Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(4):520-529. doi:10.7150/ijms.80047 This issue Cite
Research Paper
ZNF460-regulated COMMD7 Promotes Acute Myeloid Leukemia Proliferation Via the NF-κB Signaling Pathway
1. Central Laboratory of Yongchuan Hospital, Chongqing Medical University, Chongqing 402160, China
2. Key Laboratory of Laboratory Medical Diagnostics, Ministry of Education, Department of Laboratory Medicine, Chongqing Medical University, Chongqing 400016, China
Received 2022-10-20; Accepted 2023-1-10; Published 2023-2-21
Abstract
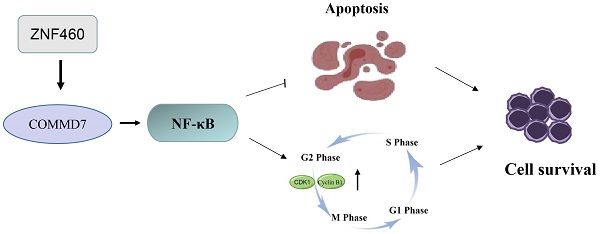
Acute myeloid leukemia (AML) is a malignancy of the hematological system, for which there remains an urgent need for new therapeutic and diagnostic targets. COMM domain containing 7 (COMMD7) is a recently-identified oncogene linked to poor prognosis in AML. COMMD7 regulates multiple signaling pathways, including nuclear factor-kappa B (NF-κB) signaling. Here, we report that COMMD7 is highly expressed in the AML cell lines KG1a and U937 and that its inhibition by shRNA reduced proliferation, promoted apoptosis and facilitated cell cycle arrest in the G2/M phase in relation to depression of the NF-κB pathway. Furthermore, zinc finger protein 460 (ZNF460) is overexpressed in AML and regulates COMMD7. We found that knockdown of ZNF460 downregulated the expression of COMMD7 while the NF-κB pathway was also inhibited. In addition, we noticed that knockdown of ZNF460 reduced proliferation and increased apoptosis rate of AML cells and that the cell cycle was blocked in the G2/M phase. In brief, our results revealed a critical effect of the ZNF460-COMMD7-NF-κB axis for the proliferation of AML cells. Therefore, COMMD7 may be a possible therapeutic target for AML.
Keywords: Acute myeloid leukemia, COMMD7, ZNF460, NF-κB pathway, Proliferation
Introduction
Acute myeloid leukemia (AML) is a clonally aggressive proliferative disorder of the primitive cells of the hematopoietic lineage, characterized by unlimited proliferation, blocked differentiation and apoptosis [1, 2]. AML is the most commonly diagnosed form of acute leukemia in adults [3]. Although significant advances in chemotherapy and other drug treatments have been made in recent years, somewhat improving clinical outcomes, AML remains poorly treatable, with a 5-year survival rate of only around 28% owing to high recurrence rates and drug resistance [4-6]. It is therefore essential to elucidate the molecular mechanisms by which AML pathogenesis occurs in order to improve existing treatment strategies and to identify new therapeutic targets.
COMM domain-containing protein 7 (COMMD7), a member of the COMMD family, is a highly conserved protein with a copper metabolism gene MURR1 (COMM) domain. The COMMD series of proteins are associated with the regulation of a wide range of biological functions [7, 8]. COMMD7 has been previously reported to play a role in a variety of solid tumors [9, 10]. It is highly expressed in hepatocellular carcinoma (HCC) where it represses apoptosis and prevents cell cycle arrest by regulating the NF-κB pathway [11, 12]. Other studies have found that COMMD7 is overexpressed in pancreatic ductal adenocarcinoma (PDAC) cell lines and correlates with a poor prognosis. Anti-tumor effects can be achieved by suppressing COMMD7 in PDAC cells [13]. Recently, bioinformatics analysis has been used to predict that high levels of COMMD7 may be implicated in poor prognosis of AML [14]. This implies that targeting COMMD7 may be a potential therapeutic strategy for AML, but its exact mode of expression, biological function and pathogenic mechanisms in AML remain poorly understood. Therefore, the biological function and use of COMMD7 as a potential target for AML therapy deserve further investigation.
Zinc finger proteins (ZNF) form the largest family of transcription factors, with zinc finger structural domains playing an essential part in modulating cell differentiation and embryonic development [15, 16]. Recently, increasing reports suggest that ZNF can function as an oncogene in cancer [17-19]. In previous studies, excessive expression of ZNF460 was implicated in adverse outcomes of colon cancer and enhanced invasive capacity. ZNF460 transcriptionally upregulates circMTO1 to promote oral squamous cell carcinoma [20]. Here, we found that ZNF460 is associated with COMMD7 and regulates its expression, but the exact biological function of ZNF460 is unknown. Therefore, ZNF460 is worth investigating as a potential therapeutic target for AML or as a regulatory molecule of COMMD7.
The current study aimed to explore the impact of COMMD7 and ZNF460 in AML development. It was shown that COMMD7 and ZNF460 are highly expressed in AML cells and promote AML proliferation via activation of the NF-κB pathway.
Materials and Methods
Cell culture
Acute myeloid leukemia cell lines NB4, THP1, U937, MV-411and KG1a were from the American Type Culture Collection (ATCC, USA). AML cells were cultured in RPMI-1640 medium (#C11875500BT, Gibco, USA) with 10% fetal bovine serum (#900-108, Gemini, USA) and 1% penicillin-streptomycin (#C0222, Beyotime, China) at 37°C, in 5% CO2 [21].
Cell proliferation assays
The viability of cells was measured with a Cell counting kit-8 (CCK-8) kit (#CA1210, Solarbio, Beijing, China). Infected cells at 5 x 103 per well were seeded into 96-well plates. 10 μL of CCK-8 reagent was added to the wells every 24 h and incubated for a further 2 h protected from light, followed by measurement of optical density (OD) at 450 nm using a microplate reader (BioTeck, CA, USA) [22].
qRT-PCR
Total RNA from AML cells was isolated using TRIzol reagent (#9108, Takara, Japan), then reverse transcribed into cDNA with PrimeScript RT regent Kits (#RR600, Takara, Japan). RT-qPCR was carried out with SYBR® Premix Ex Taq™ II kits (#RR820A, Takara) in a CFX Connect™ RT-qPCR System (Bio-Rad, USA). All primers were synthesised by Sangon Biotech (Shanghai, China). Primer sequences are given in Table S1. The number of cycles at which the fluorescence intensity increment reaches a threshold in each sample tube was taken as the Ct value. The Ct values of the gene to be tested and the internal reference gene were read separately for each specimen. Using β-actin as the internal reference, the relative expression of the gene to be tested was further calculated for each specimen using 2-ΔΔCt [23].
Western blotting
Cells were washed thrice using PBS and then lysed for half an hour in a RIPA lysis buffer (#P0013B, Beyotime, China) containing protease inhibitor (#ST505, Beyotime, China) followed by centrifugation at 12,000 rpm for 30 min at 4°C. A BCA kit (#P0010, Beyotime) was then used to determine the protein concentration. Separation of equivalent quantities of total protein samples by SDS-PAGE at a suitable concentration. The protein was subsequently transferred to polyvinylidene difluoride (PVDF) membranes (#BS-PES-45, Millipore, USA). Following blocking for 2 h, appropriate primary antibodies were incubated with the blots for at least 16 h at 4°C. The antibodies employed were as follows: Cleaved-Caspase-3 (#9961,Cell Signaling Technology, USA), PARP (#A5037, Bimake, USA), NF-κB p50/105 (#A5600, Bimake), Bcl2 (#A5010, Bimake), Phospho-CDK1 (#R26267, ZENBIO,China), Caspase3 (#9622, Cell Signaling Technology), NF-κB p65 (#A5075,Bimake), Cleaved-PARP (#A5034, Bimake), Bax (#5023, Cell Signaling Technology), Cyclin B1 (#R23324, ZENBIO, China), MMP9 (#R380831, ZENBIO), ZNF460 (#TD4628, Abmart, China), p38 (#8690, Cell Signaling Technology), COMMD7 (#GTX112076, GeneTex, USA), p21 (#A5163, Bimake), CDK1 (#R23884, ZENBIO), c-Myc (Wanleibio, China), p53 (#A5722, Bimake). GAPDH (#60004-1-Ig, Proteintech, China). The secondary antibody (Biosharp, China) was then incubated with the blot for 1 h at 25℃. Quantification was done using the ECL kit (#WBKLS0500, Millipore, USA) [24].
Cell infection
AML cells with stable knockdown of COMMD7 and ZNF460 were constructed using recombinant lentivirus (Genechem, Shanghai, China). AML cells were infected with lentivirus and infection enhancer for 48 h. Preliminary determination of infection efficiency by observation of cellular GFP fluorescence, followed by selection of cells with 2 µg/ml of puromycin (#HY-15695, MCE, China) for two weeks was carried out. Follow-up experiments were performed using selected cells. The sequences of shRNA used are listed in Table S2[25].
Isolation of peripheral blood mononuclear cells
Peripheral blood was drawn from healthy individuals into anticoagulated blood collection tubes. Peripheral blood mononuclear cells were isolated with human whole blood mononuclear cell separation medium (#LDS1075, TBD, Tianjin, China) [26].
Flow cytometry
To detect apoptosis, 1×106 AML cells were collected and washed thrice with PBS and resuspended in 1 ml PBS, then stained using Annexin V APC-A and DAPI kit. Apoptosis was detected by flow cytometry (CytoFLEX; Beckman, USA) and data analyzed by CytExpert software (Beckman Coulter, USA) [27].
To assess the cell cycle, 1 × 106 cells were treated with 75% ethanol at 4°C overnight. Following centrifugation, the cell supernatant was discarded and RNase A was added in the dark, incubated at 37°C for 25 min and finally Propidium Iodide (PI, Millipore) was added. Cells were plated in the dark at 4°C for 25 min and the cell cycle was assessed by FACSCaliburTM Flow cytometry (BD Biosciences) [28].
Co-immunoprecipitation assay
Cells were washed thrice, followed by lysis with IP-Western blot lysis buffer for 30 min. Subsequently cells were centrifugd for 30 min and the supernatant aspirated. Protein A/G Magnetic Beads were washed with PBST thrice and incubated with anti-ZNF460 and anti-IgG (#4414, CST) separately with Protein A/G Magnetic Beads (#16-662, MCE, China) on an inverted mixer for 1 h at 25℃. Beads were washed thrice and subsequently incubated with the supernatant at 4°C for at least 16 h. The next day, proteins were detected by western blotting [29].
Bioinformatics analysis
UCSC and JASPAR were employed to predict transcription factors that might affect COMMD7. Original data of The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) were downloaded from UCSC XENA. R 4.1.1 was used to process raw data and subsequent analysis. The GEPIA database was used to analyze the correlation between ZNF460 and COMMD7 [30, 31].
Statistical analysis
All data are from three independent replicate experiments and are presented as mean ± SD. All statistical analyses were performed using unpaired Student's t-tests and one-way ANOVA using GraphPad software (prism8). p < 0.05 was regarded as statistically significant (*).
Results
COMMD7 is highly expressed and promotes proliferation of AML cells
It was reported that COMMD7 was predicted to be highly expressed in AML and was implicated in poor survival prospects in previous studies [14]. In an effort to explore the relevance of COMMD7 in AML, we first quantified its expression in peripheral blood mononuclear cells (PBMCs) and the AML cell lines NB4, THP1, KG1a, U937 and MV-411. qRT-PCR analysis and western blotting demonstrated markedly higher levels of COMMD7 expression in AML cell lines compared to PBMCs, especially in KG1a and U937 (Fig. 1A). To further study the role of COMMD7 in AML, we knocked it down with lentivirus in U937 and KG1a cells and compared the knockdown efficiency of the three shRNAs at the mRNA and protein levels by qRT-PCR (Fig. 1B) and western blotting (Fig. 1C). We then chose shRNA#3 for the following experiments. Subsequently, we performed CCK-8 assay to examine the impact of COMMD7 on the proliferation of AML cells. The results showed that silencing COMMD7 suppressed the proliferation of KG1a and U937 cells (Fig. 1D). These findings indicate that hyper-expression of COMMD7 facilitates the proliferation of AML cells.
Knockdown of COMMD7 promotes apoptosis and G2/M arrest via the NF-κB pathway
Although our results above suggested that knockdown of COMMD7 inhibited proliferation, it was not known exactly how this is achieved. We considered two possibilities: pro-apoptosis and cell cycle arrest. First, AML cells were examined for their apoptosis rate after knockdown of COMMD7. Flow cytometry (FCM) showed that knockdown of COMMD7 led to a significant increase in apoptosis in KG1a and U937 cells (Fig. 2A). Western blotting revealed a marked increase in the expression of Bax, Cleaved-PARP and Cleaved-caspase3, which promote apoptosis, accompanied by a reduction in Bcl2, an apoptosis-suppressing protein (Fig. 2B). Next, the effect of knocking down COMMD7 on the cell cycle was measured by FCM. The outcomes displayed that the proportion of cells in G2/M phase was markedly elevated in both KG1a and U937 cell lines (Fig. 2C). Next, based on the above results, we examined the crucial cyclin-dependent kinases (CDKs) and other molecules that relate to the G2/M phase. Western blotting detected a decrease in Cyclin B1, CDK1 and p38 and an increase in p21, p53 and phosphorylated CKD1 (p-CDK1) (Fig. 2D). This implies that a decrease in CyclinB1/CDK1 activity induces cell cycle arrest in the G2/M phase. This finding is in line with the aforementioned FCM data. Previous work has documented that hyper-expression of COMMD7 activates NF-κB p65 in HCC, so we proceeded to investigate whether the biological function of COMMD7 in AML is achieved through activation of the NF-κB pathway [32]. By western blotting, we observed that knockdown of COMMD7 led to a reduction in the phosphorylation level of p65 and the expression of NF-κB p50. An increase in p105, a precursor of p50, was also detected. Some additional proteins were reduced to some extent as well, such as c-Myc and MMP9 (Fig. 2E). In conclusion, knockdown of COMMD7 in AML cells inhibits proliferation, promotes apoptosis and blocks the cell cycle, an effect likely achieved by the suppression of the activation of the NF-κB pathway.
ZNF460 interacts with COMMD7 and promotes AML cell proliferation
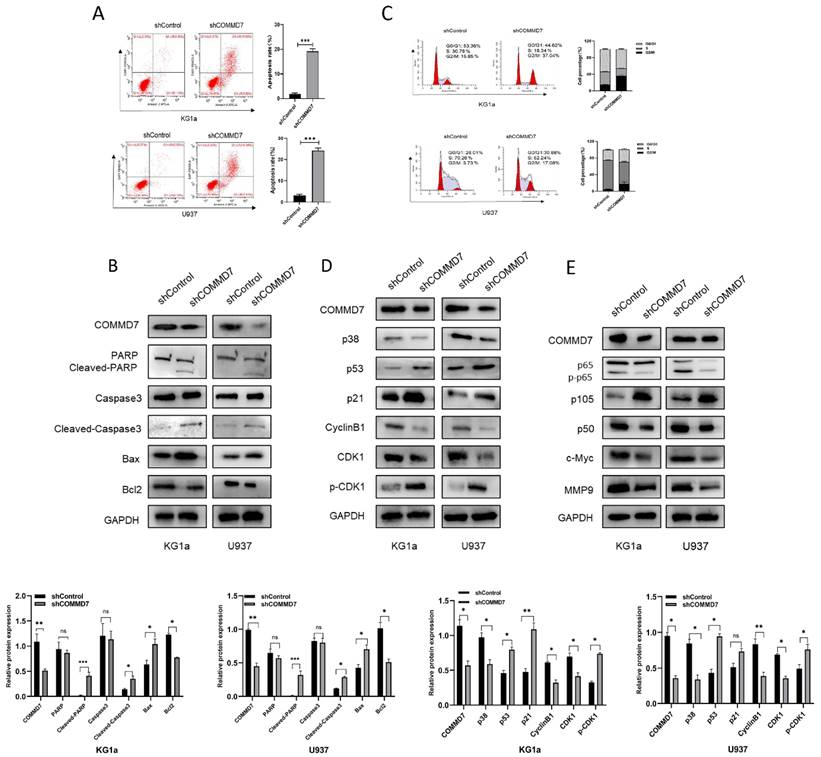
We next explored the upstream regulatory mechanisms of COMMD7 action in AML cells. By means of the bioinformatics tools UCSC and JASPAR we identified transcription factor ZNF460 as upstream of COMMD7 (Fig. 3A). We then analyzed the correlation between ZNF460 and COMMD7 expression using the bioinformatics tool GEPIA and found that they were highly correlated (Fig. 3B). Next, we analyzed the expression of ZNF460 by normal samples in the TCGA and GTEx databases and corresponding AML samples in the former and found that it was highly expressed in AML (Fig. 3C). The qRT-PCR and western blotting results confirmed these predictions (Fig. 3D). Interactions between ZNF460 and COMMD7 were next investigated using co-immunoprecipitation (Co-IP) assays. Immunoprecipitation with anti-ZNF460 antibody pulled down endogenous COMMD7 in both KG1a and U937 (Fig. 3E). To investigate the biological function of ZNF460, we knocked it down in these cell lines. We found that using shZNF460#1, with the best knock-down efficiency, resulted in downregulation not only of ZNF460 but also COMMD7, at both the mRNA and protein levels (Fig. 3F-G). Next, we used the CCK-8 assay to detect the effect of knocking down ZNF460 on the proliferation of KG1a and U937 cells and found that their growth was significantly inhibited. Taken together, these data suggest that the highly expressed ZNF460 upregulates COMMD7 and promotes AML cell proliferation.
COMMD7 is highly expressed and promotes proliferation of AML cells. The mRNA and protein levels of COMMD7 in NB4, KG1a, THP1, U937, MV-411 and PBMCs (A). Different COMMD7 shRNAs were employed into KG1a and U937 cells. Knock-down efficiency was detected by qRT-PCR (B) and western blotting (C). Knockdown of COMMD7 inhibited KG1a and U937 cells proliferation. Cell viability was analyzed by the CCK-8 assay (D).
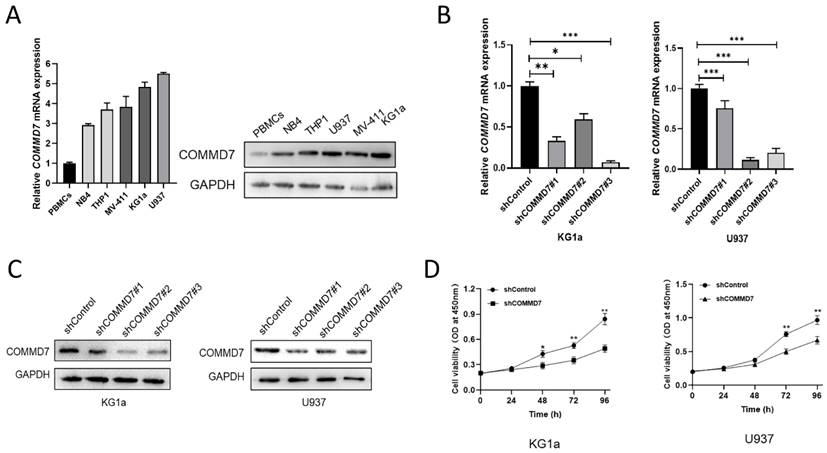
Knockdown of COMMD7 promotes apoptosis and G2/M arrest via the NF-κB pathway. Apoptosis rates of KG1a and U937 were measured by flow cytometry by using annexin V/PI kit (A). Expression of apoptosis-related proteins was tested by western blotting (B). Cell cycle was determined by flow cytometry (C). Western blotting was employed to assayed the cell cycle-associated proteins (D). NF-κB pathway-related protein expression was measured by immunoblotting (E).
The effect of ZNF460 downregulation on apoptosis and cell cycle in AML cells is mediated via the NF-κB pathway
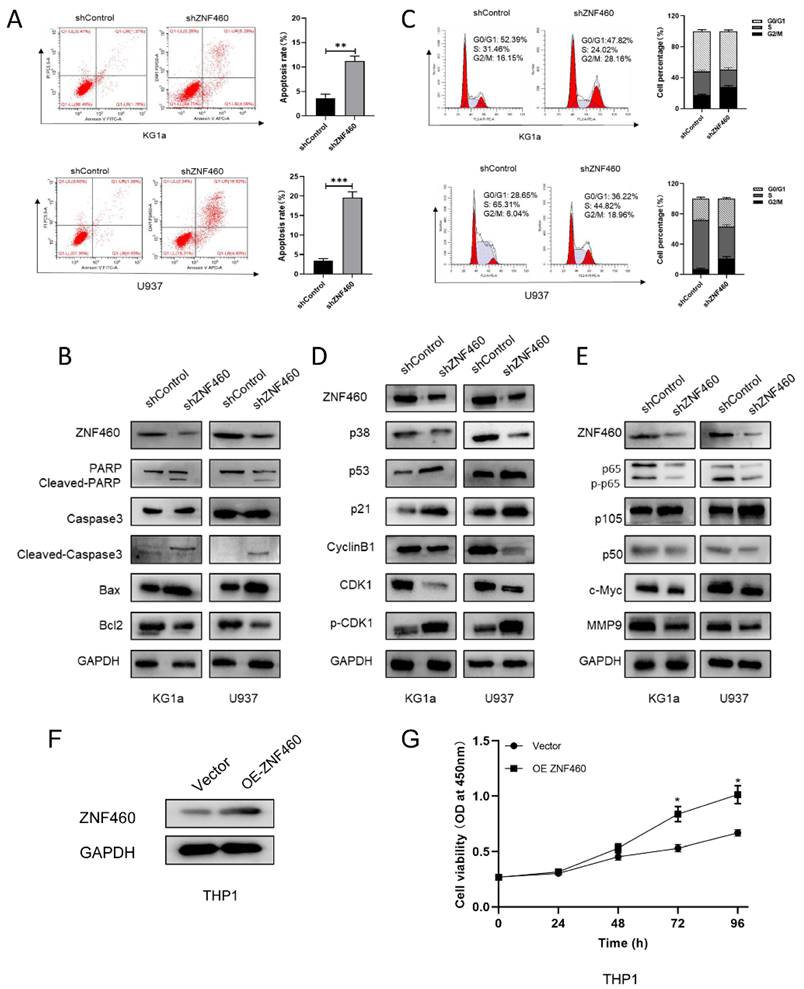
To explore the manner by which knocking down ZNF460 inhibits proliferation, we assayed apoptosis and the cell cycle as before. The outcomes suggested that the silencing of ZNF460 promoted apoptosis in AML cells (Fig. 4A). Western blotting documented that knockdown of ZNF460 resulted in an increase in Cleaved-caspase3, Bax and Cleaved-PARP, along with a corresponding decrease in some apoptosis-inhibiting proteins, such as Bcl2 (Fig. 4B). Cell cycle assays using FCM showed that knockdown of ZNF460 also leads to cell cycle blockade at the G2/M phase (Fig. 4C). Therefore, we monitored the expression of the relevant proteins by western blotting. The findings suggested that there was a reduction in the expression of Cyclin B1 and CDK1 and an increase in p-CDK1, implying a decrease in CDK1/Cyclin B1 activity. Corresponding changes were seen for other molecules, such as p53 and p21, which inhibit the Cyclin B1/CDK1 complex (Fig. 4D). Finally, we explored the mechanisms by which ZNF460 plays a role in AML and, given the above results, we examined the regulation of the NF-κB pathway by knocking down ZNF460. The findings also suggested a decrease in p65 and phosphorylated p65 (p-p65), implying that both the expression and the activity of p65 were suppressed. A decrease in p50 and an increase in its precursor p105 could also be detected. c-Myc and MMP9 were all downregulated to some extent (Fig. 4E). In a further study of the definitive role of ZNF460 in AML proliferation, we overexpressed ZNF460 in THP1 cells, a leukemic cell line with low expression of ZNF460. Western blotting showed successful overexpression of ZNF460 (Fig. 4F). Finally, we analyzed the effect of overexpression of ZNF460 on proliferation using CCK-8 assay and found that the proliferation of THP1 cells was promoted to some extent following the overexpression of ZNF460 (Fig. 4G). In conclusion, knockdown of ZNF460 inhibits AML cell proliferation by promoting apoptosis and cell cycle arrest.
ZNF460 interacts with COMMD7 and promotes AML cell proliferation. Potential transcription factors of COMMD7 promoter were projected through linking UCSC database with JASPAR website (A). Correlation between ZNF460 and COMMD7 was analyzed by GEPIA (B). Expression of ZNF460 in normal and AML groups (C). Expression of ZNF460 in AML cell lines was assessed by qRT-PCR and western blotting (D). Protein extracted from KG1a and U937 cells was co-immunoprecipitated with anti-ZNF460 antibody and blotted with anti-ZNF460 and anti-COMMD7 antibodies (E). Knockdown efficiency of ZNF460 in KG1a and U937 cells and expression of COMMD7 after knockdown of ZNF460 were detected by qRT-PCR (F) and western blotting (G). CCK-8 assays were used to assess cell proliferation (H).
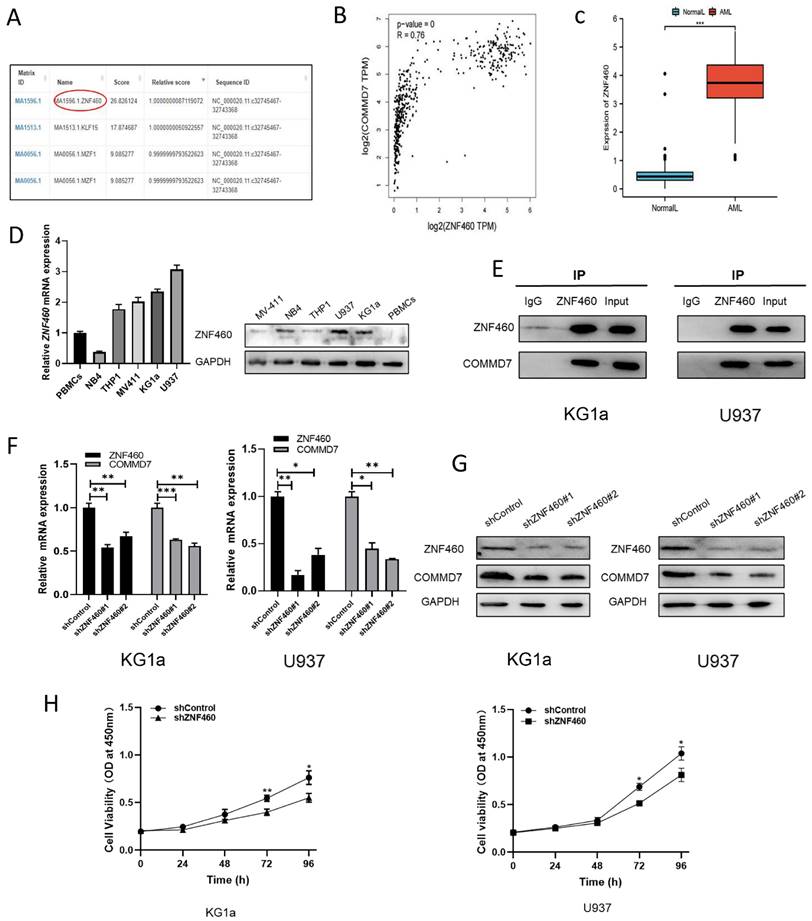
The effect of ZNF460 downregulation on apoptosis and cell cycle in AML cells is mediated via the NF-κB pathway. Apoptosis rates of KG1a and U937 were measured by flow cytometry by using annexin V/PI kit (A). Expression of apoptosis-related proteins was tested by western blotting (B). Cell cycle was determined by flow cytometry (C). Western blotting was employed to assayed the cell cycle-associated proteins (D). NF-κB pathway-related protein expression was measured by immunoblotting (E). Western blotting was used to detect overexpression of ZNF460 (F). CCK-8 assay was used to assess cell proliferation (G).
Discussion
AML is the most common form of acute leukemia in adults, caused by radiation, genetic mutations or chemicals [33]. Treatment with chemotherapy, standard intensive 7+3 therapy, stem cell transplantation and targeted drugs is commonly used, but there are many cases of poor patient prognosis with low five-year survival rates and drug resistance [34, 35]. In addition, the heavy burden on patients and the severe side effects of treatment are also serious problems [36, 37]. The molecular mechanisms by which AML develops are not thoroughly understood, and there is still a lack of effective targets for AML treatment [38, 39]. Therefore, finding a promising therapeutic target is a pressing issue. In this report, the involvement of ZNF460 and COMMD7 in AML was investigated for the first time. The findings confirmed the hypothesis that the ZNF460-COMMD7-NF-κB axis affects AML cell proliferation and provide a new approach for the modulation of the NF-κB signaling pathway.
Mounting evidence points to an influential role for COMMD7 in promoting tumor progression [40]. In a recent report, it was shown that knocking down COMMD7 in HCC stem cells inhibited their proliferation, migration and invasion in vitro, and also tumor progression in vivo [32]. Recently, Cao and colleagues found that abnormal expression of COMMD7 has an important role in gastric cancer. Either targeting miR-514a-5p by Linc00852 to regulate COMMD7 expression or directly knocking out COMMD7 heightened the sensitivity of GC cells to DPP, thus facilitating GC therapy [41]. In the present study, we characterized COMMD7 as an emerging regulatory molecule for the development of AML. Our results documented that after knockdown of COMMD7, proliferation of KG1a and U937 cells was inhibited, apoptosis was elevated, and the cell cycle was arrested in the G2/M phase. In line with previous studies, COMMD7 has an activatory effect on the NF-κB pathway, but the mechanism by which COMMD7 functions in AML is not clear. In their study, Hawe et al. noted that COMMD7 can be disturbed by genetic sequence variants and abnormal DNA methylation, which can affect white blood cell counts. This is consistent with the abnormal white cell count in AML and may suggest a promising potential mechanism involving COMMD7 in AML [42]. This deserves further investigation. Taken together, our findings suggest that COMMD7 facilitates AML development.
A growing number of studies are identifying ZNFs as proto-oncogenes for many different tumors, associated with poor disease prognosis. Previous studies have shown that certain ZNFs have multiple effects in promoting tumor proliferation, migration and invasion. For example, LINC00857 regulated by ZNF460 promotes migration, invasion and epithelial mesenchymal transition of pancreatic cancer cells by upregulating CLDN12 expression through sponging miR-150-5p and recruiting SRSF1 [43]. Hao and co-workers have shown that hyperexpression of ZNF460 is related to adverse prognosis and that it facilitates cell migration in colon cancer via the JAK2/STAT3 signaling pathway [44]. Nevertheless, the exact levels of expression and molecular functionality of ZNF460 in AML remain unclear. Here, we report for the first time that ZNF460 is overexpressed in AML cell lines, and that it mobilizes the NF-κB pathway. In addition, we found that ZNF460 functions to boost AML by upregulating COMMD7. However, the exact mechanism involved is still not clear. Nonetheless, the above findings demonstrate that ZNF460 is a promising biomarker and therapeutic target in AML.
Taken together, our study found that COMMD7 and ZNF460 were overexpressed in AML cells and that this was associated with their proliferation. However, due to limited conditions, we did not detect the expression of ZNF460 or COMMD7 in primary cells ex vivo in our study and will refine it in the future. They have a role in boosting growth and progression through the cell cycle, in inhibiting apoptosis and activating the NF-κB pathway. In previous studies, COMMD7 was considered to be a regulatory protein of the NF-κB pathway, but the mechanisms involved were not clarified [8, 45, 46]. We have attempted to investigate this issue and have used bioinformatics to screen for ZNF460, a transcription factor that regulates COMMD7, showing that ZNF460 upregulates the mRNA and protein expression of COMMD7. Taken together, these data suggest that ZNF460 and COMMD7 may be therapeutic targets for AML, and we will proceed with in-depth studies on the specific regulatory mechanisms of ZNF460 on COMMD7 and abnormal DNA methylation.
Conclusions
In conclusion, we found that aberrant high expression of COMMD7 promotes AML cell proliferation and cell cycle progression, inhibits apoptosis and activates the NF-κB signaling pathway. ZNF460 up-regulates COMMD7 to promote AML development.
Abbreviations
AML: acute myeloid leukemia; COMMD7: COMM domain containing 7; Co-IP: co-immunoprecipitation; PVDF: polyvinylidene difluoride; qRT-PCR: quantitative real-time PCR; ZNF460: Zinc finger proteins 460; HCC: hepatocellular carcinoma; NF-κB: nuclear factor-kappa B; PDAC: pancreatic ductal adenocarcinoma; PBS: Phosphate Buffered Saline; CDKs: cyclin-dependent kinase; CCK-8: Cell counting kit-8; GTEx: Genotype-Tissue Expression; TCGA: Cancer Genome Atlas; PBMCs: peripheral blood mononuclear cells; p-CDK1: phosphorylated CKD1; p-p65: phosphorylated p65; OD: optical density; FCM: Flow cytometry.
Supplementary Material
Supplementary tables.
Acknowledgements
Funding
This research was supported by grants from the National Natural Science Foundation of China (Grant Number 81772280) and Key Technology Innovation Special of Key Industries of the Chongqing Science and Technology Bureau (Grant Number csct2022ycjh-bgzxm0034).
Author contributions
Xin Shao: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Writing - original draft.
Liang Zhong: Conceptualization, Project administration, Resources, Supervision.
Xuan Chu: Investigation, Methodology, Validation.
Peng Wan: Investigation, Methodology.
Ziwei Zhou: Investigation, Methodology.
Shuyu Chen: Investigation, Methodology.
Meng Wang: Software, Visualization.
Hongyan Zhang: Validation.
Beizhong Liu: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing - review & editing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Short NJ, Rytting ME, Cortes JE. Acute myeloid leukaemia. Lancet. 2018;392:593-606
2. Dohner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136-52
3. Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 2019;36:70-87
4. Chen X, Xie H, Wood BL, Walter RB, Pagel JM, Becker PS. et al. Relation of clinical response and minimal residual disease and their prognostic impact on outcome in acute myeloid leukemia. J Clin Oncol. 2015;33:1258-64
5. Newell LF, Cook RJ. Advances in acute myeloid leukemia. BMJ. 2021;375:n2026
6. Xu XQ, Wang JM, Gao L, Qiu HY, Chen L, Jia L. et al. Characteristics of acute myeloid leukemia with myelodysplasia-related changes: A retrospective analysis in a cohort of Chinese patients. Am J Hematol. 2014;89:874-81
7. Maine GN, Burstein E. COMMD proteins: COMMing to the scene. Cell Mol Life Sci. 2007;64:1997-2005
8. Maine GN, Burstein E. COMMD proteins and the control of the NF kappa B pathway. Cell Cycle. 2007;6:672-6
9. Zheng L, Deng CL, Wang L, Huang XB, You N, Tang YC. et al. COMMD7 is correlated with a novel NF-kappaB positive feedback loop in hepatocellular carcinoma. Oncotarget. 2016;7:32774-84
10. Esposito E, Napolitano G, Pescatore A, Calculli G, Incoronato MR, Leonardi A. et al. COMMD7 as a novel NEMO interacting protein involved in the termination of NF-kappaB signaling. J Cell Physiol. 2016;231:152-61
11. Zheng L, You N, Huang X, Gu H, Wu K, Mi N. et al. COMMD7 Regulates NF-kappaB Signaling Pathway in Hepatocellular Carcinoma Stem-like Cells. Mol Ther Oncolytics. 2019;12:112-23
12. Zheng L, Liang P, Li J, Huang XB, Liu SC, Zhao HZ. et al. ShRNA-targeted COMMD7 suppresses hepatocellular carcinoma growth. PLoS One. 2012;7:e45412
13. You N, Li J, Gong Z, Huang X, Wang W, Wang L. et al. COMMD7 functions as molecular target in pancreatic ductal adenocarcinoma. Mol Carcinog. 2017;56:607-24
14. Li K, Chen L, Zhang H, Wang L, Sha K, Du X. et al. High expression of COMMD7 is an adverse prognostic factor in acute myeloid leukemia. Aging (Albany NY). 2021;13:11988-2006
15. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860-921
16. Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53
17. Sobocinska J, Molenda S, Machnik M, Oleksiewicz U. KRAB-ZFP Transcriptional Regulators Acting as Oncogenes and Tumor Suppressors: An Overview. Int J Mol Sci. 2021 22
18. Liu Y, Zhang X, Blumenthal RM, Cheng X. A common mode of recognition for methylated CpG. Trends Biochem Sci. 2013;38:177-83
19. Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32:63-70
20. Zou C, Li X, Lv X, Wu S, Song J, Tang Z. et al. Circular RNA mitochondrial translation optimization 1 homologue (CircMTO1) induced by zinc finger protein 460 (ZNF460) promotes oral squamous cell carcinoma progression through the microRNA miR-320a / alpha thalassemia/mental retardation, X-linked (ATRX) axis. Bioengineered. 2021;12:9585-97
21. Baust JM, Buehring GC, Campbell L, Elmore E, Harbell JW, Nims RW. et al. Best practices in cell culture: an overview. In vitro Cell Dev Biol Anim. 2017;53:669-72
22. James N, Kini S, Pai S, Shenoy N, Kabekkodu SP. Comparative Evaluation of Corneal Storage Medias Used as Tooth Avulsion Medias in Maintaining the Viability of Periodontal Ligament Cells Using the Cell Counting Kit-8 Assay. Clin Cosmet Investig Dent. 2022;14:87-94
23. Jozefczuk J, Adjaye J. Quantitative real-time PCR-based analysis of gene expression. Methods Enzymol. 2011;500:99-109
24. Mahmood T, Yang PC. Western blot: technique, theory, and trouble shooting. N Am J Med Sci. 2012;4:429-34
25. He X, He Q, Yu W, Huang J, Yang M, Chen W. et al. Optimized protocol for high-titer lentivirus production and transduction of primary fibroblasts. J Basic Microbiol. 2021;61:430-42
26. Fuss IJ, Kanof ME, Smith PD, Zola H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol. 2009 Chapter 7: 7 1 -7 1 8
27. Crowley LC, Marfell BJ, Scott AP, Waterhouse NJ. Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry. Cold Spring Harb Protoc. 2016. 2016
28. Darzynkiewicz Z, Huang X, Zhao H. Analysis of Cellular DNA Content by Flow Cytometry. Curr Protoc Immunol. 2017;119:5 7 1-5 7 20
29. Lin JS, Lai EM. Protein-Protein Interactions: Co-Immunoprecipitation. Methods Mol Biol. 2017;1615:211-9
30. Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R. et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260-D6
31. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
32. You N, Li J, Huang X, Wu K, Tang Y, Wang L. et al. COMMD7 promotes hepatocellular carcinoma through regulating CXCL10. Biomed Pharmacother. 2017;88:653-7
33. Short NJ, Konopleva M, Kadia TM, Borthakur G, Ravandi F, DiNardo CD. et al. Advances in the Treatment of Acute Myeloid Leukemia: New Drugs and New Challenges. Cancer Discov. 2020;10:506-25
34. Dohner H, Wei AH, Lowenberg B. Towards precision medicine for AML. Nat Rev Clin Oncol. 2021;18:577-90
35. Kadia TM, Ravandi F, Cortes J, Kantarjian H. New drugs in acute myeloid leukemia. Ann Oncol. 2016;27:770-8
36. Liu H. Emerging agents and regimens for AML. J Hematol Oncol. 2021;14:49
37. Yang X, Wang J. Precision therapy for acute myeloid leukemia. J Hematol Oncol. 2018;11:3
38. Carter JL, Hege K, Yang J, Kalpage HA, Su Y, Edwards H. et al. Targeting multiple signaling pathways: the new approach to acute myeloid leukemia therapy. Signal Transduct Target Ther. 2020;5:288
39. Prada-Arismendy J, Arroyave JC, Rothlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31:63-76
40. de Bie P, van de Sluis B, Burstein E, Duran KJ, Berger R, Duckett CS. et al. Characterization of COMMD protein-protein interactions in NF-kappaB signalling. Biochem J. 2006;398:63-71
41. Cao S, Fu B, Cai J, Zhang D, Wang C, Wu H. Linc00852 from cisplatin-resistant gastric cancer cell-derived exosomes regulates COMMD7 to promote cisplatin resistance of recipient cells through microRNA-514a-5p. Cell Biol Toxicol. 2022
42. Hawe JS, Wilson R, Schmid KT, Zhou L, Lakshmanan LN, Lehne BC. et al. Genetic variation influencing DNA methylation provides insights into molecular mechanisms regulating genomic function. Nat Genet. 2022;54:18-29
43. Zhang Y, Fang Y, Ma L, Xu J, Lv C, Deng L. et al. LINC00857 regulated by ZNF460 enhances the expression of CLDN12 by sponging miR-150-5p and recruiting SRSF1 for alternative splicing to promote epithelial-mesenchymal transformation of pancreatic adenocarcinoma cells. RNA Biol. 2022;19:548-59
44. Hao T, Xu J, Fang S, Jiang J, Chen X, Wu W. et al. Overexpression of ZNF460 predicts worse survival and promotes metastasis through JAK2/STAT3 signaling pathway in patient with colon cancer. J Cancer. 2021;12:3198-208
45. Bartuzi P, Hofker MH, van de Sluis B. Tuning NF-kappaB activity: a touch of COMMD proteins. Biochim Biophys Acta. 2013;1832:2315-21
46. Singla A, Chen Q, Suzuki K, Song J, Fedoseienko A, Wijers M. et al. Regulation of murine copper homeostasis by members of the COMMD protein family. Dis Model Mech. 2021 14
Author contact
![]() Corresponding author: Dr. Beizhong Liu. Email: liubeizhongedu.cn
Corresponding author: Dr. Beizhong Liu. Email: liubeizhongedu.cn

 Global reach, higher impact
Global reach, higher impact