Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(1):87-101. doi:10.7150/ijms.78590 This issue Cite
Research Paper
Prognostic and Immune Infiltration Value of Proteasome Assembly Chaperone (PSMG) Family Genes in Lung Adenocarcinoma
1. Graduate Institute of Cancer Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei 11031, Taiwan.
2. Department of Emergency Medicine, Kaohsiung Medical University Hospital, Kaohsiung Medical University, Kaohsiung 80708, Taiwan.
3. Graduate Institute of Clinical Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 80708, Taiwan.
4. Ph.D. Program for Cancer Molecular Biology and Drug Discovery, College of Medical Science, Taipei Medical University, Taipei 11031, Taiwan.
5. Department of Statistics, Faculty of Science and Technology, PGRI Adi Buana University, East Java, Surabaya 60234, Indonesia.
6. TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei 11031, Taiwan.
#Equal contributors.
Received 2022-9-2; Accepted 2022-11-19; Published 2023-1-1
Abstract
The complexity of lung adenocarcinoma (LUAD) including many interacting biological processes makes it difficult to find therapeutic biomarkers for treatment. Previous studies demonstrated that PSMG (proteasome assembly chaperone) family members regulate the degradation of abnormal proteins. However, transcript expressions of this gene family in LUAD still need to be more fully investigated. Therefore, we used a holistic bioinformatics approach to explore PSMG genes involved in LUAD patients by integrating several high-throughput databases and tools including The Cancer Genome Atlas (TCGA), and Kaplan-Meier plotter database. These data demonstrated that PSMG3 and PSMG4 were expressed at significantly higher levels in neoplastic cells than in normal lung tissues. Notably, increased expressions of these proteins were correlated with poor prognoses of lung cancer patients, which probably confirmed their fundamental roles in the staging of LUAD tumors. Meanwhile, it was also indicated that there were positive correlations between PSMG family genes and the immune response, metabolism of ubiquinone, cell cycle regulatory pathways, and heat shock protein 90 (HSP90)/phosphatidylinositol 3-kinase (PI3K)/Wnt signaling. Experimental data also confirmed that the knockdown of PSMG4 in LUAD cell lines decreased cell proliferation and influenced expressions of downstream molecules. Collectively, this study revealed that PSMG family members are novel prognostic biomarkers for LUAD progression, which also provide new therapeutic targets of LUAD patients.
Keywords: PSMG family genes, Lung cancer, HSP90/PI3K/Wnt
Introduction
As the most fatal malignancy worldwide, primary lung cancer generally ranks first in both incidence and mortality of cancers. Recent statistics show that fatal cases of lung cancer consistently accounted for approximately one-fourth of all cancer deaths, almost double the number of fatal cases caused by second-place colorectal cancer [1]. According to the traditional classification, lung cancer is broadly grouped into two main types: small cell lung cancer (SCLC) - the aggressive one associated with previous smoking that makes up to 15% of total cases - and non-SCLC (NSCLC) - the less aggressive but more-prevalent one that accounts for the majority (up to 85%) of the remainder. Both of them are characterized by distinct cell morphology and topology which lead to significant differences in tailored treatments and diverse disease prognoses [2]. Among the three main subtypes of NSCLC which can be distinguished by distinct histopathological characteristics, lung adenocarcinoma (LUAD) appears to be the most common type of primary lung cancer found in lifelong non-smokers and individuals with a history of smoking as well. Despite recent advances in diagnostic approaches, including imaging-based screening tests, sputum cytology, and modern biopsy techniques, up to 75% of patients are detected in later stages, when metastases are present. Tracking numbers provided by the Surveillance, Epidemiology, and End Results (SEER) database reveal that prognoses of lung cancer are far more troubling than other leading cancer sites (colon, breast, and prostate), with overall 5-year relative survival peaking at 26% but plummets to 8% or even lower in cases of distant metastases. In contrast, patients diagnosed at early stages with localized tumors not only benefit from surgical resection but also from multiple choices of treatment options, with cure rates of as high as 64%. Therefore, early detection of lung cancer in asymptomatic patients is of utmost concern in an effort to improve the disease's dismal outcomes.
Recent genomic studies have confirmed the presence of tumor-harboring somatic mutations and alterations of specific genes in individuals with LUAD, most notably mutation of the epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK), which paved the way for novel classes of drugs known as targeted kinase inhibitors. In addition, activating mutations in certain genes such as KRAS, BRAF, ERBB2, and PIK3CA; translocations found in RET, ROS1, and ALK; and fusions detected in the NRTK1/2/3 genes have been target subjects of a significant number of on-going clinical trials [3, 4]. However, specific types of mutation, such as loss-of-function mutations and deletions in tumor suppressor genes, have yet to be therapeutically exploited [5-7]. Therefore, extensive knowledge of carcinogenesis driver gene alterations will become even more critical to guide the clinical care of LUAD patients. In an attempt to extend current knowledge of details of how molecular signaling is involved in this malignant disease, this study was designed to seek novel biomarkers that can contribute to early detection and prognostic evaluation of LUAD.
The 26S proteasome is a large ATP-dependent protease complex built from a 20S catalytic core particle (CP) responsible for protein degradation and one or two 19S regulatory particles (RPs) essential for ubiquitin recognition. The 20S CP is a combination of two outer α-rings and two inner β-rings stacked in a given order of αββα, each composed of seven homologous subunits respectively referred to as α1-α7 and β1-β7 [8]. Apart from the most prominent role in clearing malfunctioned and damaged proteins, recent studies further elucidated how the human ubiquitin-proteasome system takes control of cell-cycle progression, cellular survival, apoptosis, and activation of nuclear factor (NF)-κB, all of which are emerging hallmarks of cancer accelerated by abnormal proteolytic activity in a genetic mutation-independent context [9-13]. To date, three proteasome inhibitors have been brought to clinical use to treat myelomas and leukemia [14]. As an organelle made up of multi-subunits, proteasomes lack the ability to spontaneously assemble [15, 16]. Initiation and proper formation to create complete and biologically active proteasomes are thus guided and monitored by proteasome assembly chaperones (PSMGs), which are dedicated to preventing aberrant dimerization and ensuring proper incorporation among subcomplexes [16-20]. Given that proteasomes are crucial for intracellular protein degradation and turnover, disorders of proteasome assembly are frequently linked to cellular dysfunction and diminished abilities to respond to proteotoxic stresses, and these disorders have also been well-documented during the past few decades [21]. Being part of the human chaperone complex, four members of the proteasome assembly chaperone family genes, referred to as PSMG1~4, were previously reported to be associated with multiple disorders [22-25]. More specifically in terms of malignancies, earlier studies revealed that PSMG1 was related to increasing susceptibility to inflammatory bowel disease which can lead to colon cancer-related diseases [26, 27], whereas a co-expression relationship of NUP37 with PSMG1 was proposed to play a specific role in breast cancer [28]. PSMG3 is characterized as an oncogenic driver factor in various types of cancer, remarkably breast cancer and cervical squamous cell carcinoma [29]. PSMG4 variants, on the other hand, were some of the differential proteomics identified in Epstein-Barr virus-associated gastric cancer. Since relationships between the PSMG gene family and cancers have barely been described thus far, our study aimed to better elucidate roles of the PSMG gene family in LUAD using multi-approach bioinformatics analyses.
By leveraging high-throughput screening analyses performed in public databases, we previously reported that certain family genes encoded for proteasome complexes are associated with poor prognoses and progressive proliferation of various cancer types [30-32]. In this study, differential expression analysis of PSMG family members was first examined at the transcriptome level in a LUAD cohort of The Cancer Genome Atlas (TCGA), and later confirmed at the proteome level by immunohistochemical (IHC) staining of LUAD specimens. The survival significance of each PSMG gene was evaluated through corresponding estimated Kaplan-Meier curves of overall survival (OS). Next, the significance of PSMG family genes were investigated through functional enrichment analyses, by which coexpressed gene interaction networks, relevant biological processes, and functional annotations were revealed. Additionally, extensive analyses were also performed to clarify the family's relationships with immune infiltration levels. Finally, wet lab validation was conducted to confirm the roles of the most significant PSMG genes in relation to LUAD tumor proliferation and their influence on downstream signaling pathways.
Materials and Methods
Cell culture and RT-qPCR
The human lung alveolar type II epithelial cell line (A549), kindly gifted of Prof. Chiou-Feng Lin from Taipei Medical University (TMU; Taipei, Taiwan) [33]. Cells were cultured in Dulbecco's modified Eagle medium (DMEM) (90-113-PB, Corning, USA) supplemented with 10% fetal bovine serum (Avantor, USA) plus 1% penicillin/streptomycin (Corning), and maintained at 37 °C in a humidified incubator in 5% CO2. PSMG4 gene silencing was generated using a small hairpin (sh)RNA knockdown vector system and lipofectamine 2000 (Life Technologies Inc., Carlsbad, CA) transfection was carried out according to the manufacturer's protocol as we previously described [34, 35]. All shRNA vectors harboring puromycin and enhanced green fluorescent proteins, including two human PSMG4 shRNA, and a non-target control (pLKO.1) shRNA against luciferase (shLuc), were constructed by the National RNAi Core Facility (Academia Sinica, Taiwan; https://rnai.genmed.sinica.edu.tw). Stable clones expressing pLKO_TRC005 were selected by constant treatment with puromycin (2 µg/mL) from 72 hours lipofectamine transfection. A non-target control (pLKO.1) shRNA against luciferase (shLuc) was employed as an expression control. On day 28 post-transfection, the efficacy of gene silencing was further confirmed by comparing quantitative reverse-transcription polymerase chain reaction (RT-qPCR) of two groups of shPSMG4-transfected A549 cells versus pLKO.1(vector control)-transfected cells. Total RNAs from stable clones of the A549 cell line were extracted using the GENEzol™ TriRNA Pure Kit (Geneaid Biotech, Taiwan) following the manufacturer's protocol, whereas complementary (c)DNA was subsequently reverse-transcribed using a PrimeScript Synthesis Kit (Takara Bio, Japan). An RT-qPCR was performed using cDNAs as templates and GoalBio SYBR green master mix (Hycell International, Taiwan) on the Roche Light Cycler 96 platform. Primer pair sequences targeting PSMG4, HSP90AA1, and 18S ribosomal (r)RNA were constructed by MDBio (MDBio, Taiwan). Relative fold changes in expression of the PSMG4 and HSP90AA1 genes were calculated by the delta-delta Ct (2-∆∆Ct) method after being normalized against the Ct value of 18S rRNA as the housekeeping gene. All experiments were performed in triplicate. RT-qPCR results are presented as the mean ± standard deviation [36-38].
Colony-formation assay
A549 cells of three experimental groups were seeded at the same density (1000 cells/well) in six-well plates for 2~3 weeks until macroscopic colonies had formed. After that, the medium was discarded, and cells underwent absolute methanol fixation in 20 min at room temperature, followed by short-term incubation with 2% methylene blue staining in 30 min at room temperature. The number of colonies formed in each well was counted under a low-magnification (×100) light microscope. Experiments were performed in triplicate. Results were represented as the mean ± SD of triplicate data [39-41].
UALCAN, Cancer Cell Line Encyclopedia (CCLE), and Kaplan-Meier analysis
TCGA database is the largest and most widely used public resource providing gene expression profiling, gene methylation profiling, copy number variation profiling, single-nucleotide polymorphism (SNP) genotyping, genome-wide DNA methylation profiling, microRNA profiling, and exon sequencing. Among the many computational tools that have been developed to assist scientists in performing specific analyses based on TCGA database, UALCAN (http://ualcan.path.uab.edu) is an interactive web tool capable of calculating the relative expression of a queried gene across a specific tumor or various tumor sub-groups against normal samples. Relevant analyses are based on either individual cancer stages, tumor grades, or other clinicopathological features. In our study, we examined alterations in terms of the transcriptome of four PSMG family genes in 515 primary LUAD samples versus 59 normal samples, along with different clinicopathological characteristics and stages. Next, we used CCLE to further explore the gene expression level of all PSMG members on a broader scale [42]. This web-based application gives users access to more than 1000 human cancer cell lines. Additionally, independent LUAD cancer cell lines were subjected to the integrated RNA-Seq Aligned Reads method to plot the corresponding expression of each PSMG gene. The KMplot survival database (https://kmplot.com/) was leveraged to explore which PSMG family members are novel prognostic biomarkers [43]. The comparisons between two groups of patients were performed with 95% confidence intervals (CIs) of the hazard ratio (HR) and fixed log-rank p-value as we previously described [44-46].
Tumor Immunological Estimation Resource (TIMER) database analysis
The latest version (2.0) of TIMER, available at http://timer.cistrome.org/, is generally acknowledged as an efficient tool for conducting systematic analyses of host immune infiltrates among various cancer types and associated diseases [47]. In other words, using the DiffExp module and default parameters, this webserver assists in estimating the abundances of six immune cell types grouped into two distinct lineages in the tumor microenvironment (TME): B cells, cluster of differentiation-positive (CD4+) T cells, and CD8+ T cells that belong to lymphoid lineage cells, together with neutrophils, macrophages, and dendritic cells (DCs) that belong to myeloid lineage cells. Finally, scatterplots were employed to show the associations, with the x-axis representing PSMG gene expression levels and the y-axis showing markers for immune cells that infiltrate lung tumors.
Functional enrichment analysis of PSMG target genes
The InteractiVenn tool was selected to generate a one-way Venn diagram that presents the overlap and number of genes linked with expressions of PSMG target genes across two given datasets obtained from TCGA databases (available at the cBioPortal platform). This online web tool (available at http://www.bioinformatics.com.cn/srplot), together with the MetaCore platform to investigate the potential pathways and involved networks, as well as an online platform (http://www.bioinformatics.com.cn/ en?keywords=heatmap) was conducted for data visualization. A p-value of <0.05 was set as the cutoff value [48-50].
Results
PSMG family members play important roles in developmental stages of LUAD
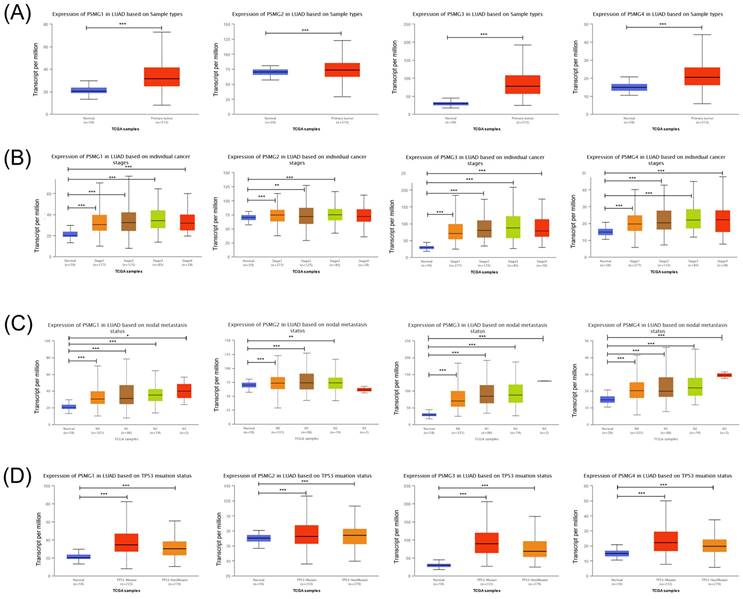
Prior studies discovered six PSMG family members in Homo species as well as the significant roles in cancer progression of some of them. We wanted to further identify PSMG family gene signatures in relationship with LUAD development. Results demonstrated that compared to normal tissues, all PSMG family genes had high expression levels in LUAD tissues. Surprisingly, we found that PSMG4 had positive correlations with the tumor stage, especially with the highest expression in stage 4 LUAD. PSMG1, PSMG3, and PSMG4 had positive correlations with the nodal metastasis status. In addition, since the TP53 mutation was correlated with LUAD development, we also found that PSMG1, PSMG3, and PSMG4 had the highest expression in TP53 mutant LUAD. It was suggested that PSMG family member overexpression may promote tumor growth, and overexpression of PSMG1, PSMG3, and PSMG4 may further promote the development of cancer metastasis. Therefore, we decided to perform further bioinformatics analyses on these family genes in LUAD (Figure 1).
Associations of PSMG family gene interpretations in cancer cell lines with clinicopathological parameters of LUAD
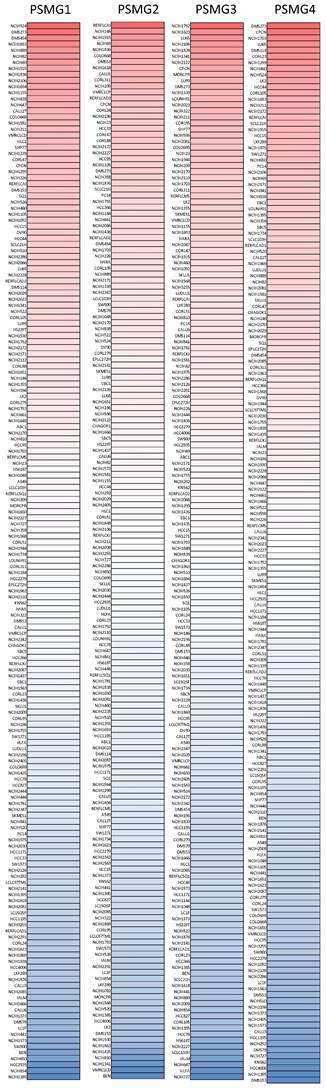
After properly examining differences in PSMG family gene expressions between neoplastic and normal tissues, we next performed a comprehensive analysis of PSMG family genes in lung cancer cell lines via the CCLE dataset. These data also indicated that PSMGs were overexpressed at the messenger (m)RNA level measured in the lung cancer cell lines (Figure 2).
Relationships between disease prognostication and PSMG gene expression levels measured in tumor specimens and survival statuses
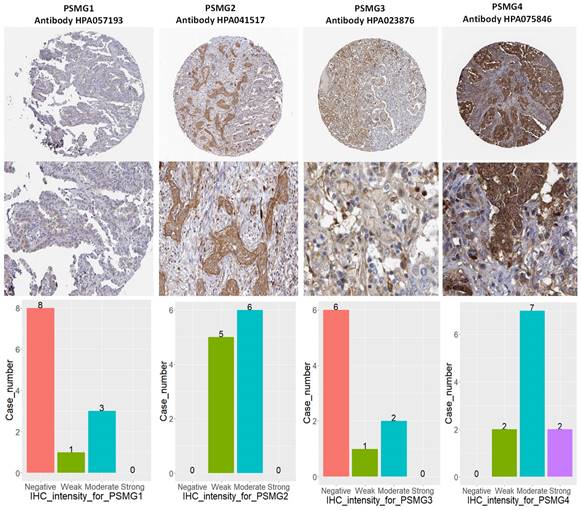
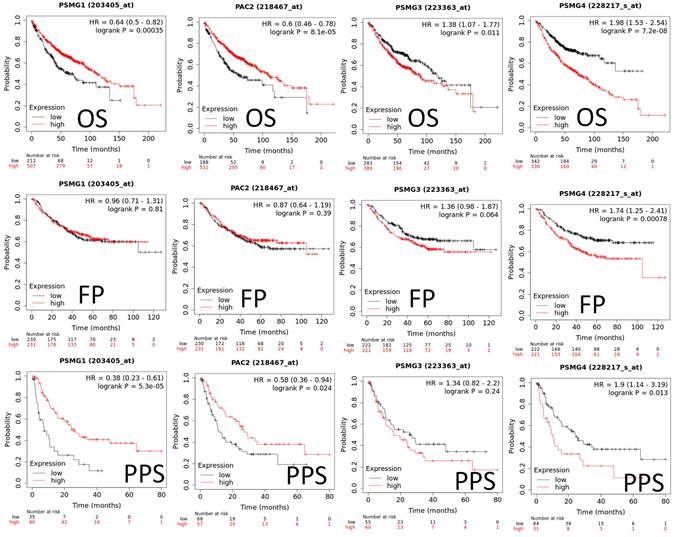
Since samples from LUAD patients included different expressions of PSMG family members, we further explore how these target genes take part in particular aspects of tumor progression prior to investigating the clinical relevance. Hence, the intensities of antibodies indicated in clinical LUAD specimens were extracted from the Human Protein Atlas database. IHC images revealed densely and moderately distributed PSMG family protein expressions in LUAD samples (Figure 3). In addition, when we performed a required analysis on the TIMER database, PSMG member genes also indicated relevance to the immune infiltration profiles of LUAD, which included six various tumor-infiltrating immune cell types grouped into two separate groups: B cells, CD4+ T cells, and CD8+ T cells that belong to lymphoid lineage cells, together with neutrophils, macrophages, and DCs that belong to myeloid lineage cells (Figure 4). The KM plotter database also indicated that most PSMG members were associated with different LUAD survival statuses. High expressions of PSMG1 and PSMG2 in LUAD were correlated with good OS, FP, and PPS. In contrast, high expressions of PSMG3 and PSMG4 were correlated with poor OS, FP, and PPS (Figure 5). These survival data imply that PSMG3 and PSMG4 have oncogenic roles in LUAD progression (Supplementary Figure S1).
Pathway and network analysis of PSMG family member genes
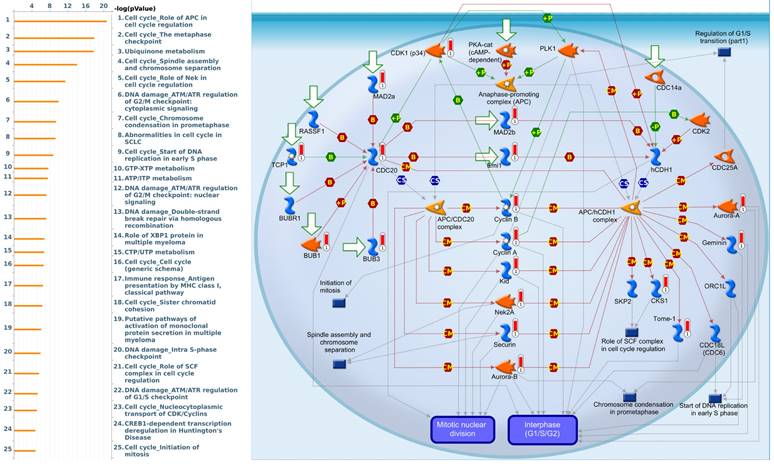
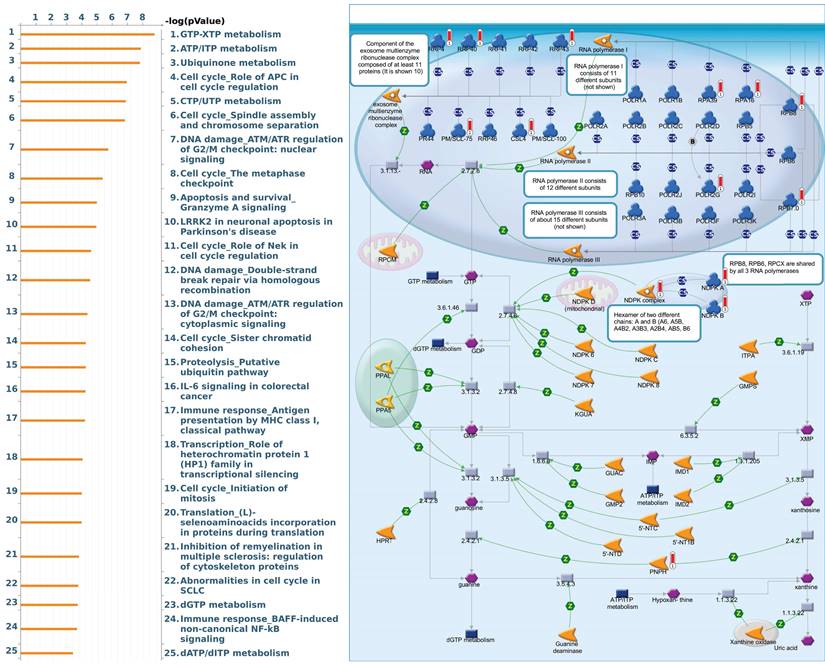
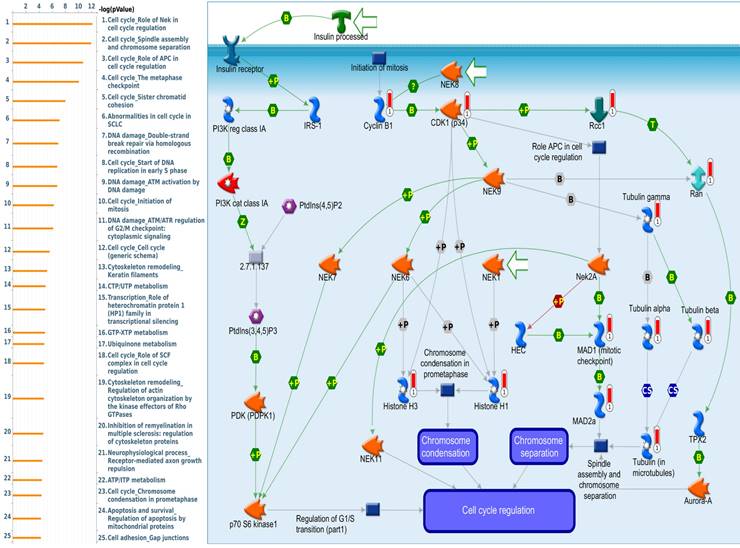
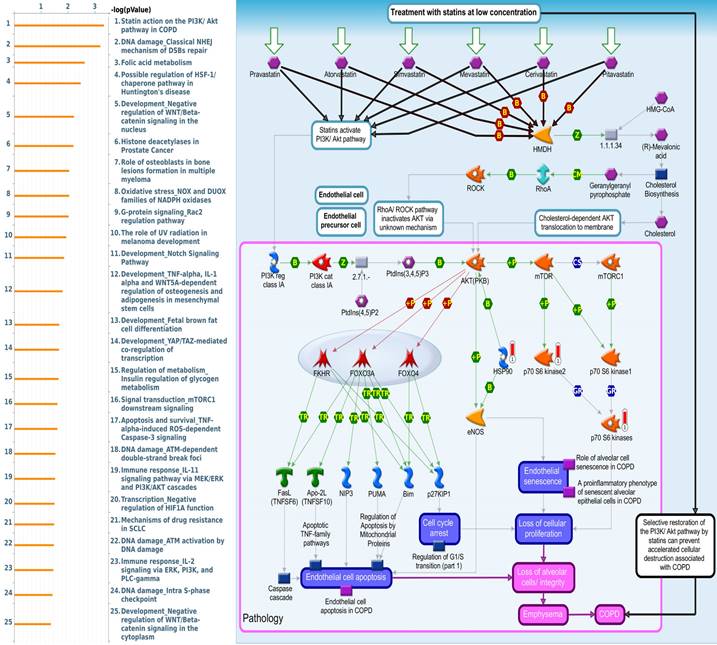
Since there is still limited potential information to refine the full picture of regulated pathways available to PSMG family genes, the GeneGo MetaCore platform was conducted to the aforementioned coexpression patterns of PSMG family genes. We obtained PSMG1 co-expression profiles in LUAD from both the LUAD TCGA [51] and CPTAC datasets [52]. As a result, annotations of almost all biological processes obtained from GeneGo Metacore showed that genes coexpressed with PSMG1 regulated cell cycle-related pathways such as “Cell cycle_Role of APC in cell cycle regulation”, “Cell cycle_The metaphase checkpoint”, “Ubiquinone metabolism”, “Cell cycle_Spindle assembly and chromosome separation”, and “Cell cycle_Role of Nek in cell cycle regulation” (Figure 6; Supplementary Table S1). PSMG2 was involved in cancer metabolism-related pathways, including “GTP-XTP metabolism”, “ATP/ITP metabolism”, “Ubiquinone metabolism”, “Cell cycle_Role of APC in cell cycle regulation”, and “CTP/UTP metabolism” (Figure 7; Supplementary Table S2). PSMG3 was involved in pathways related to “Cell cycle_Role of Nek in cell cycle regulation”, “Cell cycle_Spindle assembly and chromosome separation”, “Cell cycle_Role of APC in cell cycle regulation”, “Cell cycle_The metaphase checkpoint”, and “Cell cycle_Sister chromatid cohesion” (Figure 8; Supplementary Table S3). PSMG4 was involved in pathways related to “Statin action on the PI3K/Akt pathway in COPD”, “DNA damage_Classical NHEJ mechanism of DSBs repair”, “Folic acid metabolism”, “Possible regulation of HSF-1/chaperone pathway in Huntington's disease”, and “Development_Negative regulation of WNT/Beta-catenin signaling in the nucleus” (Figure 9; Supplementary Table S4).
Transcriptional expression of PSMG (proteasome assembly chaperone) family members in lung adenocarcinoma (LUAD). (A) Boxplot of PSMG transcriptomic levels recorded in LUAD patients (red) compared to healthy individuals (blue). Box plots of variations in transcriptomic levels of PSMG1~4 recorded in normal individuals and LUAD patients (B) from stages 1 to 4 (C) grouped into four subgroups (N0~N3) based on the regional lymph node metastasis status (D) with and without the TP53 mutation. A t-test was applied considering p<0.05 as a significant difference.
Expression degree of PSMG (proteasome assembly chaperone) family members among common types of lung adenocarcinoma (LUAD) cell lines in a heatmap. Varying degrees of upregulated expressions (red color), downregulated expressions (blue color), and no changes in expression of PSMG1~4, measured in different LUAD cell lines.
IHC intensity patterns of PSMG1~4 (proteasome assembly chaperone1~4) recorded in lung adenocarcinoma (LUAD) patient tissues. (A) PSMG1~4 protein expressions of LUAD tissue staining patterns, along with corresponding healthy tissues. (B) Bar chart of case numbers grouped according to the level of staining intensity for PSMG1~4 in LUAD tissues.
PSMG4 mRNA and protein in LUAD cell lines
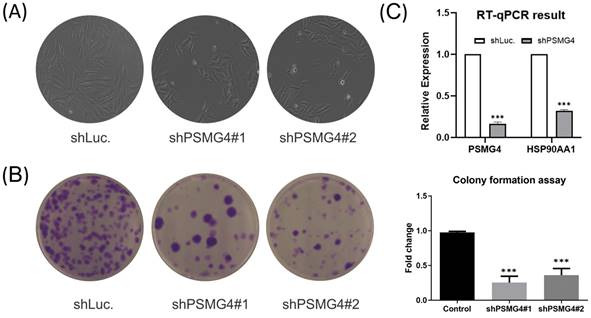
The above bioinformatics analysis indicated that PSMG4 expression was higher in tumor samples compared to normal samples, and further promoted development advance in the tumor stage via the heat shock protein 90 (HSP90)/phosphatidylinositol 3-kinase (PI3K)/AKT/Wnt signaling pathway. Therefore, among PSMG family members, we chose PSMG4 for further study. Intriguingly, in the absence of PSMG4 in A549 cells, cells changed to a more-cuboidal (epithelial-like) morphology compared to shLuciferase control cells (Figure 10A). Lipofectamine transfection of PSMG4-shRNA was performed to inhibit PSMG4 expression in LUAD cells. The suppressive efficacy was confirmed by a qPCR (Figure 10B). Meanwhile, expression levels of an HSP90-related marker (HSP90AA1) decreased after PSMG4 expression was downregulated (Figure 10B). We investigated long-term cell proliferation by a colony-formation assay, and found that the cellular proliferation in PSMG4-silenced LUAD cells had decreased (Figure 10C).
Discussion
Recent epidemiologic studies indicated that lung cancer remains at the top of fatal malignancies, despite remarkable improvements that have been made in medical and surgical approaches. As a matter of fact, shortages of highly sensitive screening tests, delays in early screening, and high probabilities of drug and chemoresistance have resulted in increased risks of metastasis and relapse, as well as meager survival rates for lung cancer patients. Therefore, identifying specific and highly sensitive biomarkers for lung cancer is urgent, with the goal of formulating effective treatments known as personalized medicine [53].
To date, the rapid growth of microarray and high-throughput sequencing techniques has generated enormous amounts of data available in either web-based platforms or comprehensive tools that allow us to properly monitor tumor progression at various levels, including genomics, epigenomics, transcriptomics, proteomics, and metabolomics [54, 55]. The underlying pathogenesis of tumors in general and lung cancer, in particular, has thus been reported to be dominated by specific somatic genomic alterations. Notably, KRAS, BRAF, ERBB2, PIK3CA, RET, ROS1, ALK, and NRTK1/2/3 have been proposed as prognostic markers, making substantial contributions to genotype-directed therapies for advanced lung cancer. By applying for advances in high-throughput screening on cancer transcriptomic profiling, alterations in transcriptome patterns of gene families encoded for human proteasomes were discovered to be significantly associated with several types of malignancies, among which PSMG gene expressions were found to be involved not only in multi-stage tumor progression but also in other tumor-related aspects. However, since previous research has yet to elaborate on the roles of PSMG family genes in LUAD, this study can serve as the first and foremost work that specifically examined the roles of PSMG individuals in LUAD.
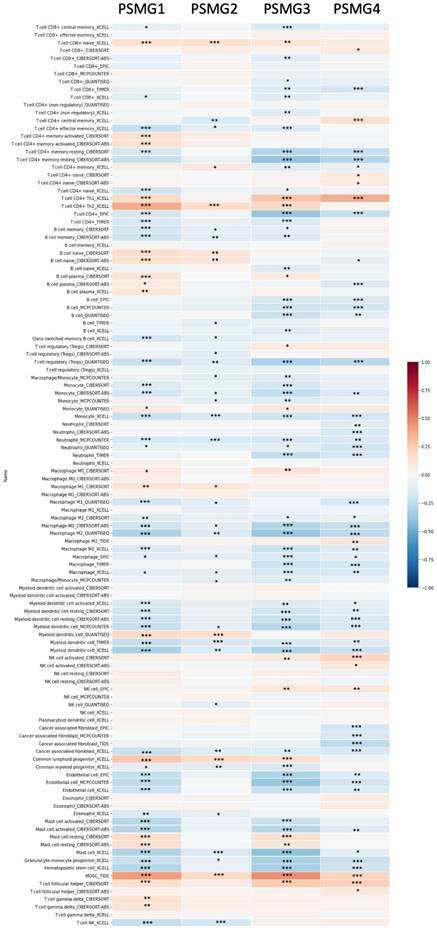
Correlations between gene expressions of PSMG1~4 (proteasome assembly chaperone 1~4) and immune infiltration levels among lung adenocarcinoma (LUAD) patients in TCGA cohorts through data mining from the TIMER database. Using the "Purity Adjustment" option of TIMER, correlations between expression levels of six major immune cell populations and four subtypes of breast cancer were evaluated based on seven cell type quantification algorithms (xCell, CIBERSORT, CIBERSORT abs. mode, EPIC, MCP-counter, TIMER, and quanTIseq). Results are presented as a Pearson correlation coefficient (r) that ranges from -1 (negative correlation: blue) to +1 (positive correlation: red). Three statistical significance levels were reported, including * p < 0.05, ** p < 0.01, and *** p< 0.001.
Predicted survival analyses according to expression levels of PSMG1~4 (proteasome assembly chaperone 1~4) in lung adenocarcinoma (LUAD) patients. The two survival curves respectively present percentages of LUAD survivors over time, in patients with low (black curves) and high (red curves) expressions of PSMG1~4. Elevated mRNA levels of the PSMG1 and PSMG2 genes resulted in good prognoses, whereas elevated mRNA levels of PSMG3 and PSMG4 showed the contrary. (HR, hazard ratio; OS, overall survival; FP, first progression; PPS, post-progression survival; p < 0.05 was considered statistically significant).
Significantly associated signaling pathways regulated by top genes coexpressed with PSMG1 (proteasome assembly chaperone 1), as predicted by MetaCore. Top 10% coexpressed genes with PSMG1 shared from TCGA databases were selected for gene expression network predictions using "biological processes" of MetaCore. The involved pathways were ranked in order of decreasing -log[p values]. Regulatory pathways for the cell cycle are among the most significant ones.
Significantly associated signaling pathways regulated by top genes coexpressed with PSMG2 (proteasome assembly chaperone 2), as predicted by MetaCore. The top 10% coexpressed genes with PSMG2 shared from TCGA databases were selected for gene expression network predictions using "biological processes" of MetaCore. The involved pathways were ranked in order of decreasing -log[p values]. Regulatory pathways for energy metabolism and cell division progression were among the most significant ones.
PSMG1 and PSMG2, previously referred to as PAC1 and PAC2, are two evolutionarily conserved, ubiquitously expressed chaperone proteins promoting proper assembly of the α-ring of the 20S CP human proteasome [56]. PSMG1 assembles in a complex with PSMG2 since intact PSMG1-PSMG2 heterodimers help to stabilize the assembly of the heteroheptameric α-ring and prevent accumulation of non-productive α-ring dimers [57]. The previous literature confirmed that PSMG1 plays significant roles in increasing susceptibility to inflammatory bowel diseases, a key factor leading to a higher risk of colorectal cancer, which can be explained by the close location of rs2836878 to PSMG1 supporting their function in the ubiquitin-proteasome system [58-61]. In addition, targeting PSMG1 caused by miR-484 inhibition led to reductions in cell migration and invasion in prostate cancer [62]. In this study, analytical results confirmed elevated expression of the PSMG1 transcriptome in LUAD tissues compared to normal ones. Furthermore, incremental expression of the PSMG1 gene was recorded in advanced stages of LUAD, in both individuals and nodal statuses, and especially in patients bearing the TP53 mutation, suggesting that a regulatory axis exists between PSMG1 and other cellular factors that drive the progression of LUAD. Subsequent findings also designated the “Cell cycle_Role of APC in cell cycle regulation” as a dominant pathway presented in the coexpression gene network of PSMG1. The findings mentioned above are consistent with previous knowledge of PSMG1's roles; however, survival analysis showed no signs of a relationship between elevated levels of the PSMG1 transcriptome and FP rates of patients with LUAD; and a limited number of IHC staining samples came back positive for PSMG1, demonstrating the minor presence of this gene in biopsies of LUAD patients.
Significantly associated signaling pathways regulated by top genes coexpressed with PSMG3 (proteasome assembly chaperone 3), as predicted by MetaCore. The top 10% coexpressed genes with PSMG3 shared from TCGA databases were selected for building a gene expression network using "biological processes" of MetaCore. The involved pathways were ranked in order of decreasing -log[p values]. Regulatory pathways for cell division progression were among the most significant ones.
The distinct roles in human diseases of PSMG2, on the other hand, were largely unknown, apart from the monogenic inheritance of CANDLE (chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature) syndrome that was previously reported to occur of the PSMG2 mutations [63]. As specified by the MetaCore enrichment pathway analysis, GTP, XTP, ATP, ITP, and ubiquinone-related metabolism signaling are among PSMG2 coexpressed pathways with the most significant p values; however, there were no noticeable changes recorded in gene expression levels either in later stages, nodal statuses, or in the TP53-mutated population. Similarly, a subsequent immune infiltration analysis also did not signify substantial correlations between expression patterns of specific immune cells and PSMG2 transcriptomics. Such observations suggest that PSMG2 seems to only play a minor role in LUAD progression.
PSMG3 and PSMG4, alternatively known as PAC3 and PAC4, are mainly regulate the recruitment of the β-ring [64]. Given that malfunctions of these assembly chaperones cause the accumulation of imperfectly assembled or misassembled complexes of proteasomal subunits, knockdown experiments involving PSMG3 and PSMG4 resulted in the accumulation of abnormal α-subunit oligomers [65]. Elevated PSMG3 levels found in the plasma of glioblastoma multiforme (GBM) patients help distinguish those with sarcoidosis and healthy patients [50]. In addition, a significant number of lncRNA-PSMG3 and miRNA-PSMG3 axis regulatory mechanisms were reported to be associated with malignant diseases, including lung cancer [66-69]. On the contrary, limited literature has described the relationship between PSMG4 and human diseases, especially tumors. In our study, expressions of PSMG3 and PSMG4 were both found to progressively remain elevated toward increasing advanced stages of individuals, nodal statuses, and also in LUAD patients with the TP53 mutation. The survival analysis indicated that upregulation of PSGM3 and PSMG4 resulted in poor prognoses, in terms of OS in general and both FP and PPS in particular.
Subsequent analyses of PSMG4, in particular, showed strong consistency with earlier studies. First, abundant expression of the PSMG4 protein was found in LUAD tissues through IHC staining, compared to the normal adjacent lung tissues. Second, regulatory pathways predicted based on the top genes coexpressed with PSMG4 suggested that the top three most significant pathways included “Statin action on the PI3K Akt pathway in COPD”, “DNA damage_Classical NHE mechanism of DSBs repair”, and “Folic acid metabolism”, which may partly explain why PSMG4 plays a key role in LUAD. To be more specific, DNA damage, particularly DNA double-strand breaks (DSBs), is highly deleterious since a single unrepaired DSB is sufficient to trigger cellular senescence or apoptosis, and DSB-repair and associated sub-pathways play key roles in suppressing tumorigenesis. However, the relationship between folic acid intake and lung cancer risk remains controversial due to biased evidence among genders and ages [70-72]. Most remarkable of all, there is ample evidence that dysregulation of cholesterol homeostasis, accumulation of cholesteryl ester-rich lipid droplets in lung tissues, and NSCLC are intimately linked. Interestingly, statin treatment was observed to show antitumorigenic activities against lung cancer by suppressing AKT and the Braf/mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase 1/2 (ERK1/2) pathways [73-75]. Moreover, numerous experiments on the NSCLC-derived A459 cell line revealed that p-Akt inhibition helped increase apoptosis and reduce radio-resistance [75-77], and bioinformatics data also presented DNA methylation expression levels of PSMG4 in lung cancer (Supplementary Figure S2). Third, our wet lab validation indicated that silencing of the PSMG4 gene not only helped attenuate proliferation of tumor cells, but also significantly reduced the mRNA level of heat shock protein 90 (HSP90AA1) - a cytosolic protein that belongs to the HSP90 family, which was in agreement with the downstream signaling pathway predicted by MetaCore (Figure 9).
Significantly associated signaling pathways regulated by top genes coexpressed with PSMG4 (proteasome assembly chaperone 4), as predicted by MetaCore. The top 10% coexpressed genes with PSMG4 shared from TCGA databases were selected for gene expression network predictions using "biological processes" of MetaCore. The involved pathways were ranked in order of decreasing -log[p values]. “Statin action on the PI3K Akt pathway in COPD”, “DNA damage_Classical NHE mechanism of DSBs repair”, and “Folic acid metabolism” were among the most significant ones.
PSMG4 (proteasome assembly chaperone 4) expression in lung adenocarcinoma (LUAD) cell lines. (A) Bright-field microscopic images of A549-shLuc as the control, along with two A549-shPSMG4 counterparts, using two-dimensional cell culture modes. Spindle-like morphology of the A549-shLuc control became epithelial-like after PSMG4 silencing. (B) RT-qPCR showing decreased mRNA expression levels of HSP90AA1--an important regulator of autophagy involved in downstream signaling pathways regulated by PSMG4. Data were normalized against 18S rRNA as an internal control using the 2-ΔΔCt method. Statistical significance was determined by Student's t-test and error bars represent the SD (* p < 0.05, ** p < 0.01) of duplicate determinations. (C) Colony-formation assay. Representative micrographs (left) and quantification (right) of crystal violet-stained clones confirmed the influence of PSMG4 on attenuating A549 cell proliferation. The long-term proliferation of two A549 shPSMG4-knockdown cell lines markedly declined compared to A549-shLuc as the control.
As a chaperone protein well-known for its role in stabilizing various growth factor receptors (such as tumor necrosis factor receptors (TNFRs) and NF-κB) and various signaling molecules found overexpressed in tumor cells (including PI3K and AKT proteins), inhibition of HSP90 was reported to downregulate the PI3K/AKT pathway, thus resulting in apoptosis of tumor cells via downregulation of the antiapoptotic protein, BCL-w [78-81]. In addition, HSP90 also participates in a significant number of key processes in de novo angiogenesis which is necessary for tumor growth under hypoxia, and promotes tumor metastasis [82-84]. Most interestingly, the relationship of the HSP90 protein and intrinsic resistance to immunotherapies by tumors has been of interest since previous studies indicated that specific inhibition of HSP90 helps drive potent T-cell responses in the TME, specifically reducing surface expressions of several immune checkpoint proteins (such as programmed death ligand 1 (PD-L1) and PD-L2), reducing the number of regulatory T (Treg) cells, and stimulating “killer” T cells (CD8+ cytotoxic T-lymphocytes (CTLs)) and “helper” T cells (CD4+) [85-87], thereby triggering an immunosuppressive TME and relieving tumor immune escape. Similarly, such immune infiltration patterns of PSMG4 were recorded in LUAD patients via a TIMER analysis, which neatly supports the strong biological associations between PSMG4 and HSP90AA1 in the context of lung cancer.
Taken together, our study proposed that elevated expression of PSMG4 leads to dismal outcomes for LUAD patients, whereas inhibition of PSMG4 resulted in a reduced cell proliferation rate, along with a marked decline in the expression level of the HSP90AA1 protein. Additionally, similar immune infiltration patterns between PSMG4 and HSP90AA1 also suggested that PSMG4 may play a certain role in the immune-suppressive TME which is common among lung cancer patients. PSMG4 can thus possibly serve as an independent risk factor and immuno-prognostic factor for LUAD patients. However, detailed signaling pathways are mandated to figure out the immune crosstalk, hence allowing better guidance for clinical immunotherapy and providing accurate personalized treatment plans for LUAD patients.
Conclusions
To summarize, our study proposed PSMG4 as a potential prognostic marker and therapeutic target of LUAD. Knockdown of PSMG4 in a LUAD cell line reduced cell proliferation and influenced expressions of downstream molecules, including HSP90-related genes. The current study revealed that PSMG4-related signaling may potentially be targeted for the treatment of LUAD. Further in-depth experiments are mandated to extensively explore PSMG4's key roles in the underlying pathogenesis and immune crosstalk in LUAD progression and metastatic spread that participate in cancer development.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This research was supported by grants from the Ministry of Science and Technology (MOST) of Taiwan (MOST-109-2320-B-038-009-MY2 and MOST-107-2320-B-037-011-MY3), from Taipei Medical University Hospital (109TMU-TMUH-19 and 111TMU-TMUH-08), and from Kaohsiung Medical University Hospital (KMUH109-9R82 and KMUH110-0M75). This work was financially supported by the Higher Education Sprout Project of the Ministry of Education (MOE) in Taiwan. The authors truly appreciate the professional English editing by Mr. Daniel P. Chamberlin from the Office of Research and Development at Taipei Medical University. The authors acknowledge the statistical/computational/technical support of the Clinical Data Center, Office of Data Science, Taipei Medical University, Taiwan.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: a cancer journal for clinicians. 2022;72:7-33
2. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A. et al. Non-Small Cell Lung Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. Journal of the National Comprehensive Cancer Network: JNCCN. 2022;20:497-530
3. Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nature medicine. 2012;18:349-51
4. Westphalen CB, Krebs MG, Le Tourneau C, Sokol ES, Maund SL, Wilson TR. et al. Genomic context of NTRK1/2/3 fusion-positive tumours from a large real-world population. npj Precision Oncology. 2021;5:69
5. Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869-73
6. Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM. et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer research. 2002;62:3659-62
7. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069-75
8. Wu W, Sahara K, Hirayama S, Zhao X, Watanabe A, Hamazaki J. et al. PAC1-PAC2 proteasome assembly chaperone retains the core α4-α7 assembly intermediates in the cytoplasm. 2018; 23: 839-48.
9. Adams J. The proteasome: structure, function, and role in the cell. Cancer Treatment Reviews. 2003;29:3-9
10. Kumatori A, Tanaka K, Inamura N, Sone S, Ogura T, Matsumoto T. et al. Abnormally high expression of proteasomes in human leukemic cells. Proceedings of the National Academy of Sciences. 1990;87:7071-5
11. Shah SA, Potter MW, McDade TP, Ricciardi R, Perugini RA, Elliott PJ. et al. 26S proteasome inhibition induces apoptosis and limits growth of human pancreatic cancer. Journal of cellular biochemistry. 2001;82:110-22
12. MacLaren AP, Chapman RS, Wyllie AH, Watson CJ. p53-dependent apoptosis induced by proteasome inhibition in mammary epithelial cells. Cell Death & Differentiation. 2001;8:210-8
13. Sasaki K, Hamazaki J, Koike M, Hirano Y, Komatsu M, Uchiyama Y. et al. PAC1 Gene Knockout Reveals an Essential Role of Chaperone-Mediated 20S Proteasome Biogenesis and Latent 20S Proteasomes in Cellular Homeostasis. 2010; 30: 3864-74.
14. Park JE, Miller Z, Jun Y, Lee W, Kim KB. Next-generation proteasome inhibitors for cancer therapy. Translational Research. 2018;198:1-16
15. Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:12-36
16. Budenholzer L, Cheng CL, Li Y, Hochstrasser M. Proteasome Structure and Assembly. Journal of Molecular Biology. 2017;429:3500-24
17. Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nature Reviews Molecular Cell Biology. 2018;19:697-712
18. Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol. 2018;19:697-712
19. Schnell HM, Walsh RM Jr, Rawson S, Kaur M, Bhanu MK, Tian G. et al. Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis. Nat Struct Mol Biol. 2021;28:418-25
20. Dimitrakopoulos FD, Kottorou AE, Kalofonou M, Kalofonos HP. The Fire Within: NF-κB Involvement in Non-Small Cell Lung Cancer. Cancer research. 2020;80:4025-36
21. Carlisle C, Prill K, Pilgrim D. Chaperones and the Proteasome System: Regulating the Construction and Demolition of Striated Muscle. Int J Mol Sci. 2017;19:32
22. Kao TJ, Wu CC, Phan NN, Liu YH, Ta HDK, Anuraga G. et al. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging (Albany NY). 2021;13:17970
23. Chiao CC, Liu YH, Phan NN, An Ton NT, Ta HDK, Anuraga G. et al. Prognostic and Genomic Analysis of Proteasome 20S Subunit Alpha (PSMA) Family Members in Breast Cancer. Diagnostics (Basel). 2021 11
24. Xuan DTM, Wu CC, Kao TJ, Ta HDK, Anuraga G, Andriani V. et al. Prognostic and immune infiltration signatures of proteasome 26S subunit, non-ATPase (PSMD) family genes in breast cancer patients. Aging (Albany NY). 2021;13:24882-913
25. Wang CY, Li CY, Hsu HP, Cho CY, Yen MC, Weng TY. et al. PSMB5 plays a dual role in cancer development and immunosuppression. Am J Cancer Res. 2017;7:2103-20
26. Waterman M, Xu W, Stempak JM, Milgrom R, Bernstein CN, Griffiths AM. et al. Distinct and overlapping genetic loci in Crohn's disease and ulcerative colitis: correlations with pathogenesis. Inflammatory bowel diseases. 2011;17:1936-42
27. Latiano A, Palmieri O, Latiano T, Corritore G, Bossa F, Martino G. et al. Investigation of multiple susceptibility loci for inflammatory bowel disease in an Italian cohort of patients. PloS one. 2011;6:e22688
28. Li K, Liu T. Evaluation of Oncogene NUP37 as a Potential Novel Biomarker in Breast Cancer. Frontiers in oncology. 2021;11:669655
29. Man S, Li X, Zhu W. miR-4417 targets lncRNA PSMG3-AS1 to suppress cell invasion and migration in cervical squamous cell carcinoma. Oncol Lett. 2021;22:502
30. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166-80
31. Huang T-C, Lee P-T, Wu M-H, Huang C-C, Ko C-Y, Lee Y-C. et al. Distinct roles and differential expression levels of Wnt5a mRNA isoforms in colorectal cancer cells. PloS one. 2017;12:e0181034
32. Cheng L-C, Chao Y-J, Overman MJ, Wang C-Y, Phan NN, Chen Y-L. et al. Increased expression of secreted frizzled related protein 1 (SFRP1) predicts ampullary adenocarcinoma recurrence. Scientific reports. 2020;10:1-16
33. Tseng PC, Chen CL, Lee KY, Feng PH, Wang YC, Satria RD. et al. Epithelial-to-mesenchymal transition hinders interferon-γ-dependent immunosurveillance in lung cancer cells. Cancer Lett. 2022;539:215712
34. Kan J-Y, Yen M-C, Wang J-Y, Wu D-C, Chiu Y-J, Ho Y-W. et al. Nesfatin-1/Nucleobindin-2 enhances cell migration, invasion, and epithelial-mesenchymal transition via LKB1/AMPK/TORC1/ZEB1 pathways in colon cancer. Oncotarget. 2016;7:31336
35. Hsu H-P, Wang C-Y, Hsieh P-Y, Fang J-H, Chen Y-L. Knockdown of serine/threonine-protein kinase 24 promotes tumorigenesis and myeloid-derived suppressor cell expansion in an orthotopic immunocompetent gastric cancer animal model. Journal of Cancer. 2020;11:213
36. Weng TY, Huang SS, Yen MC, Lin CC, Chen YL, Lin CM. et al. A novel cancer therapeutic using thrombospondin 1 in dendritic cells. Mol Ther. 2014;22:292-302
37. Hung YH, Chan YS, Chang YS, Lee KT, Hsu HP, Yen MC. et al. Fatty acid metabolic enzyme acyl-CoA thioesterase 8 promotes the development of hepatocellular carcinoma. Oncol Rep. 2014;31:2797-803
38. Shahi P, Wang CY, Lawson DA, Slorach EM, Lu A, Yu Y. et al. ZNF503/Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression. Proc Natl Acad Sci U S A. 2017;114:3169-74
39. Weng TY, Wu HF, Li CY, Hung YH, Chang YW, Chen YL. et al. Homoharringtonine induced immune alteration for an Efficient Anti-tumor Response in Mouse Models of Non-small Cell Lung Adenocarcinoma Expressing Kras Mutation. Sci Rep. 2018;8:8216
40. Cho CY, Lee KT, Chen WC, Wang CY, Chang YS, Huang HL. et al. MST3 promotes proliferation and tumorigenicity through the VAV2/Rac1 signal axis in breast cancer. Oncotarget. 2016;7:14586-604
41. Huang HL, Chen WC, Hsu HP, Cho CY, Hung YH, Wang CY. et al. Argininosuccinate lyase is a potential therapeutic target in breast cancer. Oncol Rep. 2015;34:3131-9
42. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603-7
43. Lánczky A, Győrffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med Internet Res. 2021;23:e27633
44. Lin YY, Wang CY, Phan NN, Chiao CC, Li CY, Sun Z. et al. PODXL2 maintains cellular stemness and promotes breast cancer development through the Rac1/Akt pathway. Int J Med Sci. 2020;17:1639-51
45. Phan NN, Liu S, Wang CY, Hsu HP, Lai MD, Li CY. et al. Overexpressed gene signature of EPH receptor A/B family in cancer patients-comprehensive analyses from the public high-throughput database. Int J Clin Exp Pathol. 2020;13:1220-42
46. Wang C-Y, Chang Y-C, Kuo Y-L, Lee K-T, Chen P-S, Cheung CHA. et al. Mutation of the PTCH1 gene predicts recurrence of breast cancer. Scientific reports. 2019;9:1-14
47. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer research. 2017;77:e108-e10
48. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4
49. Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169
50. Chen L, Wang G, Xu Z, Lin K, Mu S, Pan Y. et al. Overexpression of LncRNA PSMG3-AS1 Distinguishes Glioblastomas from Sarcoidosis. J Mol Neurosci. 2020;70:2015-9
51. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511: 543-50.
52. Gillette MA, Satpathy S, Cao S, Dhanasekaran SM, Vasaikar SV, Krug K. et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell. 2020;182:200-25.e35
53. Jiang W, Cai G, Hu PC, Wang Y. Personalized medicine in non-small cell lung cancer: a review from a pharmacogenomics perspective. Acta Pharm Sin B. 2018;8:530-8
54. Pettini F, Visibelli A, Cicaloni V, Iovinelli D, Spiga O. Multi-Omics Model Applied to Cancer Genetics. Int J Mol Sci. 2021;22:5751
55. Zhao Y, Gao Y, Xu X, Zhou J, Wang H. Multi-omics analysis of genomics, epigenomics and transcriptomics for molecular subtypes and core genes for lung adenocarcinoma. BMC Cancer. 2021;21:257
56. Vidal-Taboada JM, Lu A, Pique M, Pons G, Gil J, Oliva R. Down syndrome critical region gene 2: expression during mouse development and in human cell lines indicates a function related to cell proliferation. Biochemical and biophysical research communications. 2000;272:156-63
57. Hirano Y, Hendil KB, Yashiroda H, Iemura S-i, Nagane R, Hioki Y. et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381-5
58. Wagner J, Catto-Smith AG, Cameron DJ, Kirkwood CD. Pseudomonas infection in children with early-onset Crohn's disease: an association with a mutation close to PSMG1. Inflammatory bowel diseases. 2013;19:E58-9
59. Wagner J, Sim WH, Ellis JA, Ong EK, Catto-Smith AG, Cameron DJS. et al. Interaction of Crohn's disease susceptibility genes in an Australian paediatric cohort. PloS one. 2010;5:e15376-e
60. Samadder NJ, Valentine JF, Guthery S, Singh H, Bernstein CN, Leighton JA. et al. Family History Associates With Increased Risk of Colorectal Cancer in Patients With Inflammatory Bowel Diseases. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2019;17:1807-13.e1
61. Taylor CC, Millien VO, Hou JK, Massarweh NN. Association Between Inflammatory Bowel Disease and Colorectal Cancer Stage of Disease and Survival. The Journal of surgical research. 2020;247:77-85
62. Lee D, Tang W, Dorsey TH, Ambs S. miR-484 is associated with disease recurrence and promotes migration in prostate cancer. Biosci Rep. 2020 40
63. Ebstein F, Poli Harlowe MC, Studencka-Turski M, Krüger E. Contribution of the Unfolded Protein Response (UPR) to the Pathogenesis of Proteasome-Associated Autoinflammatory Syndromes (PRAAS). Front Immunol. 2019;10:2756
64. Hirano Y, Kaneko T, Okamoto K, Bai M, Yashiroda H, Furuyama K. et al. Dissecting β-ring assembly pathway of the mammalian 20S proteasome. The EMBO journal. 2008;27:2204-13
65. Satoh T, Yagi-Utsumi M, Okamoto K, Kurimoto E, Tanaka K, Kato K. Molecular and Structural Basis of the Proteasome α Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer. Int J Mol Sci. 2019 20
66. Zhang J, Huang J, Chen W, Hu Z, Wang X. miR-143-3p Targets lncRNA PSMG3-AS1 to Inhibit the Proliferation of Hepatocellular Carcinoma Cells. Cancer Manag Res. 2020;12:6303-9
67. Cui Y, Fan Y, Zhao G, Zhang Q, Bao Y, Cui Y. et al. Novel lncRNA PSMG3-AS1 functions as a miR-143-3p sponge to increase the proliferation and migration of breast cancer cells. Oncol Rep. 2020;43:229-39
68. Man S, Li X, Zhu W. miR-4417 targets lncRNA PSMG3-AS1 to suppress cell invasion and migration in cervical squamous cell carcinoma. Oncol Lett. 2021;22:502
69. Yue N, Ye M, Zhang R, Guo Y. MiR-449b-5p targets lncRNA PSMG3-AS1 to suppress cancer cell proliferation in lung adenocarcinoma. BMC Pulmonary Medicine. 2020;20:152
70. Zhang YF, Zhou L, Zhang HW, Hou AJ, Gao HF, Zhou YH. Association between folate intake and the risk of lung cancer: a dose-response meta-analysis of prospective studies. PloS one. 2014;9:e93465
71. Stanisławska-Sachadyn A, Borzyszkowska J, Krzemiński M, Janowicz A, Dziadziuszko R, Jassem J. et al. Folate/homocysteine metabolism and lung cancer risk among smokers. PloS one. 2019;14:e0214462-e
72. Durda K, Kąklewski K, Gupta S, Szydłowski M, Baszuk P, Jaworska-Bieniek K. et al. Serum folate concentration and the incidence of lung cancer. PloS one. 2017;12:e0177441-e
73. Iksen Pothongsrisit S, Pongrakhananon V. Targeting the PI3K/AKT/mTOR Signaling Pathway in Lung Cancer: An Update Regarding Potential Drugs and Natural Products. Molecules. 2021
74. Hoppstädter J, Dembek A, Höring M, Schymik HS, Dahlem C, Sultan A. et al. Dysregulation of cholesterol homeostasis in human lung cancer tissue and tumour-associated macrophages. eBioMedicine. 2021;72:103578
75. Hanai J-i, Doro N, Sasaki AT, Kobayashi S, Cantley LC, Seth P. et al. Inhibition of lung cancer growth: ATP citrate lyase knockdown and statin treatment leads to dual blockade of mitogen-activated protein Kinase (MAPK) and Phosphatidylinositol-3-kinase (PI3K)/AKT pathways. Journal of Cellular Physiology. 2012;227:1709-20
76. Sanli T, Liu C, Rashid A, Hopmans SN, Tsiani E, Schultz C. et al. Lovastatin sensitizes lung cancer cells to ionizing radiation: modulation of molecular pathways of radioresistance and tumor suppression. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:439-50
77. Del Curatolo A, Conciatori F, Cesta Incani U, Bazzichetto C, Falcone I, Corbo V. et al. Therapeutic potential of combined BRAF/MEK blockade in BRAF-wild type preclinical tumor models. J Exp Clin Cancer Res. 2018;37:140 -
78. Lurje G, Lenz HJ. EGFR Signaling and Drug Discovery. Oncology. 2009;77:400-10
79. Sawai A, Chandarlapaty S, Greulich H, Gonen M, Ye Q, Arteaga CL. et al. Inhibition of Hsp90 down-regulates mutant epidermal growth factor receptor (EGFR) expression and sensitizes EGFR mutant tumors to paclitaxel. Cancer research. 2008;68:589-96
80. Mohsin SK, Weiss HL, Gutierrez MC, Chamness GC, Schiff R, DiGiovanna MP. et al. Neoadjuvant Trastuzumab Induces Apoptosis in Primary Breast Cancers. Journal of Clinical Oncology. 2005;23:2460-8
81. Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature Reviews Cancer. 2005;5:761-72
82. Fontana J, Fulton D, Chen Y, Fairchild TA, McCabe TJ, Fujita N. et al. Domain mapping studies reveal that the M domain of hsp90 serves as a molecular scaffold to regulate Akt-dependent phosphorylation of endothelial nitric oxide synthase and NO release. Circulation research. 2002;90:866-73
83. Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends in Biochemical Sciences. 2006;31:164-72
84. Eustace BK, Sakurai T, Stewart JK, Yimlamai D, Unger C, Zehetmeier C. et al. Functional proteomic screens reveal an essential extracellular role for hsp90α in cancer cell invasiveness. Nature Cell Biology. 2004;6:507-14
85. Graner MW. Chapter Eight - HSP90 and Immune Modulation in Cancer. In: Isaacs J, Whitesell L, editors. Advances in Cancer Research: Academic Press. 2016 p. 191-224
86. Zavareh RB, Spangenberg SH, Woods A, Martínez-Peña F, Lairson LL. HSP90 Inhibition Enhances Cancer Immunotherapy by Modulating the Surface Expression of Multiple Immune Checkpoint Proteins. Cell chemical biology. 2021;28:158-68.e5
87. Song K-H, Oh SJ, Kim S, Cho H, Lee H-J, Song JS. et al. HSP90A inhibition promotes anti-tumor immunity by reversing multi-modal resistance and stem-like property of immune-refractory tumors. Nature Communications. 2020;11:562
Author contact
![]() Corresponding authors: Chih-Yang Wang (E-mail: chihyangedu.tw) and Meng-Chi Yen (E-mail: yohococom).
Corresponding authors: Chih-Yang Wang (E-mail: chihyangedu.tw) and Meng-Chi Yen (E-mail: yohococom).

 Global reach, higher impact
Global reach, higher impact