Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(2):254-261. doi:10.7150/ijms.79409 This issue Cite
Research Paper
m6A modifications of circular RNAs in ischemia-induced retinal neovascularization
1. Department of Ophthalmology, The Second Xiangya Hospital of Central South University, Changsha, Hunan 410011, China.
2. Hunan Clinical Research Center of Ophthalmic Disease, The Second Xiangya Hospital of Central South University, Changsha, Hunan 410011, China.
3. National Clinical Research Center for Metabolic Diseases, The Second Xiangya Hospital of Central South University, Changsha, Hunan 410011, China.
4. Department of Ophthalmology, Kurume University School of Medicine, Kurume, Fukuoka 830-0011, Japan.
Received 2022-9-29; Accepted 2023-1-12; Published 2023-1-22
Abstract
Ischemia-induced pathological neovascularization in the retina is a leading cause of blindness in various age groups. The purpose of the current study was to identify the involvement of circular RNAs (circRNAs) methylated by N6-methyladenosine (m6A), and predict their potential roles in oxygen-induced retinopathy (OIR) in mice. Methylation assessment via microarray analysis indicated that 88 circRNAs were differentially modified by m6A methylation, including 56 hyper-methylated circRNAs and 32 hypo-methylated circRNAs. Gene ontology enrichment analysis predicted that the enriched host genes of the hyper-methylated circRNAs were involved in cellular process, cellular anatomical entity, and protein binding. Host genes of the hypo-methylated circRNAs were enriched in the regulation of cellular biosynthetic process, the nucleus, and binding. According to the Kyoto Encyclopedia of Genes and Genomes analysis, those host genes were involved in the pathways of selenocompound metabolism, salivary secretion, and lysine degradation. MeRIP-qPCR verified significant alterations in m6A methylation levels of mmu_circRNA_33363, mmu_circRNA_002816, and mmu_circRNA_009692. In conclusion, the study revealed the m6A modification alterations in OIR retinas, and the findings above shed light on the potential roles of m6A methylation in circRNA regulatory functions in the pathogenesis of ischemia-induced pathological retinal neovascularization.
Keywords: circRNA, m6A, oxygen-induced retinopathy, retinal neovascularization, RNA methylation
Introduction
As the shared pathogenesis of proliferative diabetic retinopathy (PDR), retinopathy of prematurity (ROP), and other retinal vasculopathies, ischemia-induced pathological retinal neovascularization is a common cause of irreversible blindness globally in various age groups [1-3]. Therefore, effective intervention has become crucial to reducing the severe vision loss caused by retinal neovascularization.
As the most prevalent mRNA modification in mammals [4], N6‑methyladenosine (m6A) RNA methylation exerts potential epigenetic functions in various physiological and pathological processes [5-7]. We recently revealed the profiles of altered m6A epitranscriptomes of mRNAs and long non-coding RNAs in the retinas of oxygen-induced retinopathy (OIR) [8], a classical mouse model for retinal neovascular diseases [9]. Several studies suggested the regulatory roles of m6A modifications in retinal neovascular diseases. For example, it has been reported that YTHDF2 induces mRNA instability of ITGB1 in an m6A‑dependent manner, and attenuated the development of diabetic retinopathy [10]. Yao et al. [11] indicated that the m6A writer enzyme METTL3 is a promising strategic target for the treatment of pathological neovascularization. Therefore, the potential role of m6A methylation in the pathogenesis of retinal neovascularization is worth exploring.
Circular RNA (circRNA) is a closed RNA that lacks 5' and 3' ends [12]. Recent studies have revealed important physiological and pathological functions of circRNAs, and their potential as novel molecular biomarkers and therapeutic targets [13]. In the vitreous humour of PDR, a total of 131 circRNAs were dysregulated compared to the controls [14]. Besides, Li et al. [15] revealed altered exosomal circRNAs in the serum of PDR. In OIR mouse model, 539 circRNAs were significantly changed in the retinal neovascularization group, indicating the possible involvement of circRNA-associated ceRNA networks [16]. Significant alteration of circRNAs in peripheral blood mononuclear cells in premature infants suggests they may also be biomarkers and/or therapeutic targets for ROP [17]. Numerous studies have revealed profile alterations in m6A-modified circRNAs in many diseases, including cerebral infarction [18], severe acute pancreatitis [19], acute myeloid leukemia [20], and oral squamous cell carcinoma [21]. Interestingly, Huang et al. [22] recently demonstrated that circRNA circFAT1 enhanced autophagy and reduced pyroptosis in glucose-stressed retinal pigment epithelial cells, and reported the binding of circFAT1 and m6A reader YTHDF2.
To date, m6A modification of circRNAs in retinal neovascularization remains unknown, and requires further investigation. In the current study, the epitranscriptomic profile of m6A-modified circRNAs was established via microarray analysis in the OIR model in mice, followed by validation with MeRIP-qPCR. The potential functions of those circRNAs with altered m6A methylation levels were predicted by further bioinformatics analyses.
Materials & Methods
Establishment of OIR mouse model
C57BL/6J mice obtained from the SJA Laboratory Animal Center (Changsha, Hunan, China) were used in the study. The OIR model was established as previously described [8, 9]. Pups were exposed to hyperoxic conditions (75%) on postnatal day (P) 7, and moved back to room air at P12. Pups in the control group were exposed to room air continuously. At P17, the retinas of mice in both groups were harvested for use in subsequent experiments. Four retinas from two mice were mixed to constitute one sample, and three samples were assessed for microarray analysis of each group. The animal experiments were in accordance with the ARVO Statement and the procedures have been approved by the Institutional Animal Care and Use Committee of the Second Xiangya Hospital of Central South University.
RNA extraction and m6A immunoprecipitation
Total RNA was extracted from retinal tissues using TRIzol (Invitrogen, Carlsbad, CA, USA), the RNA purity and concentration were determined by a NanoDrop ND-1000 spectrophotometer. Either a Bioanalyzer 2100 or Mops electrophoresis was used to assess RNA integrity.
Total RNA and m6A spike-in control mixture was mixed with immunoprecipitation (IP) buffer, added the anti-m6A rabbit polyclonal antibody (Synaptic Systems, Goettingen, Germany), and then the preparation was incubated at 4°C for 2 h. For each sample, 20 μL Dynabeads M-280 sheep anti-rabbit IgG suspension (Invitrogen) was used, and the preparation was blocked with 0.5% BSA (4°C, 2 h). After washing, they were resuspended in the prepared mixture containing total RNA and antibody and incubated at 4°C for 2 h. After washing, the enriched RNA was eluted with elution buffer at 50°C for 1 h, and then acid phenol-chloroform and ethanol precipitation were used for RNA extraction.
Epitranscriptomic microarray analysis of circRNAs
IP and Sup RNAs were digested with RNase R (Epicentre, Inc.) to remove linear RNAs and enrich circRNAs. The Super RNA Labeling Kit (Arraystar, Rockville, MD, USA) was used to label Sup RNAs with Cy3, and IP RNAs with Cy5, and then preparations were purified via the RNeasy Mini Kit. The labeled cRNA mixture was fragmented, hybridized on an m6A‑circRNA Epitranscriptomic Microarray slide (Arraystar) following the manufacturer's instructions, and scanned by a microarray scanner G2505C (Agilent Technologies, Santa Clara, CA, USA).
Agilent Feature Extraction software (V11.0.1.1) was used to analyze images of captured array. Average of the log2-scaled spike-in RNA intensities were used for normalization of the IP and Sup raw intensities. m6A quantities were calculated based on normalized IP amounts, and m6A methylation levels were calculated as the percentage of modification in the input according to normalized intensities. Raw data from the microarray analysis were deposited in the database of Gene Expression Omnibus (No. GSE213362). The associations between circRNA m6A methylation and expression levels were conducted by the intersection of the current m6A microarray data and the expression profile of circRNAs in OIR retinas in accordance with a previous study by our research group [16].
Methylated RNA immunoprecipitation-qPCR
Methylated RNA immunoprecipitation (MeRIP)-qPCR analysis was conducted to validate the microarray data. SuperScriptTM III Reverse Transcriptase (Invitrogen) was used to synthesize the first-strand cDNA from IP RNA, and the system of QuantStudioTM 5 Real‑Time PCR (Applied Biosystems, Foster City, CA, USA) with 2X PCR master mix (Arraystar) was used to conduct RT-qPCR. The IP fraction in the input was calculated as MERIP/input (%) as described in Xing et al. [23]. Data are presented as means ± SEM. Primer sequences are displayed in Table 1.
Primer sequences of the circRNAs validated by MeRIP-qPCR
| CircRNA | Forward/reverse primers | Tm (°C) | Product length (bp) |
|---|---|---|---|
| mmu_circRNA_33363 | F: 5' GTGATCTCAGTATTCAAAGGTGT 3' R: 5' CTCATGGTTTCCTATGGTATT 3' | 60 | 191 |
| mmu_circRNA_002816 | F: 5' AGAAGCCCATAAGTAAACAAGG 3' R: 5' AAAATTCTCCACTTTATCCGTT 3' | 60 | 88 |
| mmu_circRNA_009692 | F: 5' CAAACCTCACAAAGCACAATATC 3' R: 5' CGAAGAACCTGAACCTGCTG 3' | 60 | 73 |
| mmu_circRNA_27462 | F: 5' ATCCGAAGACTCTGTGAAACTGT 3' R: 5' CTTGTTTACTTATGGGCTTCTTCA 3' | 60 | 126 |
Bioinformatics analyses
Analyses of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to reveal the functional annotation and the pathways involved. The network of circRNA-miRNA-mRNA was constructed under the competing endogenous RNA (ceRNA) hypothesis.
Statistical analysis
The differences between the two groups were compared by Student's t-test. Significant differences in m6A-methylated circRNAs between the two groups in microarray analyses were determined by fold change ≥ 1.5 and p < 0.05. For MeRIP-qPCR data, p < 0.05 was deemed to indicate statistical significance.
Results
OIR alters the profile of m6A methylation levels in circRNAs
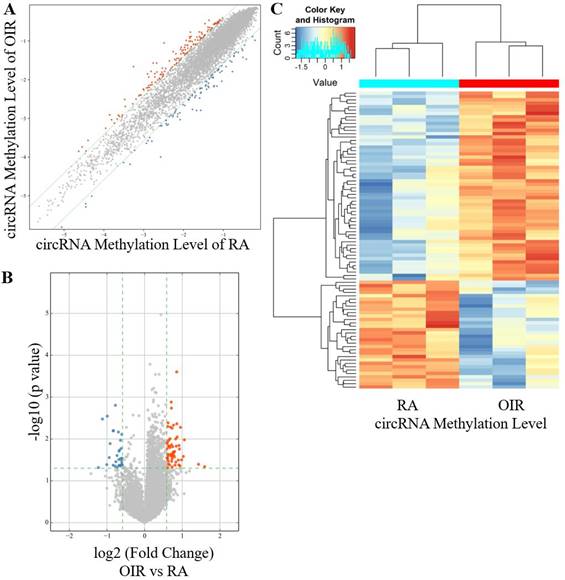
A total of 88 circRNAs were differentially modified by m6A methylation in the retinas of the OIR group and the control group, including 56 hyper-methylated circRNAs and 32 hypo-methylated circRNAs in the OIR group (Figure 1A-1C). The top ten hyper-methylated and hypo-methylated circRNAs are listed in Table 2.
Top 10 hyper-methylated and hypo-methylated circRNAs in OIR retinas
| CircRNA | Regulation | Fold change | p value | Chrom | CircRNA type |
|---|---|---|---|---|---|
| mmu_circRNA_002170 | hyper | 3.020529 | 0.045852 | 15 | intergenic |
| mmu_circRNA_007369 | hyper | 2.701580 | 0.040396 | 15 | exonic |
| mmu_circRNA_42751 | hyper | 2.079537 | 0.010493 | 8 | sense overlapping |
| mmu_circRNA_40551 | hyper | 2.040813 | 0.026124 | 6 | exonic |
| mmu_circRNA_29908 | hyper | 1.977367 | 0.031570 | 16 | exonic |
| mmu_circRNA_42252 | hyper | 1.935942 | 0.005281 | 7 | exonic |
| mmu_circRNA_43770 | hyper | 1.918786 | 0.043799 | 9 | intronic |
| mmu_circRNA_32467 | hyper | 1.898660 | 0.017431 | 19 | exonic |
| mmu_circRNA_010066 | hyper | 1.884827 | 0.014267 | 2 | exonic |
| mmu_circRNA_38600 | hyper | 1.877949 | 0.041738 | 5 | exonic |
| mmu_circRNA_21859 | hypo | 2.340441 | 0.048265 | 10 | intronic |
| mmu_circRNA_33363 | hypo | 2.174187 | 0.003338 | 2 | exonic |
| mmu_circRNA_33864 | hypo | 2.007061 | 0.041051 | 2 | exonic |
| mmu_circRNA_22928 | hypo | 1.989066 | 0.002853 | 11 | exonic |
| mmu_circRNA_26912 | hypo | 1.916923 | 0.027862 | 13 | exonic |
| mmu_circRNA_19661 | hypo | 1.887089 | 0.012903 | 1 | exonic |
| mmu_circRNA_35922 | hypo | 1.784253 | 0.042428 | 3 | exonic |
| mmu_circRNA_19595 | hypo | 1.784158 | 0.006434 | 1 | exonic |
| mmu_circRNA_26987 | hypo | 1.777264 | 0.006297 | 13 | exonic |
| mmu_circRNA_27462 | hypo | 1.718866 | 0.044697 | 14 | exonic |
Associations between circRNA m6A methylation and expression levels
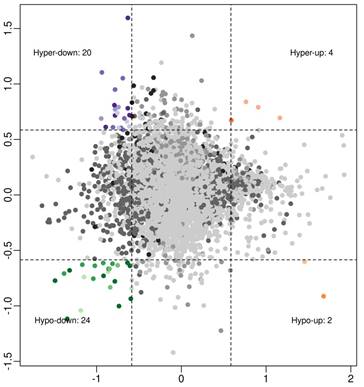
To investigate associations between circRNA expression and m6A methylation levels, circRNAs were intersected with altered m6A methylation and differentially expressed circRNAs by a previous study [16]. Using a threshold of ≥ 1.5-fold change, four interaction modes were identified (Figure 2), including 4 hyper-methylated and upregulated circRNAs, 20 hyper‑methylated and downregulated circRNAs (6 circRNAs met the significance threshold of p < 0.05 in both methylation and expression), 2 hypo-methylated and upregulated circRNAs, and 24 hypo‑methylated and downregulated circRNAs (10 circRNAs met the significance threshold of p < 0.05 in both methylation and expression).
Predictions of involved functions and pathways in host genes of the altered m6A-modified circRNAs
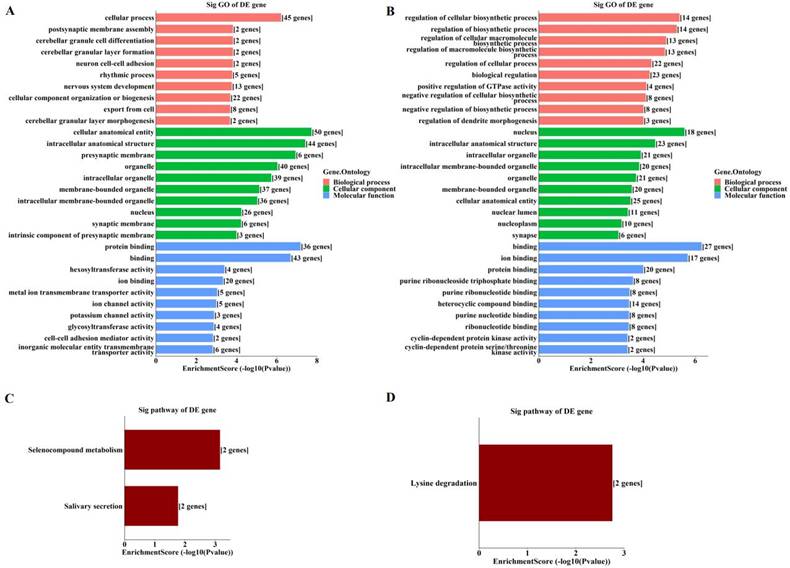
By GO enrichment analyses, host genes of the enriched hyper‑methylated circRNAs were involved in cellular process, cellular anatomical entity, and protein binding (Figure 3A). Those of the enriched hypo-methylated circRNAs were involved in the regulation of cellular biosynthetic process, the nucleus, and binding (Figure 3B). KEGG analyses identified only three pathways, with host genes of hyper-methylated circRNAs involved in selenocompound metabolism and salivary secretion (Figure 3C), and those of hypo-methylated circRNAs involved in lysine degradation (Figure 3D).
CircRNA m6A methylation levels in retinas of OIR and room air control groups. (A) Scatter plot showing correlation distribution of circRNA m6A methylation level in OIR mice and room air (RA) controls. The red and blue dots represent the hyper- and hypo-methylated circRNAs. The gray lines indicate the default alteration of 1.5-fold. (B) Volcano plot showing significantly hyper-methylated and hypo-methylated circRNAs in OIR retinas compared to RA controls. The red and blue dots represent the hyper- and hypo-methylated circRNAs with statistical significance (fold change ≥ 1.5 and p < 0.05). (C) Heatmap of hierarchical clustering showing differentially methylated circRNAs in OIR retinas compared to RA controls (n = 3 per group).
Association analysis of circRNA m6A methylation levels and expressions in OIR retinas: 4 hyper-up circRNAs, 20 hyper-down circRNAs, 2 hypo-up circRNAs, and 24 hypo-down circRNAs (fold change ≥ 1.5).
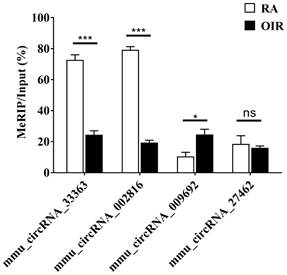
Verification of altered methylation levels by MeRIP-qPCR
m6A methylation levels of four selected circRNAs assessed via MeRIP-qPCR are shown in Figure 4. The m6A methylation levels of mmu_circRNA_33363 and mmu_circRNA_002816 significantly decreased in OIR group, and m6A methylation level of mmu_circRNA_009692 was significantly increased (p < 0.05). The m6A methylation level of mmu_circRNA_27462 was slightly downregulated, but of no statistical significance (p > 0.05). The downward trend was consistent with the microarray analysis.
ceRNA analysis and further predictions
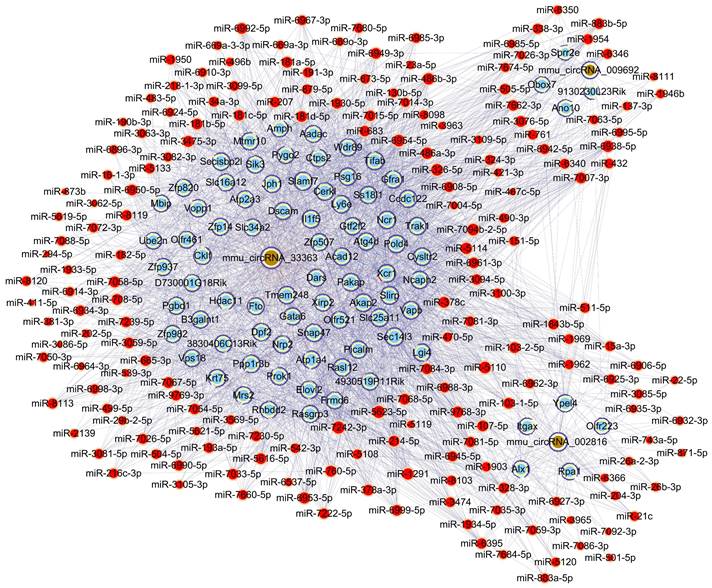
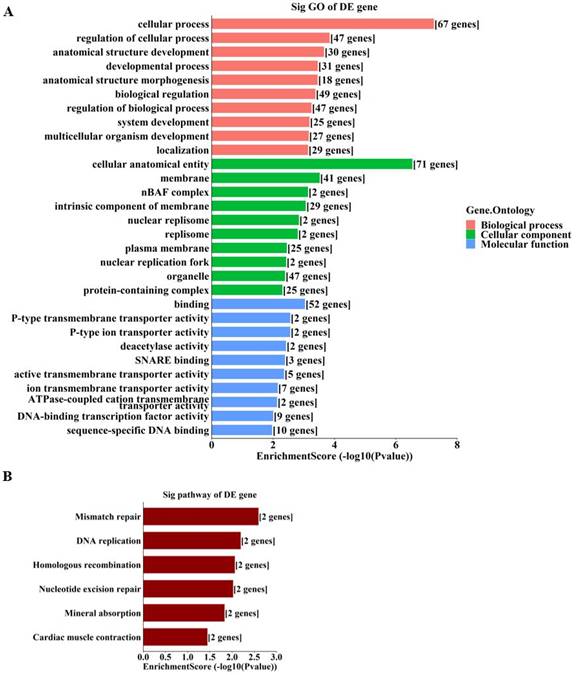
To further unravel the mechanisms of the above-described circRNAs, a circRNA-miRNA-mRNA network was constructed under ceRNA hypothesis. The network was composed of 272 nodes (3 circRNAs, 185 miRNAs, and 84 mRNAs) and 2697 edges (2613 directed edges and 84 undirected edges) (Figure 5). Based on the involved mRNAs, GO analysis identified the top enriched terms, including “cellular process”, “cellular anatomical entity”, and “binding” (Figure 6A). KEGG pathway analysis identified the top pathways involved, including mismatch repair, DNA replication, and homologous recombination (Figure 6B).
Enrichment analyses of host genes of significantly altered m6A-modified circRNAs in OIR retinas. (A, B) GO analysis of host genes of significantly hyper-methylated (A) and hypo-methylated (B) circRNAs. (C, D) KEGG analysis of host genes of significantly hyper-methylated (C) and hypo-methylated (D) circRNAs. The bar plots display the top enrichment scores of the significant enrichment terms.
M6A methylation levels of mmu_circRNA_33363, mmu_circRNA_002816, mmu_circRNA_009692, and mmu_circRNA_27462 determined by MeRIP-qPCR. The data were presented as mean ± SEM (n = 3 per group). *p < 0.05; ***p < 0.001; ns, not significant.
Discussion
M6A modification, as an essential epigenetic regulator, has been suggested to involve in vision function and pathogenesis of ophthalmic diseases [24]. Recently, with the wide application of high-throughput sequencing technologies, there is accumulating evidence that crosstalk between m6A modifications and circRNAs contributes to multifaceted physiological and pathological processes [25, 26]. Therefore, a better understanding of m6A-modified circRNAs in the pathogenesis of retinal neovascularization is warranted.
The current study revealed the epitranscriptomic profile of m6A-modified circRNAs in OIR retinas. A total of 88 circRNAs with significantly altered m6A modifications were identified (Figure 1). The study also identified enriched biological functions via GO analysis, and the pathways involved via KEGG analysis, based on the host genes of hyper-methylated and hypo-methylated circRNAs (Figure 3). These data and predictions shed light on the potential functions of m6A modifications in circRNA regulation of retinal neovascularization. For example, “lysine degradation” was an enriched pathway of the host genes of hypo-methylated circRNAs. Dong et al. [27] demonstrated the presence and involvement of acrolein-lysine adduct in fibrovascular membrane in PDR patients. In our previous study, lysine was significantly upregulated in retinal tissues of mice with OIR [28]. Lysine degradation occurs mainly via saccharopine formation and the pipecolic acid pathway, and involves interaction with subcellular compartmentalization and enzyme deficiencies [29]. However, the specific roles of lysine degradation and the mechanisms involved require further investigation.
It has been suggested that understanding ceRNA crosstalk provides extraordinarily essential views of the regulatory roles of circRNAs [30]. A recent study indicated that m6A modification promotes the capability of binding of circALG1—an overexpressed circRNA in colorectal cancer—to miR-342-5p, enhancing ceRNA regulation, which may facilitate the development of a novel therapeutic method targeting colorectal cancer [31]. Li et al. [32] reported that m6A‑dependent upregulation of circMETTL3 aggravated breast cancer progression by acting as a ceRNA via the circMETTL3/miR-31-5p/CDK1 axis. In the current study, MeRIP-qPCR results indicated significant alteration of m6A methylation levels of mmu_circRNA_33363, mmu_circRNA_002816, and mmu_circRNA_009692 (Figure 4). Therefore, a circRNA-miRNA-mRNA network was constructed under hypothesis of ceRNA based on these three validated circRNAs (Figure 5). GO analysis based on the 84 mRNAs involved showed that the majority of the enriched genes were involved in cellular process, cellular anatomical entity, and binding (Figure 6A); indicating the potential importance of m6A-modified circRNAs in retinal neovascularization. Recent studies showed the involvement of RNA editing modifications in retinal and vascular pathogenesis [33, 34], which indicated that the differential edited genes were enriched in pathways associated with angiogenesis, inflammation and apoptosis. It is also interesting to conduct the integrated analysis of multi-omics of the m6A methylation with other kinds of RNA modifications to further explore the cellular mechanisms in ischemia-induced retinopathy.
Construction of the circRNA-miRNA-mRNA network under ceRNA hypothesis based on three validated circRNAs with significantly altered m6A methylation levels. Light blue nodes represent mRNAs, red nodes represent miRNAs, and brown nodes represent circRNAs with significantly altered m6A methylation levels. Edges with T-shaped arrows and those without arrows indicate directed and undirected relationships, respectively.
Enrichment analyses of involved genes from the ceRNA network. A. GO analysis of involved genes from the ceRNA network. B. KEGG analysis of involved genes from the ceRNA network. The bar plots display the top enrichment scores of the significant enrichment terms.
In summary, the present study investigated the m6A modification alterations of circRNAs in the retinas of OIR. The possible relevant functions and pathways were also predicted by bioinformatics analyses. These provide novel insight into the involvement of m6A methylation in circRNA regulatory functions in OIR. Further investigations are needed to reveal the m6A epitranscriptomic profiles of circRNAs in clinical samples from retinal neovascular diseases, and to identify the specific roles of these m6A-modified circRNAs in the pathological process of ischemia-induced retinal neovascularization.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 82271110 and 82171087), the Natural Science Foundation of Hunan Province (No. 2022JJ30869), the Science and Technology Innovation Program of Hunan Province (No. 2021SK53526), the Scientific Research Project of Hunan Provincial Health Commission (No. 202207022574, 202207022581), the Fundamental Research Funds for the Central Universities of Central South University (No. 2021zzts1068), and the New Technology Incubation Funds in Ophthalmology.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ryu J. New Aspects on the Treatment of Retinopathy of Prematurity: Currently Available Therapies and Emerging Novel Therapeutics. Int J Mol Sci. 2022 23
2. Youngblood H, Robinson R, Sharma A, Sharma S. Proteomic Biomarkers of Retinal Inflammation in Diabetic Retinopathy. Int J Mol Sci. 2019 20
3. Belamkar AV, Jabbehdari S, Harris A, Hajrasouliha AR. Clinical implications of retinal oximetry in retinal vein occlusion: a review. Acta Ophthalmol. 2022;100:624-31
4. Dierks D, Garcia-Campos MA, Uzonyi A, Safra M, Edelheit S, Rossi A. et al. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat Methods. 2021;18:1060-7
5. Xu Z, Xie T, Sui X, Xu Y, Ji L, Zhang Y. et al. Crosstalk Between Histone and m(6)A Modifications and Emerging Roles of m(6)A RNA Methylation. Front Genet. 2022;13:908289
6. Zhou H, Mao L, Xu H, Wang S, Tian J. The functional roles of m(6)A modification in T lymphocyte responses and autoimmune diseases. Cytokine Growth Factor Rev. 2022;65:51-60
7. Yang C, Dong Z, Ling Z, Chen Y. The crucial mechanism and therapeutic implication of RNA methylation in bone pathophysiology. Ageing Res Rev. 2022;79:101641
8. Peng Y, Wang Z, Li B, Tan W, Zou J, Li Y. et al. N(6)-methyladenosine modifications of mRNAs and long noncoding RNAs in oxygen-induced retinopathy in mice. Exp Eye Res. 2022;220:109114
9. Connor KM, Krah NM, Dennison RJ, Aderman CM, Chen J, Guerin KI. et al. Quantification of oxygen-induced retinopathy in the mouse: a model of vessel loss, vessel regrowth and pathological angiogenesis. Nat Protoc. 2009;4:1565-73
10. Qi Y, Yao R, Zhang W, Cui Q. KAT1 triggers YTHDF2-mediated ITGB1 mRNA instability to alleviate the progression of diabetic retinopathy. Pharmacol Res. 2021;170:105713
11. Yao MD, Jiang Q, Ma Y, Liu C, Zhu CY, Sun YN. et al. Role of METTL3-Dependent N(6)-Methyladenosine mRNA Modification in the Promotion of Angiogenesis. Mol Ther. 2020;28:2191-202
12. Liu X, Zhang Y, Zhou S, Dain L, Mei L, Zhu G. Circular RNA: An emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J Control Release. 2022;348:84-94
13. Hanineva A, Park KS, Wang JJ, DeAngelis MM, Farkas MH, Zhang SX. Emerging roles of circular RNAs in retinal diseases. Neural Regen Res. 2022;17:1875-80
14. He M, Wang W, Yu H, Wang D, Cao D, Zeng Y. et al. Comparison of expression profiling of circular RNAs in vitreous humour between diabetic retinopathy and non-diabetes mellitus patients. Acta Diabetol. 2020;57:479-89
15. Li X, Wang J, Qian H, Wu Y, Zhang Z, Hu Z. et al. Serum Exosomal Circular RNA Expression Profile and Regulative Role in Proliferative Diabetic Retinopathy. Front Genet. 2021;12:719312
16. Cao M, Zhang L, Wang JH, Zeng H, Peng Y, Zou J. et al. Identifying circRNA-associated-ceRNA networks in retinal neovascularization in mice. Int J Med Sci. 2019;16:1356-65
17. Li Y, Zhou H, Huang Q, Tan W, Cai Y, Wang Z. et al. Potential biomarkers for retinopathy of prematurity identified by circular RNA profiling in peripheral blood mononuclear cells. Frontiers in Immunology. 2022 13
18. Li Y, Li H, Luo Y, Li X, Chen Z, Zhang W. et al. The Alteration Profiles of m(6)A-Tagged circRNAs in the Peri-Infarct Cortex After Cerebral Ischemia in Mice. Front Neurosci. 2022;16:869081
19. Wu J, Yuan XH, Jiang W, Lu YC, Huang QL, Yang Y. et al. Genome-wide map of N(6)-methyladenosine circular RNAs identified in mice model of severe acute pancreatitis. World J Gastroenterol. 2021;27:7530-45
20. Issah MA, Wu D, Zhang F, Zheng W, Liu Y, Chen R. et al. Expression profiling of N(6)-methyladenosine modified circRNAs in acute myeloid leukemia. Biochem Biophys Res Commun. 2022;601:137-45
21. Zhao W, Liu J, Wu J, Ma X, Wang X, Zhang L. et al. High-throughput microarray reveals the epitranscriptome-wide landscape of m(6)A-modified circRNA in oral squamous cell carcinoma. BMC Genomics. 2022;23:611
22. Huang C, Qi P, Cui H, Lu Q, Gao X. CircFAT1 regulates retinal pigment epithelial cell pyroptosis and autophagy via mediating m6A reader protein YTHDF2 expression in diabetic retinopathy. Exp Eye Res. 2022;222:109152
23. Xing M, Deng M, Shi Y, Dai J, Ding T, Song Z. et al. Identification and characterization of N6-methyladenosine circular RNAs in the spinal cord of morphine-tolerant rats. Front Neurosci. 2022;16:967768
24. Li X, Ma B, Liao M, Li L, Zhang X, Du M. et al. Potential Impact of N6-Methyladenosine RNA Methylation on Vision Function and the Pathological Processes of Ocular Diseases: New Discoveries and Future Perspectives. Front Biosci (Landmark Ed). 2022;27:207
25. Ma L, He LN, Kang S, Gu B, Gao S, Zuo Z. Advances in detecting N6-methyladenosine modification in circRNAs. Methods. 2022;205:234-46
26. Qin S, Zhang Q, Xu Y, Ma S, Wang T, Huang Y. et al. m(6)A-modified circRNAs: detections, mechanisms, and prospects in cancers. Mol Med. 2022;28:79
27. Dong Y, Noda K, Murata M, Yoshida S, Saito W, Kanda A. et al. Localization of Acrolein-Lysine Adduct in Fibrovascular Tissues of Proliferative Diabetic Retinopathy. Curr Eye Res. 2017;42:111-7
28. Zhou Y, Tan W, Zou J, Cao J, Huang Q, Jiang B. et al. Metabolomics Analyses of Mouse Retinas in Oxygen-Induced Retinopathy. Invest Ophthalmol Vis Sci. 2021;62:9
29. Leandro J, Houten SM. The lysine degradation pathway: Subcellular compartmentalization and enzyme deficiencies. Mol Genet Metab. 2020;131:14-22
30. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-52
31. Lin C, Ma M, Zhang Y, Li L, Long F, Xie C. et al. The N(6)-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol Cancer. 2022;21:80
32. Li Z, Yang HY, Dai XY, Zhang X, Huang YZ, Shi L. et al. CircMETTL3, upregulated in a m6A-dependent manner, promotes breast cancer progression. Int J Biol Sci. 2021;17:1178-90
33. Scimone C, Alibrandi S, Donato L, Alafaci C, Germano A, Vinci SL. et al. Editome landscape of CCM-derived endothelial cells. RNA Biol. 2022;19:852-65
34. Donato L, Scimone C, Alibrandi S, Scalinci SZ, Rinaldi C, D'Angelo R. et al. Epitranscriptome Analysis of Oxidative Stressed Retinal Epithelial Cells Depicted a Possible RNA Editing Landscape of Retinal Degeneration. Antioxidants (Basel). 2022 11
Author contact
![]() Corresponding author: Yun Li, MD, PhD, Department of Ophthalmology, The Second Xiangya Hospital of Central South University, Changsha, Hunan 410011, China. Telephone: +86-731-85292175; E-mail: yun.liedu.cn.
Corresponding author: Yun Li, MD, PhD, Department of Ophthalmology, The Second Xiangya Hospital of Central South University, Changsha, Hunan 410011, China. Telephone: +86-731-85292175; E-mail: yun.liedu.cn.

 Global reach, higher impact
Global reach, higher impact