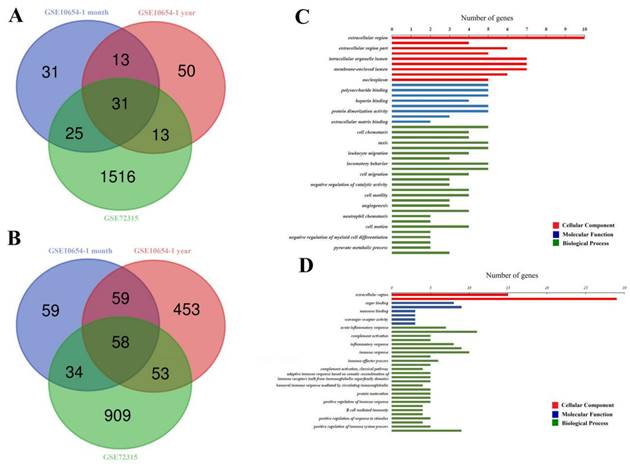
Thirty-one overlapping upregulated DEGs (100 in GSE10654-1-month-old subgroup, 107 in GSE10654-1-year-old subgroup, and 1585 in GSE72315, Fig. 1A) and 58 overlapping downregulated DEGs (210 in GSE10654-1-month-old subgroup, 623 in GSE10654-1-year-old subgroup, 1054 in GSE72315, Fig. 1B) were identified. Significant GO terms were exhibited in Figure 1C and D.
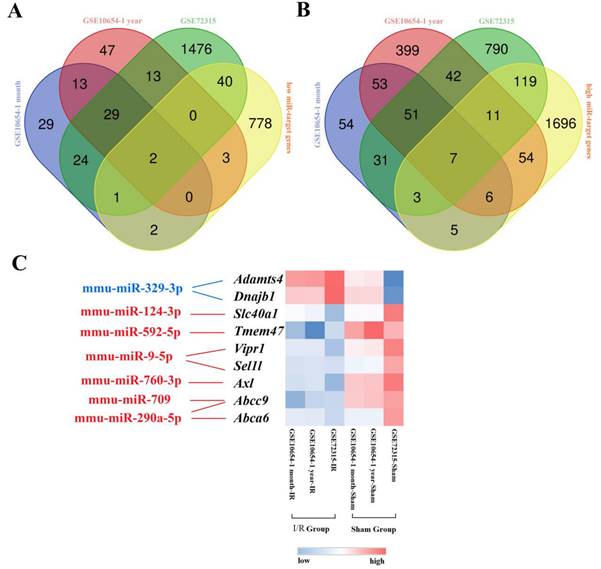
To identify aberrantly expressed miRNA-gene networks following I/R injury, GSE72315 was used for DEMiR screening. Sixty upregulated and 26 downregulated DEMiRs were detected. We used miRTarBase to subsequently identify 2688 upregulated and 1539 downregulated miRT-DEGs (Supplementary Materials). Finally, seven low DEMiR-high miRT-DEGs and two high DEMiR-low miRT-DEGs were obtained from overlapping DEMiRs and miRT-DEGs (Fig. 2A, B). In total, seven novel miRNAs, including mmu-miR-329-3p, mmu-miR-124-3p, mmu-miR-592-5p, mmu-miR-9-5p, mmu-miR-760-3p, mmu-miR-709, and mmu-miR-290a-5p, were thought to possibly regulate the expression of nine genes, including Adamts4, Dnajb1, Slc40a1, Tmem47, Vipr1, Sel1l, Axl, Abcc9, and Abca6 (Fig. 2C).
Association between age and severity of hepatic I/R injury in mice
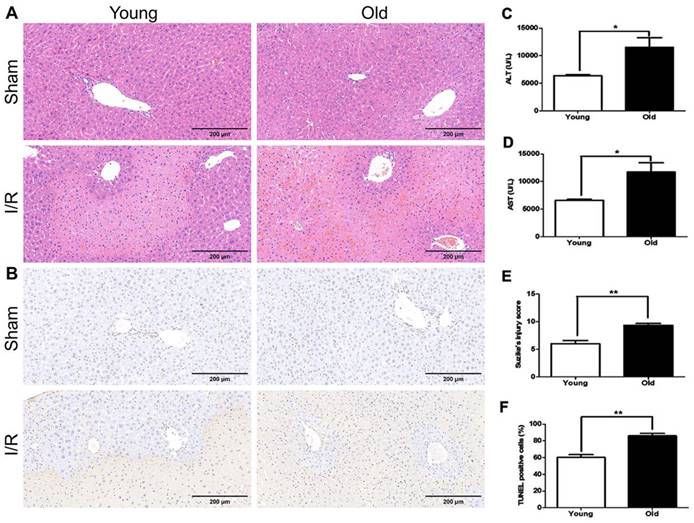
Histopathological examination revealed that the 'older mice' group had much more significant morphological alterations in the liver at 60 min after reperfusion than in younger mice, including severe damage to the morphology of the liver lobe, excessive hemorrhage and necrosis, swollen hepatocytes, and inflammatory cell infiltration (Fig. 3A, E). The number of TUNEL-positive cells increased more in older mice than in younger mice (Fig. 3B, F). In addition, the serum levels of ALT and AST were higher in older mice than in younger mice (Fig. 3C, D). Together these data suggest more significant hepatic I/R injury in older than younger mice in this model.
Differential expression of miRNAs and target genes before and after hepatic I/R injury in young and older mice
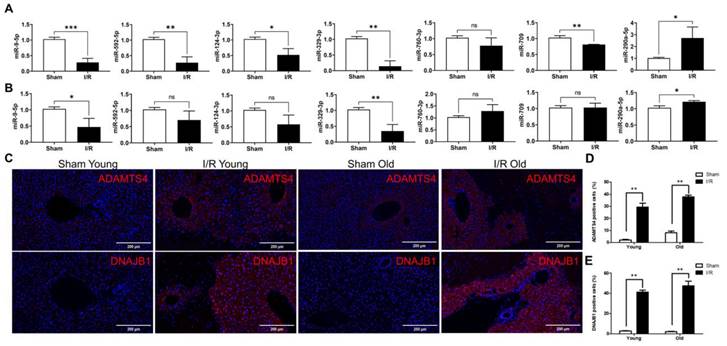
The expression of seven novel miRNAs identified from bioinformatic analysis was evaluated with RT-qPCR. In younger mice, differential expression of six miRNAs (miR-9-5p, miR-592-5p, miR-124-3p, miR-329-3p, miR-709, and miR-290a-5p) was apparent after hepatic I/R injury (Fig. 4A). In older mice the expression of only three miRNAs (miR-9-5p, miR-329-3p, and miR-290a-5p) was significantly different after hepatic I/R injury (Fig. 4B). In addition, immunofluorescence staining result revealed a significant increase in the expression of two target genes (Adamts4 and Dnajb1) of miR-329-3p after hepatic I/R injury in both young and old mice (Fig. 4C-E). And we also analyzed potential binding sites of miR-329-3p on the two target genes in Supplementary Figure 2.
Comparison of profiles of gene expression in following hepatic I/R injury. (A and B) are Venn diagrams displaying the number of overexpressed (A) or downregulated (B) DEGs from datasets GSE72315 and GSE10654. (C and D) display significant GO terms. The cellular Component (CC) is displayed in red, the Molecular Function (MF) in blue, and the Biological Process (BP) in green. P < 0.05 (acquired from DAVID function annotation tool).
Graphical representation of the aberrantly expressed miRNA-gene network following hepatic I/R injury. (A and B) display Venn diagrams with the number of low DEMiR-high miRT-DEGs (A) and high DEMiR-low miRT-DEGs (B) following I/R injury from overlapping DEMiRs and miRT-DEGs. (C) is a heatmap of miRT-DEGs constructed based on the average linkage method and the Person's distance measurement method. Genes with higher expression are shown in red, while those with lower expression are shown in blue. Downregulated miRNAs are shown in blue and upregulated miRNAs are shown in red.
Discussion
In this study, we sought to identify differentially expressed miRNA and their plausible target genes after hepatic I/R injury and made a direct comparison between younger and older mice using qPCR and immunofluorescence techniques. The findings of this study provide several avenues for the development of novel therapeutic molecules to minimize the impact of I/R injury, particularly in more vulnerable liver tissues in elderly patients.
The biological mechanisms underlying hepatic I/R injury remain unclear. In recent years, several studies implicated miRNAs in the pathway of injury. Many miRNAs are dysregulated in I/R injury, and may serve as both promising diagnostic biomarkers and potential therapeutic targets [16-18]. Here, we identified several miRNAs and target genes using bioinformatic analysis that were differentially regulated following I/R injury. Many of these have not undergone previous investigation in models of liver I/R injury and may present promising targets for future research. Some of the miRNAs we highlighted here have undergone extensive previous exploration. For example, miR-9-5p regulates the expression of Nfkb3 and plays an important role in the protective effect of sevoflurane in hepatic I/R injury [19]. miR-124-3p directly interacts with mmu_circRNA_005186 and Epha2 and is closely related to hepatic I/R injury and ischemic postconditioning (IPO) [20]. The effect of microRNA-592-5p on hippocampal neuron injury following hypoxic-ischemic brain damage in neonatal mice was also recently studied [21]. In this study, we explored these miRNAs using bioinformatic tools alone. Future studies including both in vitro and in vivo experiments are required to validate these early findings.
The association between age and severity of hepatic I/R injury in mice. Younger and older mice were subjected to hepatic I/R injury (I/R) or a sham operation (Sham). Liver and blood samples were collected 6 hours after reperfusion. (A) Representative histology of the liver as analysed with H&E staining. (B) Representative images of TUNEL staining of liver tissues. (C/D) The serum levels of ALT and AST as measured after reperfusion. (E) The pathological score of liver damage after hepatic I/R injury. (F) The quantification of TUNEL-positive cells in liver tissues. The data are expressed as mean ± SEM (*P < 0.05, **P < 0.01, young I/R versus old I/R group).
The differential expression of miRNAs and selected target genes after hepatic I/R injury in younger versus older mice. (A and B) show the seven differentially expressed miRNAs after hepatic I/R injury in younger (A) and older mice (B) using qRT-PCR. miRNA expression levels were normalized to the expression of the internal control U6. (C) Liver tissues were stained with specific antibodies linking Adamts4 and Dnajb1. All sections were counterstained with DAPI. (D and E) Quantification of cells expressing Adamts4 (D) and Dnajb1 (E) in liver tissues. The data are expressed as mean ± SEM (*P < 0.05, **P < 0.01, I/R versus Sham group).
Many studies have explored the association between older age and increased vulnerability to hepatic liver damage in I/R injury models [22, 23]. Here we used three methods to explore the association of age and the severity of injury in a hepatic I/R model in older versus younger mice. This included histopathology, a TUNEL assay, and serum AST/ALT levels. These methods are those widely used in the assessment of hepatic I/R injury [24]. We found that some miRNAs related to hepatic I/R injury exhibited differential expression after hepatic I/R injury in both younger and older mice. However, the differential expression of some miRNAs was not consistent with the predicted results of the bioinformatic analysis. This inconsistency in differential expression has been previously reported in I/R injury for both miR-9-5p and miR-124-3p [25, 26]. Our data suggested that miR-9-5p, miR-329-3p, and miR-290a-5p may play an important role in hepatic I/R injury in both young and old mice, with the differential expression of miR-329-3p observed most significantly. However, the functional implications of changes in miRNAs expression remained unclear. The predicted target genes (Adamts4 and Dnajb1) of miR-329-3p had a corresponding overexpression in hepatic I/R injury in both younger and older mice. ADAM metallopeptidase with thrombospondin type 1 motif 4 (ADAMTS4) is an inflammation-regulated enzyme, which has previously demonstrated overexpression in the ischemic brain in both human and murine models [27]. A recent comprehensive and combined genomic analysis revealed that DnaJ heat shock protein family member B1 (DNAJB1) is another key gene in the hepatic I/R injury pathway, with likely clinical significance [28]. Our data suggest that these pathways may be independent of age-related changes in the severity of I/R injury observed and require further investigation.
In summary, we identified several miRNAs and linked regulatory genes that may be linked to hepatic I/R injury using a bioinformatic analysis. We then explored the differential expression of miR-329-3p and its regulatory genes Adamts4 and Dnajb1 after hepatic I/R injury in younger and older mice, where it was upregulated in both groups. miR-329-3p may be a new therapeutic target to mitigate against I/R injury in both younger and older livers. The emergence of miR-329-3p and its regulatory genes therapeutic candidates requires further validation.
Abbreviations
I/R: ischemia/reperfusion; GO: Gene Ontology; miRNA: MicroRNA; mRNA: messenger RNA; ALT: alanine aminotransferase; AST: aspartate aminotransferase; H&E: hematoxylin and eosin; DNAJB1: DnaJ heat shock protein family member B1.
Supplementary Material
Supplementary figures.
Acknowledgements
We thank Editage (http://www.editage.cn/) for its linguistic assistance during the preparation of the manuscript.
Funding
This work was supported by funds from the National Natural Science Foundation of China (82000602). The authors gratefully acknowledge this funding support.
Availability of data and materials
The raw data of the study can be provided upon request with the maintenance of confidentiality, privacy, and anonymity of the research participants.
Author contributions
Study concept and design (BY, LW), acquisition of data (LZ, WD), analysis and interpretation of data (LZ, WD, BY, LW), drafting of the manuscript (LZ), critical revision of the manuscript for important intellectual content (BY, LW), administrative, technical, or material support, study supervision (BY, LW).
Ethics approval
All experimental procedures involving animals were approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology.
Consent for publication
All authors have contributed to, seen, and approved the final, submitted version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Peralta C, Jiménez-Castro MB, Gracia-Sancho J. Hepatic ischemia and reperfusion injury: effects on the liver sinusoidal milieu. Journal of hepatology. 2013;59:1094-106
2. Dar WA, Sullivan E, Bynon JS, Eltzschig H, Ju C. Ischaemia reperfusion injury in liver transplantation: Cellular and molecular mechanisms. Liver international: official journal of the International Association for the Study of the Liver. 2019;39:788-801
3. Ali JM, Davies SE, Brais RJ, Randle LV, Klinck JR, Allison ME. et al. Analysis of ischemia/reperfusion injury in time-zero biopsies predicts liver allograft outcomes. Liver transplantation: official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society. 2015;21:487-99
4. Kahn J, Schemmer P. Control of Ischemia-Reperfusion Injury in Liver Transplantation: Potentials for Increasing the Donor Pool. Visceral medicine. 2018;34:444-8
5. Kan C, Ungelenk L, Lupp A, Dirsch O, Dahmen U. Ischemia-Reperfusion Injury in Aged Livers-The Energy Metabolism, Inflammatory Response, and Autophagy. Transplantation. 2018;102:368-77
6. Feng S, Goodrich NP, Bragg-Gresham JL, Dykstra DM, Punch JD, DebRoy MA. et al. Characteristics associated with liver graft failure: the concept of a donor risk index. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2006;6:783-90
7. Yang D, Zhao D, Chen X. MiR-133b inhibits proliferation and invasion of gastric cancer cells by up-regulating FBN1 expression. Cancer biomarkers: section A of Disease markers. 2017;19:425-36
8. Yoshioka W, Higashiyama W, Tohyama C. Involvement of microRNAs in dioxin-induced liver damage in the mouse. Toxicological sciences: an official journal of the Society of Toxicology. 2011;122:457-65
9. Herzog K, Bandiera S, Pernot S, Fauvelle C, Jühling F, Weiss A. et al. Functional microRNA screen uncovers O-linked N-acetylglucosamine transferase as a host factor modulating hepatitis C virus morphogenesis and infectivity. 2020; 69: 380-92.
10. Wang X, Gao J, Zhou B, Xie J, Zhou G, Chen Y. Identification of prognostic markers for hepatocellular carcinoma based on miRNA expression profiles. Life sciences. 2019;232:116596
11. Huang Z, Zheng D, Pu J, Dai J, Zhang Y, Zhang W. et al. MicroRNA-125b protects liver from ischemia/reperfusion injury via inhibiting TRAF6 and NF-κB pathway. Bioscience, biotechnology, and biochemistry. 2019;83:829-35
12. Tan L, Jiang W, Lu A, Cai H, Kong L. miR-155 Aggravates Liver Ischemia/reperfusion Injury by Suppressing SOCS1 in Mice. Transplantation proceedings. 2018;50:3831-9
13. Zhang G, Wang H, Zhu K, Yang Y, Li J, Jiang H. et al. Investigation of candidate molecular biomarkers for expression profile analysis of the Gene expression omnibus (GEO) in acute lymphocytic leukemia (ALL). Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;120:109530
14. Lin J, Huang HF, Yang SK, Duan J, Qu SM, Yuan B. et al. The effect of Ginsenoside Rg1 in hepatic ischemia reperfusion (I/R) injury ameliorates ischemia-reperfusion-induced liver injury by inhibiting apoptosis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;129:110398
15. Guo A, Wang W, Shi H, Wang J, Liu T. Identification of Hub Genes and Pathways in a Rat Model of Renal Ischemia-Reperfusion Injury Using Bioinformatics Analysis of the Gene Expression Omnibus (GEO) Dataset and Integration of Gene Expression Profiles. Medical science monitor: international medical journal of experimental and clinical research. 2019;25:8403-11
16. Zhu J, Zhu F, Song W, Zhang B, Zhang X, Jin X. et al. Altered miR-370 expression in hepatic ischemia-reperfusion injury correlates with the level of nuclear kappa B (NF-κB) related factors. Gene. 2017;607:23-30
17. Li Y, Ma D, Wang Z, Yang J. MicroRNA-155 Deficiency in Kupffer Cells Ameliorates Liver Ischemia-Reperfusion Injury in Mice. Transplantation. 2017;101:1600-8
18. Selten JW, Verhoeven CJ, Heedfeld V, Roest HP, de Jonge J, Pirenne J. et al. The release of microRNA-122 during liver preservation is associated with early allograft dysfunction and graft survival after transplantation. Liver transplantation: official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society. 2017;23:946-56
19. Liao X, Zhou S, Zong J, Wang Z. Sevoflurane exerts protective effects on liver ischemia/reperfusion injury by regulating NFKB3 expression via miR-9-5p. Experimental and therapeutic medicine. 2019;17:2632-40
20. Zhang P, Ming Y, Ye Q. Comprehensive circRNA expression profile during ischemic postconditioning attenuating hepatic ischemia/reperfusion injury. 2019; 9: 264.
21. Sun LQ, Guo GL, Zhang S, Yang LL. Effects of MicroRNA-592-5p on Hippocampal Neuron Injury Following Hypoxic-Ischemic Brain Damage in Neonatal Mice - Involvement of PGD2/DP and PTGDR. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;45:458-73
22. Li Y, Ruan DY, Jia CC, Zheng J, Wang GY, Zhao H. et al. Aging aggravates hepatic ischemia-reperfusion injury in mice by impairing mitophagy with the involvement of the EIF2α-parkin pathway. Aging. 2018;10:1902-20
23. Liu R, Zhang S, Ma W, Lu H, Gao J, Gan X. et al. Age-dependent loss of induced regulatory T cell function exacerbates liver ischemia-reperfusion injury. Molecular immunology. 2018;103:251-6
24. Tan R, Tian H, Yang B, Zhang B, Dai C, Han Z. et al. Autophagy and Akt in the protective effect of erythropoietin helix B surface peptide against hepatic ischaemia/reperfusion injury in mice. Scientific reports. 2018;8:14703
25. Xiao Y, Zhang Y, Chen Y, Li J, Zhang Z, Sun Y. et al. Inhibition of MicroRNA-9-5p Protects Against Cardiac Remodeling Following Myocardial Infarction in Mice. Human gene therapy. 2019;30:286-301
26. Sørensen SS, Nygaard AB, Carlsen AL, Heegaard NHH, Bak M, Christensen T. Elevation of brain-enriched miRNAs in cerebrospinal fluid of patients with acute ischemic stroke. Biomarker research. 2017;5:24
27. Lemarchant S, Dunghana H, Pomeshchik Y, Leinonen H, Kolosowska N, Korhonen P. et al. Anti-inflammatory effects of ADAMTS-4 in a mouse model of ischemic stroke. Glia. 2016;64:1492-507
28. Huang S, Ju W, Zhu Z, Han M, Sun C, Tang Y. et al. Comprehensive and combined omics analysis reveals factors of ischemia-reperfusion injury in liver transplantation. Epigenomics. 2019;11:527-42
Author contact
![]() Corresponding authors: Lan Wang and Bo Yang. Reproductive Medicine Center, Tongji Hospital, Tongji Medicine College, Huazhong University of Science and Technology, No.1095 JieFang Avenue, Wuhan 430030, China. (L.W.); Institute of Organ Transplantation, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, No.1095 JieFang Avenue, Wuhan 430030, China. (B.Y.) Tel.: +86-027-83665283; Fax: +86-027-83665283; Email: wanglantjmu.edu.cn (L.W.); yangbotjmu.edu.cn (B.Y.)
Corresponding authors: Lan Wang and Bo Yang. Reproductive Medicine Center, Tongji Hospital, Tongji Medicine College, Huazhong University of Science and Technology, No.1095 JieFang Avenue, Wuhan 430030, China. (L.W.); Institute of Organ Transplantation, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, No.1095 JieFang Avenue, Wuhan 430030, China. (B.Y.) Tel.: +86-027-83665283; Fax: +86-027-83665283; Email: wanglantjmu.edu.cn (L.W.); yangbotjmu.edu.cn (B.Y.)

 Global reach, higher impact
Global reach, higher impact