Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(1):35-49. doi:10.7150/ijms.75625 This issue Cite
Research Paper
Glutamine synthetase regulates the immune microenvironment and cancer development through the inflammatory pathway
1. Graduate Institute of Cancer Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei 11031, Taiwan.
2. Department of Neurosurgery, Taipei Medical University Hospital, Taipei 11031, Taiwan.
3. Department of Surgery, School of Medicine, College of Medicine, Taipei Medical University, Taipei 11031, Taiwan.
4. Research Center for Cancer Biology, China Medical University, Taichung 40676, Taiwan.
5. Department of Surgery, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan 70101, Taiwan.
6. Ph.D. Program for Cancer Molecular Biology and Drug Discovery, College of Medical Science, Taipei Medical University, Taipei 11031, Taiwan.
7. Department of Statistics, Faculty of Science and Technology, PGRI Adi Buana University, East Java, Surabaya 60234, Indonesia.
8. TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei 11031, Taiwan.
*Equal contributors to this work.
Received 2022-5-30; Accepted 2022-11-3; Published 2023-1-1
Abstract
Although adjuvant tamoxifen therapy is beneficial to estrogen receptor-positive (ER+) breast cancer patients, a significant number of patients still develop metastasis or undergo recurrence. Therefore, identifying novel diagnostic and prognostic biomarkers for these patients is urgently needed. Predictive markers and therapeutic strategies for tamoxifen-resistant ER+ breast cancer are not clear, and micro (mi)RNAs have recently become a focal research point in cancer studies owing to their regulation of gene expressions, metabolism, and many other physiological processes. Therefore, systematic investigation is required to understand the modulation of gene expression in tamoxifen-resistant patients. High-throughput technology uses a holistic approach to observe differences among expression profiles of thousands of genes, which provides a comprehensive level to extensively investigate functional genomics and biological processes. Through a bioinformatics analysis, we revealed that glutamine synthetase/glutamate-ammonia ligase (GLUL) might play essential roles in the recurrence of tamoxifen-resistant ER+ patients. GLUL increases intracellular glutamine usage via glutaminolysis, and further active metabolism-related downstream molecules in cancer cell. However, how GLUL regulates the tumor microenvironment for tamoxifen-resistant ER+ breast cancer remains unexplored. Analysis of MetaCore pathway database demonstrated that GLUL is involved in the cell cycle, immune response, interleukin (IL)-4-induced regulators of cell growth, differentiation, and metabolism-related pathways. Experimental data also confirmed that the knockdown of GLUL in breast cancer cell lines decreased cell proliferation and influenced expressions of specific downstream molecules. Through a Connectivity Map (CMap) analysis, we revealed that certain drugs/molecules, including omeprazole, methacholine chloride, ioversol, fulvestrant, difenidol, cycloserine, and MK-801, may serve as potential treatments for tamoxifen-resistant breast cancer patients. These drugs may be tested in combination with current therapies in tamoxifen-resistant breast cancer patients. Collectively, our study demonstrated the crucial roles of GLUL, which provide new targets for the treatment of tamoxifen-resistant breast cancer patients.
Keywords: breast cancer, tamoxifen, glutamine synthetase (GLUL), immune microenvironment, bioinformatics
Introduction
According to the latest mortality data provided by the U.S. National Center for Health Statistics, the incidence and mortality of cancer increases with an estimated 1,9 million newly diagnosed cases and 600,000 deaths nationwide in 2022 [1]. To date, breast cancer remains one of most frequently diagnosed malignancies in the world [2]. Several studies documented heterogeneity in breast cancer in terms of great diversity in both the clinical characteristics of patients and the behaviors of tumors. As a result, clinically applicable classifications of breast cancer, which are primarily based on molecular categories and histological patterns, are still undergoing upgrades [3, 4]. Based on the expression of human epidermal growth factor receptor-2 (HER-2), and the two hormone receptors - estrogen receptor (ER) and progesterone receptor (PR), breast cancer is subcategorized into five subtypes. The estrogen-positive cancer comprises at most 80% of total patients [5, 6]. For non-metastatic estrogen-positive (ER+) breast cancer patients, tamoxifen serves as an adjuvant therapy to reduce the risk of recurrence [7, 8]. Previous clinical trials demonstrated the effect of tamoxifen therapy for patients after radical surgery, regardless of menopausal condition, the extent of metastasis, and nodal status [9, 10]. However, a large number of patients develop intrinsic resistance against tamoxifen shortly after anti-estrogen therapy [11]. To date, a significant number of alterations in molecular profiling leading to drug resistance are identified, highlighting the complexity of estrogen receptor signaling and as well as its involvement in crosstalk with other signaling pathways within breast tumors [12, 13]. There have been several reports on the mechanism of tamoxifen resistance, but they were incomplete. Among them, patients with specific cytochrome polymorphisms were reported to present a higher ratio of tamoxifen resistance and an increased probability of recurrence, which lead to the incurability of advanced ER+ breast cancer [14, 15]. On the other hand, excessive availability of glutamine not only induces the mechanistic target of rapamycin complex 1 (mTORC1) but also triggers tamoxifen-resistance when cooperating with stromal fibroblasts [16-18]. In response to this situation, existing salvage therapies for tamoxifen-resistant (TR) patients have been well-designed, including fulvestrant, cyclin-dependent kinase 4/6 (CDK4/6) inhibitors, and histone deacetylase (HDAC) inhibitors [19-21]. However, not all patients respond properly to these therapies [22-25].
Glutamate-ammonia ligase (GLUL), also named glutamine synthetase (GLNS), is the enzyme responsible for the de novo biosynthesis of glutamine from glutamate and ammonia in an ATP-dependent reaction [26]. As a conditionally crucial amino acid and crucial nutrient required in culture medium, glutamine not only serves as a major source of nitrogen for the synthesis of proteins, nucleic acids, and macromolecules but also supports the redox status to maintain amino acid homeostasis in mammalian cells in vitro [27, 28]. For those reasons, glutamine remains one of the major concerns over the last decades [29, 30]. More shreds of evidence further reveal that glutamine impacts numerous signaling pathways that contribute to tumor proliferation, activating the mammalian target of rapamycin (mTOR) kinase, and autophagy [31-34]. These findings explain the regulation of glutamine metabolism in tumors. However, explanations as to how GLUL regulates the immune microenvironment and cancer development through the inflammatory pathways are largely lacking.
Micro (mi)RNAs, a class of small single-stranded RNAs, belong to the noncoding (nc)RNA family which functions in inhibiting protein translation or degrading transcripts by binding to the 3'-untranslated region (UTR) of messenger (m)RNAs. In particular, recent studies report that miRNAs contributed to the progression of breast cancer after tamoxifen treatment. The mechanisms include the activation of estrogen receptor alpha (ERα), progression of the cell cycle, regulation of apoptosis, and controlling epithelial-to-mesenchymal transition [68-71]. For these reasons, studies of differentially expressed miRNAs (DEMs) are similar to the research of differentially expressed genes (DEGs) to improve the susceptibility of tamoxifen against breast cancer and provide novel insights for clinical doctors.
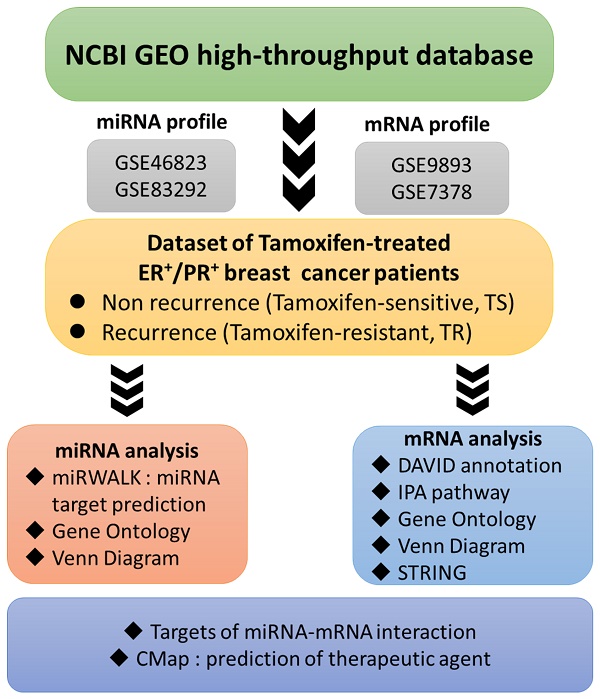
Therefore, our study aimed to provide a comprehensive understanding of the mechanisms involved in tamoxifen-resistant ER+ breast cancer using both high-throughput technology and wet-lab approaches. Microarray-based mRNA and miRNA expression profiles of tamoxifen-resistant (TR) and tamoxifen-sensitive (TS) ER+ breast cancer patients were extracted from relevant datasets available at the Gene Expression Omnibus (GEO), followed by DEMs analyses and DEGs analyses, prediction of downstream target genes, and subsequently gene regulatory network to determine potential biomarkers associated with tamoxifen resistance. Finally, wet lab experiments were performed to validate the preceding analytical results.
Material and Methods
Cell Culture and Gene Knockdown
The MCF-7 human epithelial breast cancer cell line, characterized by the presence of the ER, PR, and glucocorticoid receptors, was a kind gift from Prof. Chun Hei Antonio Cheung of National Cheng Kung University (NCKU, Taiwan). MCF-7 cells were maintained in RPMI-1640 complete medium (Corning, Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS, Avantor, USA) plus 1% penicillin/streptomycin (Corning, Corning, NY, USA), and were kept inside a humidified incubator under typical condition (37°C and 5% CO2). GLUL gene silencing was generated using a small hairpin (sh)RNA knockdown vector system. All of the shRNA vectors harboring puromycin and enhanced green fluorescent proteins, including two respective recombinant encoding human GLUL shRNA, and one non-target control (pLKO.1) shRNA against luciferase (shLuc), were constructed by the National RNAi Core Facility (Academia Sinica, Taiwan; http://rnai.genmed.sinica.edu.tw). The MCF-7 cells were seeded into six-well plates 24h prior to being incubated with three respective lipofectamine and shRNA vectors for 24h, in RPMI-1640 medium containing polybrene (8µg/mL). During an additional 48 hours of lipofectamine transfection, cells were subsequently maintained in a regular medium containing 10% FBS, and stable clones expressing shRNA were selected by constant treatment with puromycin (2µg/mL) from 72 hours onward. A non-target control (pLKO.1) shRNA against luciferase (shLuc) was employed as an expression control. At day 28 post-transfection with lipofectamine, the efficacy of gene silencing was further confirmed by a quantitative reverse-transcription polymerase chain reaction (RT-qPCR) and Western blotting in two populations of shGLUL-transfected MCF-7 cells relative to vector control group.
RT-qPCR, Western Blotting and Colony-Formation Assay
Total RNA extracted from stable clones of shGLUL-transfected MCF-7 cells was isolated and purified using a GENEzol™ TriRNA Pure Kit (Geneaid Biotech, Taiwan) prior to being subjected to complementary synthesis using a PrimeScript Synthesis Kit (Takara Bio, Japan), with triplicate determinations. The RT-qPCR was performed using GoalBio SYBR green master mix (Hycell International, Taiwan) on a Roche Light Cycler 96 platform. Primer pair sequences targeting GLUL and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were provided by Origene (Rockville, USA) and were constructed by MDBio, Inc. The relative fold changes in expression of the GLUL gene were calculated by the delta-delta Ct (2-∆∆Ct) method after being normalized against the mRNA level of GAPDH as the housekeeping gene.
Total protein extractions derived from cell lysates of stable clones of shGLUL-transfected MCF-7 and shGLUL-transfected MCF-7 cells were subjected to Western blotting following the manufacturer's protocol (Bio-Rad Laboratories, Hercules, CA, USA). For subsequent analyses, an anti-GLUL (GTX109121) rabbit polyclonal antibody (polyAb) (GeneTex, Hsinchu, Taiwan) and anti-GAPDH (GTX124502) rabbit polyAb (GeneTex) were employed as primary antibodies.
For the colony-formation assay, MCF-7 cells of the three experimental groups were respectively seeded at the low density (1000 cells/well) into six-well plates for 2~3 weeks until macroscopic colonies had formed. The medium was discarded, and cells were fixed with absolute methanol fixation (20 min at room temperature), followed by an incubation with 2% methylene blue staining. The number of proper colonies formed was counted by a stereomicroscope (×100), and experiments were performed in triplicate data.
Bioinformatics and Dataset Analyses
In this study, datasets containing molecular profiles of either ER+ and/or PR+ breast cancer patients treated with tamoxifen, regardless of their clinical outcomes (with or without recurrence), were collected from the GEO database. Differential mRNA and miRNA expression profiles of primary and relapsed ER+ breast tumors following tamoxifen treatment were extracted from two groups of datasets, GSE9893 combined with GSE7378, and GSE46823 combined with GSE83292 [35-38]. All raw data were extracted from the NCBI GEO database and subjected to the CLC Genomics Workbench v10.1 for subsequent analyses as previously described [39, 40]. In order to explore alterations in expression patterns of miRNAs and mRNAs between the TR and TS populations, the top 10% of highly DEGs in TR patients were calculated as previously described [41, 42], accompanied by false detection rate (FDR)-adjusted p-value less than 0.05. A list of significant DEGs was imported into the Ingenuity Pathway Analysis (IPA) and MetaCore platform to construct related biological networks, biological processes (BPs), and diseases, with significant cutoff points for the enrichment of a pathway or an annotated gene group set to p<0.05 as mentioned above [43]. TargetScan and miRWalk 2.0 are integrated database capable of providing more than 150 million human microRNA-target predictions. In the GSE83292 and GSE46823 datasets, the most miRNAs were downregulated or upregulated among TR ER+ breast cancer patients compared to TS ones. Finally, four down-regulated miRNAs and five common up-regulated between these two datasets, and then subjected to evaluation of the repressive strength of miRNA binding to its target mRNA, allowing producing target gene prediction. A miRmap score of > 95 was applied as the selection threshold for this analysis [44-46].
Protein Network and Gene Set Enrichment Analysis (GSEA)
The Search Tool for the Retrieval of Interacting proteins (STRING) is a huge database consolidating more than twenty billion interactions among approximately sixty-seven million proteins of nearly fourteen thousand organisms [47]. The STRING database (version 11.0) was leveraged to build up the protein-protein interacting (PPI) networks, including direct (physical) and indirect (functional) interactions based on the DEGs obtained from the previous analyses. The K-means clustering algorithm was applied to categorize proteins into different clusters. The GSEA was subsequently employed using the Bioconductor “DESeq2” and “fgsea” packages in R Studio software to identify upregulated gene sets between TR and TS breast cancer patients to determine groups of genes associated with the disease. A normalized enrichment score (NES) was first calculated, followed by the FDR with the purpose of controlling the false positive proportion. In particular, an FDR of <0.25 was set as the boundary criterion, while a nominal p-value of <0.05 and an NES of >1.5 were set as thresholds [48].
Tumor Immune Infiltration Analysis
Tumor Immune Estimation Resource (TIMER) provides comprehensive and systematic analyses of tumor-infiltrating immune cells across various cancer types, specifically six immune cell populations across 31 tumors from The Cancer Genome Atlas (TCGA). In other words, this web server helps estimate differences in infiltration levels of lymphoid lineages (including B cells, cluster of differentiation-positive (CD4+) T cells, and CD8+ T cells) along with myeloid lineages (including neutrophils, macrophages, and dendritic cells (DCs)) in the tumor microenvironment (TME) compared to adjacent normal tissues, using the DiffExp module with default parameters. Expression scatterplots between a pair of given genes in specific cancer types were constructed according to the Spearman correlation and estimated statistical significance, adjusted for tumor purity if necessary [49]. The seaborn package (Python) was used to create a heatmap as previously discussed [50, 51].
Human Protein Atlas and Connectivity Map (CMap) Analysis
Human Protein Atlas provides high-resolution images exhibiting spatial distributions of proteins in normal human tissues and various cancer types [52]. Expression profiles in breast tissues of GLUL at the protein level were examined by immunohistochemical (IHC) staining of fixed malignant tissues versus normal adjacent tissues that were labeled with antibodies against GLUL. The CMap database comprises interaction profiles of nearly 7000 human cell lines which were treated with 1300 US Food and Drug Administration (FDA)-approved compounds [53]. The final gene list of interest was subjected to a computational pipeline generated by the CMap platform to evaluate the cellular effects of given compounds. Correlations between specific compounds and breast cancer were ranked by standardized connection scores, perturbation stability, and p-values.
Statistical Analysis
Comparisons among groups of interest were performed by GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA), using a one-way analysis of variance (ANOVA) and Tukey's multiple-comparison test. A p-value of <0.05 was considered statistically significant.
Results
Differentially Expressed genes and altered pathway in TR and TS Patients
We identified the differential expressions of mRNAs and miRNAs between TR and TS patients using miRtest, and evaluated individual miRNAs and their predicted target genes in the same analysis for verifying consistency. The up and down-regulated mRNAs and miRNAs were analyzed using numerous bioinformatics tools. The IPA was used to find upstream and downstream networks of miRNAs and mRNAs, STRING to build up PPI networks, and DAVID to investigate miRNA- and mRNA- associated functions and pathways. The miRWalk2.0, miRmap, and TargetScan were used to predict targets of the upregulated and downregulated miRNAs and to screen potential miRNA-mRNA interactions (Figure 1).
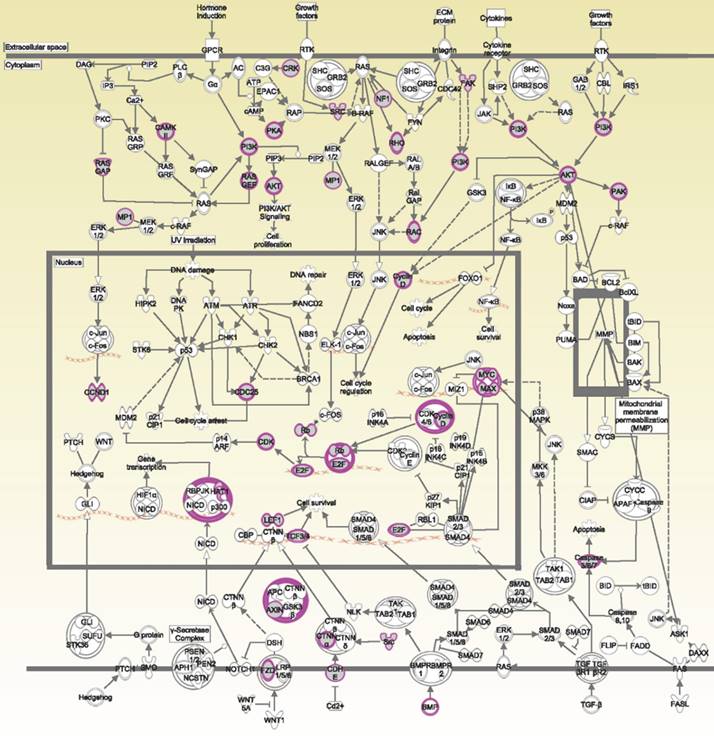
The potential genes for miRNA-mRNA interactions in TR patients were identified in the previous section, and the potential pathways and networks were explored in this section. The potential genes targeted by miRNA in GSE46823 and GSE83292 datasets were uploaded to the IPA. The canonical pathway was shown in Figure 2A and Supplementary Table S1. The “Molecular Mechanisms of Cancer” pathway was ranked as the top no.6 and included 14 target genes. Furthermore, the overlapping regulated genes in the GSE9893 and GSE7378 datasets were imported to the IPA and canonical pathways are in Figure 2B and Supplementary Table S2. Significantly enriched pathways included oncogenic pathways and regulation of the cell cycle. The “Molecular Mechanisms of Cancer” pathway was ranked in the top #11 and contained 10 upregulated genes. The detail of the “Molecular Mechanisms of Cancer” pathway was displayed in Figure 3. These oncogenic pathways play an essential role in genetic instability, carcinogenesis, and progression of ER+ TR breast cancer. Multiple oncogenes and tumor suppressor genes are contained in this canonical pathway, including G-protein-coupled receptors (GPCR) signaling, RAS/integrin signaling, AKT signaling, transforming growth factor (TGF)-β/bone morphogenetic protein (BMP) signaling, WNT signaling, Notch and Hedgehog (Hh) signaling, and death receptor signaling (the membranous receptor and intracellular part of Figure 3). These pathways were also associated with the expression of recombination signal binding protein for immunoglobulin kappa J region (RBPJ-κ), p300, histone acetyltransferase type B catalytic subunit (HAT1), and hypoxia-inducible factor 1α (HIF1α). The resulting three clusters with a core cluster containing all genes were related to cancer progression and metastasis.
Flow-chart illustrating the study design. Raw data extracted from the NCBI GEO comprising micro (mi)RNA and messenger (m)RNA profiles of ER+ breast cancer patients were categorized into two groups according to their response to tamoxifen. Abbreviations: CMap, connectivity map; DAVID, database for annotation, visualization, and integrated discovery; ER, estrogen receptor; FDA, Food and Drug Administration; IPA, Ingenuity Pathway Analysis; TR, tamoxifen-resistant; TS, tamoxifen-sensitive.
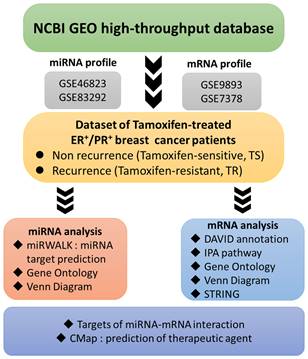
Ingenuity Pathway Analysis (IPA): top key pathways associated with differentially expressed genes in estrogen receptor-positive (ER+) tamoxifen-resistant (TR) breast cancer patients, arranged in descending order of -log(p-value). (A) IPA results of potential miRNA-targeted predicted genes in TR patients compared to TS patients. Data were collected from the GSE46823 and the GSE83292 datasets. (B) IPA results of the potential differentially expressed genes in TR patients compared to TS patients. Data were collected from the GSE9893 and the GSE7378 datasets. Abbreviations: miRNA, micro ribonucleic acid; ER+: estrogen receptor-positive. TR, tamoxifen-resistant; TS, tamoxifen-sensitive.
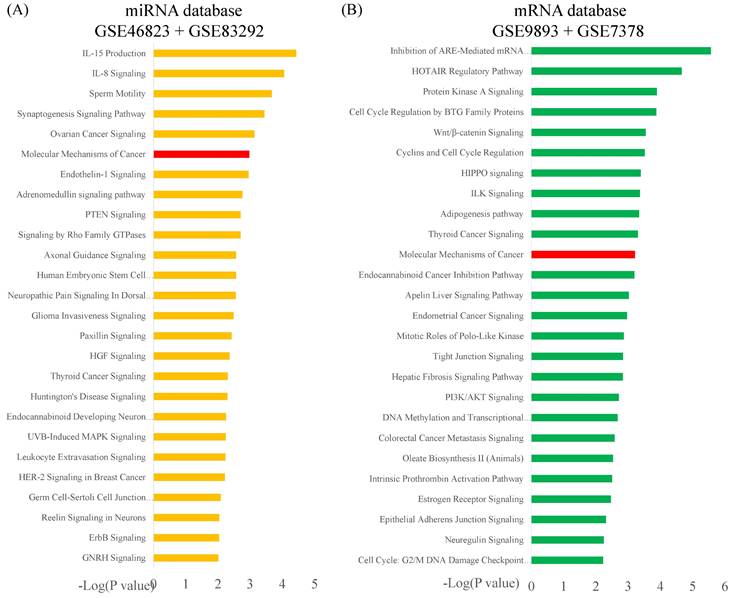
Ingenuity Pathway Analysis (IPA): molecular mechanisms of cancer associated with differentially expressed genes in estrogen receptor-positive (ER+) tamoxifen-resistant (TR) breast cancer patients. Purple boxes represent proteins involved in the common network. Abbreviations: ER+: estrogen receptor-positive. TR, tamoxifen-resistant; TS, tamoxifen-sensitive.
Gene Set Enrichment and Pathway Analysis of Tamoxifen-Treated Breast Cancer Patients
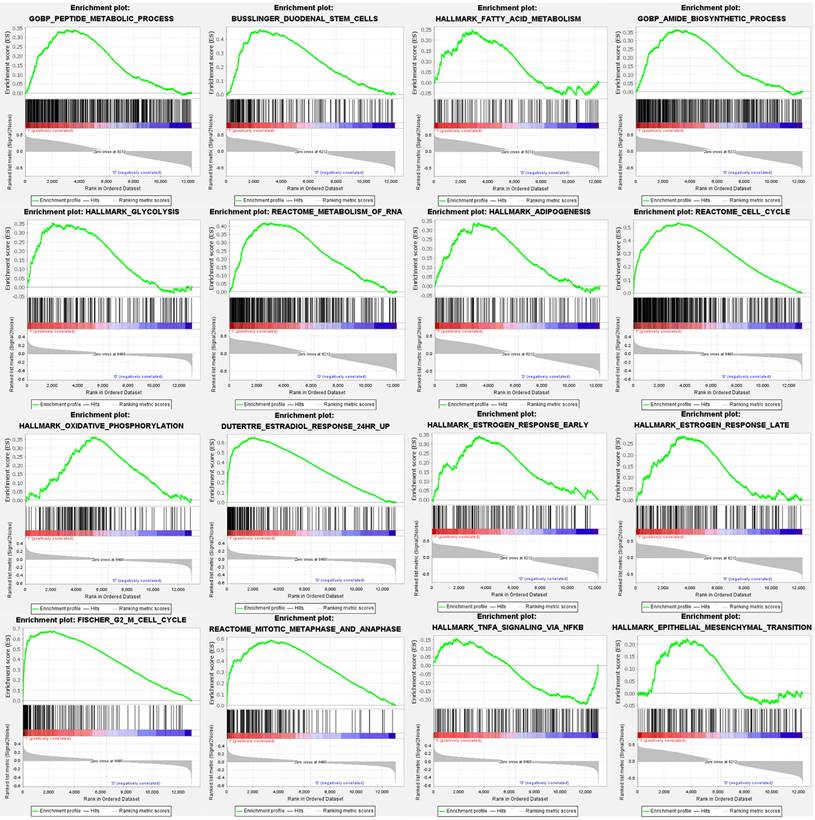
For comprehensive exploration, we utilized a public database, the Gene Set Enrichment Analysis (GSEA), to verify the importance of targeted pathways in tamoxifen-treated primary breast cancer patients compared to controls. Upregulated genes were analyzed to retrieve the Gene Ontology enrichment results and Kyoto Encyclopedia of Genes and Genomes (KEGG), including cellular components (CCs), biological process (BPs), and molecular functions (MFs). The potentially regulated networks identified included peptide metabolic process, reactome cell cycle, fatty acid metabolism, adipogenesis, stem cells, epithelial-mesenchymal transition, estradiol response, oxidative phosphorylation, estrogen response late, G2 M cell cycle, glycolysis, IL-4 signaling, TNFA signaling via nuclear factor (NF)-κB, mitotic metaphase, and anaphase (Figure 4).
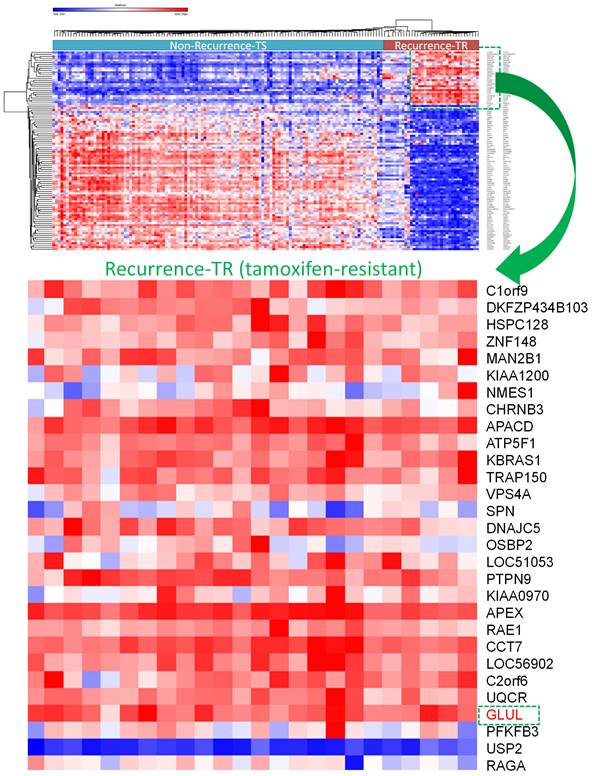
A selection of the most highly and least expressed miRNAs in TR patients compared to TS patients were extracted. There were 227 down-regulated miRNAs in the GSE46823 dataset and 157 miRNAs in the GSE83292 dataset. The intersection of these two groups obtained four down-regulated miRNAs: hsa-miR-1323, hsa-miR-711, hsa-miR-4287, hsa-miR-650. Whereas, there were 221 upregulated miRNAs in the GSE46823 dataset and 154 miRNAs in the GSE83292 dataset. The intersection of these two groups obtained five upregulated miRNAs: hsa-miR-532-5p, hsa-miR-1180, hsa-miR-152, hsa-miR-578, and hsa-miR-128. The TargetScan software was used for miRNA-targeting genes for each miRNA and these results were merged [54]. Next, two mRNA datasets (GSE9893 and GSE7378) with ER+ breast cancer patients undergoing adjuvant tamoxifen therapy were selected. The top 10% of upregulated genes in TR compared to TS patients were selected. There were 1570 upregulated genes in GSE9893 and 1842 genes in GSE7378. The intersection of these genes obtained 167 common genes, which were extracted. A Venn diagram was employed to explore the genes shared by the aforementioned miRNA-targeting genes predicted by TargetScan and 167 upregulated genes obtained from previous DEGs analysis. Final results including significantly up-regulated genes in TR ER+ breast cancer patients compared to TS ER+ ones were presented in a heatmap (Figure 5). Among them, GLUL (glutamate-ammonia ligase) was selected for further analyses (Figure 5).
Gene Set Enrichment Analysis (GSEA) enrichment curves of highly expressed gene sets in estrogen receptor-positive (ER+) tamoxifen treated breast cancer patients. Positive enrichment scores (ER) reflect gene sets enriched at the top of the ranked list, with each bar indicating one specific gene located in the ranking. In each plot, bars on the far left in red represent positive correlations with the most highly upregulated genes, while bars on the far right in blue represent positive correlations with the most highly downregulated genes. The running sum of the weighted enrichment score in each gene set is denoted by the respective green curve. Statistical significance was set as follows: false discovery rate (FDR) <0.25, normalized enrichment score (NES) >1.5 and nominal p value <0.05.
Overview of differentially expressed genes in tamoxifen-sensitive (TS) and tamoxifen-resistant (TR) estrogen receptor-positive (ER+) breast cancer patients visualized in a heatmap format. GLUL (glutamate-ammonia ligase) was one of the significantly upregulated genes in TR ER+ compared to TS ER+ breast cancer patients.
GLUL Play an Important Role in Breast Cancer Development and Involved in Immune Regulation
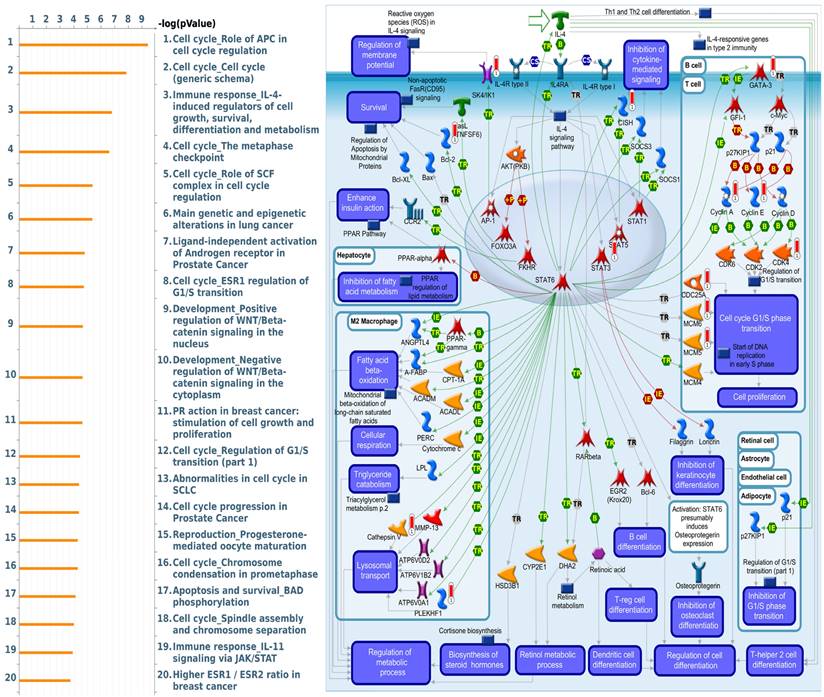
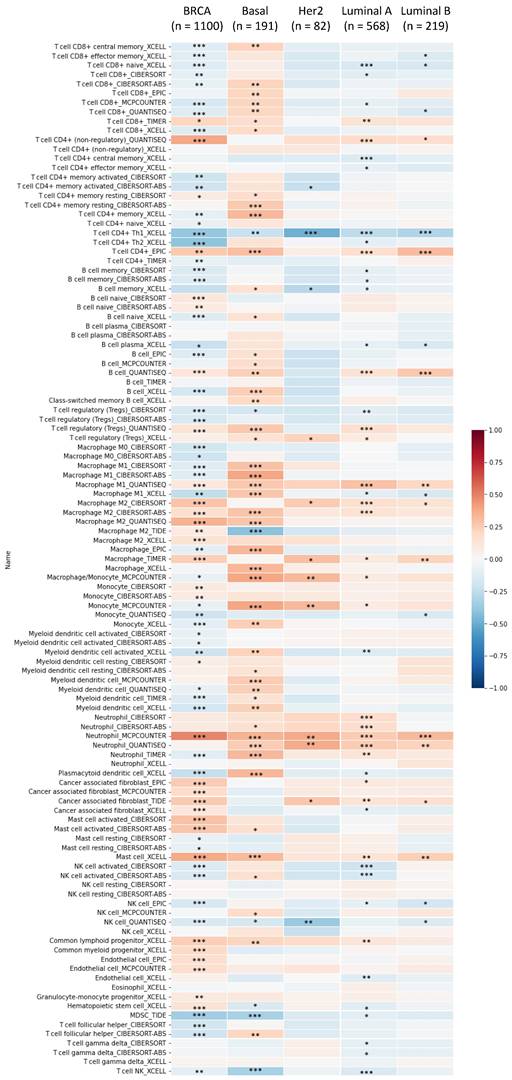
MetaCore is commonly employed to build up pathway networks from the input genes to simulate biological processes. We found interesting results related to GLUL, such as “Cell cycle_Role of APC in cell cycle regulation”, Immune response_IL-4-induced regulators of cell growth, survival, differentiation and metabolism”, “Cell cycle_role of SCF complex in cell cycle regulation”, “Cell cycle_ESR1 regulation of G1/S transition”, and “Apoptosis and survival_BAD phosphorylation”. These are immune- or cell-cycle-related pathways in the progression of breast cancer. The pathway lists and networks were respectively shown in Figure 6 and Supplementary Table S3. We found that the immune-related pathways were correlated with GLUL expression in breast cancer patients from public breast cancer patient datasets. IL-4 stimulates cell population proliferation through the activation of several cyclin-dependent kinases (CDKs), which promote the cell cycle G1/S phase transition. The increasing amount of information regarding the IL-4 pathway proved the importance of IL-4 in regulating metabolic processes. Meanwhile, an evolving tumor microenvironment is a convoluted and continuously growing entity. The configuration of the tumor microenvironment varies among tumor types. Several research reveal the important role of tumor microenvironment in cancer progression. We further evaluated the correlation coefficient between GLUL expression in breast cancer cells and activation of tumor-infiltrating immune cells, including B cells, M1 macrophages, M2 tumor-associated macrophages (M2 TAMs), neutrophils, and dendritic cells. A higher level of correlation coefficient was shown in red color, and these data suggested that GLUL expression was positively correlated with the function of immune cells (Figure 7). According to our knowledge, these data are the first to reveal the relationship between GLUL expression and subtypes of tumor-infiltrating immune cells.
Altered signaling pathways regulated by the top genes co-expressed with GLUL (glutamate-ammonia ligase) predicted by MetaCore. The top 10% of genes co-expressed with GLUL from the METABRIC and TCGA databases were subjected to a Venn diagram to get a list of 943 genes in common, which subsequently underwent “biological processes” provided by MetaCore for downstream pathway analyses. The involved pathways were ranked in order of decreasing -log[p values]. “Immune response_IL-4-induced regulators of cell growth, survival, differentiation, and metabolism” was noteworthy for its third place on the list.
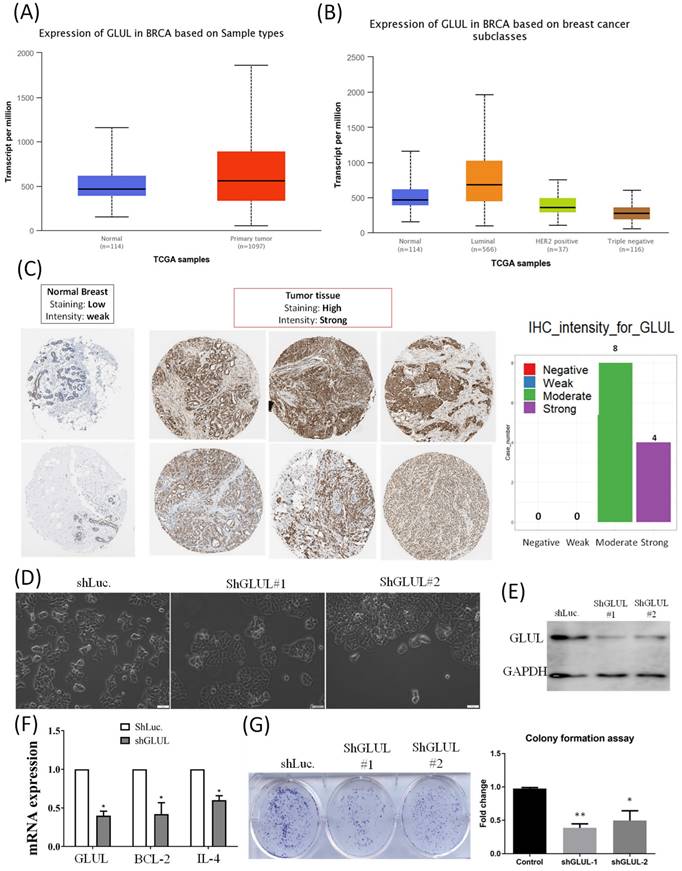
We analyzed transcripts of GLUL in the TCGA database. Expression of GLUL was higher in breast cancer samples compared to normal breast, especially in the ER+ breast cancer patients (Figure 8A, B). Expression patterns of GLUL protein in human breast cancer tissues were further confirmed by immunohistochemistry (IHC) staining provided by the Human Protein Atlas database. Staining levels of GLUL were increased in cancer specimens compared to normal breast (Figure 8C). We also investigated GLUL expression in various other types of cancer via the TCGA database (Supplementary Figure S1) and Human Protein Atlas (Supplementary Figure S2) databases. Results of bioinformatics analyses also revealed the molecular subtype specific for each cell line (Supplementary Figure S3), as well as DNA methylation levels of GLUL in breast cancer (Supplementary Figure S4). We confirmed that GLUL is important for the progression of ER+ breast cancer. Therefore, we chose the ER+ cell line- MCF-7 for further study. Lipofectamine transfection of GLUL-shRNA was performed to inhibit GLUL expression in MCF-7 cells. Intriguingly, GLUL-knockdown MCF-7 cells became more cuboidal shape in comparison with shLuciferase control cells, which is the typical morphology of epithelial-like luminal cancer (Figure 8D). The suppressive efficacy was confirmed by Western blotting (Figure 8E). Meanwhile, the expression levels of cell cycle- and immune-related markers decreased after downregulating GLUL expression, such as BCL-2 and IL-4 (Figure 8F). Specifically, IL-4 is a pleiotropic cytokine in both immune and non-immune cells regulating cell differentiation, survival, and proliferation (Supplementary Figure S5). Besides, the ability of anchorage-independent growth was suppressed in GLUL-knockdown MCF7- cells (Figure 8G). These results indicate that GLUL played a significant role in the inhibition of cellular proliferation and growth in breast epithelial cell.
Identification of Potential Inhibitory Compounds from CMap
Up- and downregulated genes determined from comparisons between TR and TS patients through a Venn diagram in Figure 2 were uploaded to a CMap database to predict potential drugs for TR patients (Figure S6A). We selected the results of a drug sensitivity test from the MCF-7 ER+ cell line in the CMap database and compared these with the gene signatures of TR patients. The top 25 compounds with negative correlations were ranked by p values. Fulvestrant ranked number 4 (from -1 to 1), and it is currently a recommended drug for metastatic ER+ breast cancer patients. Methotrexate ranked number 25 (from -1 to 1) and is currently one of the cytotoxic agents for adjuvant chemotherapy. Other compounds were potential drugs for ER+ TR patients (Figure S6B). The structures of the top six compounds were acquired from PubChem as previously described (Figure S6C). Although these findings require further investigation, these compounds have shown certain potential for TR breast cancer patients.
Discussion
The hormone receptor-positive subtype comprises the majority of cases of breast cancer, with more than four-fifths of diagnosed patients exhibiting either ER+ or PR+ statuses or both [55]. Being one of the oldest selective estrogen receptor modulators, tamoxifen has been serving as the first line of adjuvant endocrine therapy for primary and metastatic ER+ breast cancer patients [56]. However, resistance to tamoxifen therapy is possible and other therapeutic agents are required [57].
In our study, expression levels of four down-regulated miRNAs: hsa-miR-1323, hsa-miR-711, hsa-miR-4287, hsa-miR-650, as well as the five miRNAs, including hsa-miR-532-5p, hsa-miR-1180, hsa-miR-152, hsa-miR-578, hsa-miR-128, were upregulated in combined analyses of GSE46823 and GSE83292 datasets. The prediction software of miRNA-targeting genes showed potential genes in interest. In previous literature, hsa-miR-1323 widely regulate cancer progression and radiotherapy effects [58], miR-711 regulates NCI-N87 and SNU-1 cells gastric cancer lines progression by targeting CD44 [59], miR-4287 is a critical molecule in osteoarthritis development [60], miR-650 inhibits the progression of glioma by targeting FAM83F [61]. The miR-532-5p is reported to promote cancer proliferation and migration by targeting the RERG gene (RAS like estrogen-regulated growth inhibitor) and the EGFR (epidermal growth factor receptor) gene [62]. Similarly, miR-1180 participates in cell proliferation, migration and invasion by targeting the DGCR5 (DiGeorge syndrome critical region gene 5) gene, along with resistance to apoptosis by activating the nuclear factor-κB (NF-κB) signaling pathway [63, 64]. On the other hand, miR-152 is involved in regulating phosphatidylinositol 3-kinase (PI3K)/AKT and extracellular regulated protein kinases (ERK)/NF-κB signaling pathways, while miR-578 is potentially involved in angiogenesis of BRCA-related breast cancer [65, 66]. The miR-128 serves as an onco-miR to promote cancer progression in general, and is also responsible for cisplatin resistance in gastric cancer patients [67, 68]. In addition to previous findings about the significant correlation between expression of specific miRNAs and resposiveness to tamoxifen in hormone-positive breast cancer patients [69-71], our results help to identify miRNAs in interest by more relevant hypotheses.
On the other hand, upregulated mRNAs in TR compared to TS ER+ breast cancer patients obtained from the GSE9893 dataset were matched with those from the GSE7378 dataset. We found that multiple oncogenic pathways were linked with tamoxifen resistance in the present study, including WNT, GPCR, RAS, AKT, TGF-β/BMP, Notch, Hedgehog, and death receptor signaling. In particular, mutation of ESR1 gene is a well-known mechanism of tamoxifen resistance and upregulation of WNT signaling is detected in an in vitro model of tamoxifen-resistant breast cancer [72, 73].
Immune infiltration patterns of breast tumors in relation to glutamate-ammonia ligase (GLUL) estimated by TIMER visualized in a heatmap format. Using the default setting of TIMER, correlations of expression levels of six major immune cell populations with four subtypes of breast cancer were evaluated based on a partial Spearman's rho, also referred to as Pearson correlation coefficient (r) that ranges from -1 (negative correlation) to +1 (positive correlation). *P < 0.05, **P < 0.01, and ***P < 0.001.
GLUL (glutamate-ammonia ligase) mRNA and protein in breast cancer patients and cell lines. (A) Elevated GLUL expression at transcriptomic level in breast cancer compared to normal tissues. (B) Transcriptomic expression of GLUL in the different subtypes of breast cancer, which is up-regulated in the luminal subtype (ER+ cancer). (C) IHC staining of GLUL protein in breast cancer, but not in normal tissues. (D) Bright-field images of MCF7-shLuc as control, along with two GLUL-knockdown cell lines in two-dimensional cell cultures. (E) The efficacy of GLUL knockdown was further confirmed by Western blotting. (F) mRNA expression levels of downstream signaling pathways regulated by GLUL, including Bcl-2 which serves as a marker of the cell cycle, and IL-4 which is an immune-related marker. A pairwise comparison was made between the control cell line and GLUL-knockdown lines. (G) Colony-formation assay confirmed the influence of GLUL on suppressing the proliferation of MCF7 cells. The long-term proliferation of two MCF7 GLUL-knockdown cells were markedly declined compared to the MCF7-shLuc cell line as the control.
Activation of RAS/PI3K/AKT signaling blocks tamoxifen-induced anoikis [74]. Hypoxia Inducible Factor 1 Subunit Alpha (HIF-1α) or AMP-Activated Protein Kinase (AMPK) pathways induce autophagy and mediates resistance to anoikis [75, 76]. Since other pathways may also be involved in tamoxifen resistance, therfore we analyzed the interaction between miRNA-targeting genes and upregulated mRNAs and discovered GLUL plays an important role in cancer progression. GLUL protein promotes cell proliferation in breast cancer [77]. Suppression of GLUL results in drug resistance in cancer cells [78]. However, evidence about the roles of GLUL in tamoxifen resistance or recurrence of breast cancer is still absent. Meanwhile, through MetaCore and TIMER analysis revealed that “Immune response_IL-4-induced regulators of cell growth, survival, differentiation and metabolism”, and immune-related pathways were correlated with GLUL expression in breast cancer patients from TCGA and METABRIC datasets. IL-4 is a pleiotropic cytokine regulating cell differentiation, survival, and proliferation. IL-4 stimulates cell population proliferation through the activation of several Cyclin-dependent kinases (CDKs), which promote the transition of the cell cycle G1/S phase. IL-4 is also important in the regulation of metabolic processes. Specifically, IL-4/JAK/STAT6 signaling enhances the anabolic actions of insulin and is involved in the metabolism process for different types of cells. These shreds of evidence were consistent with our data and GLUL may regulate tamoxifen resistance or the recurrence of breast cancer. Meanwhile, in addition to GLUL, we also found several potential candidate genes showing high expression in tamoxifen-treated recurrence breast cancer patients, which may serve as ideal targets for the treatment of breast cancer (Figure 6), including ZNF148 [79], VPS4A [80], SPN [81], PTPN9 [82], APEX [83], RAE1 [84], CCT7 [85], USP2 [86], and RAGA [87].
Through the CMap analysis, we revealed that particular compounds with highly negative correlation is served as potential therapeutic agents for TR breast cancer patients. Fulvestrant and methotrexate are listed as potential drugs for TR patients. Fulvestrant is one of the standard salvage drugs for metastatic ER+ breast cancer [88]. Methotrexate is a cytotoxic agent and is used in combined regimens for adjuvant chemotherapy. Although new drugs are being developed, methotrexate is still a useful drug for metastatic breast cancer [89]. We also detected other compounds. Proton pump inhibitor omeprazole decreased invasion and metastasis of breast cancer cell line. The combination of sulfasalazine and MK-801 also showed antiproliferative properties in EGFR-overexpressing glioma cells [90]. MK-801 also has the same effect on the growth of pancreatic tumor xenografts in nude mice [91]. Carbachol in combination with low-dose paclitaxel suppresses the proliferation of breast cancer cells in vitro [92]. The 8-azaguanine is a potential cytotoxic drug with anticancer ability in a prediction model or in vitro study [93, 94]. Sirolimus (mTOR inhibitor), an allosteric mTORC1 inhibitor, suppresses the growth of breast cancer cells. Sirolimus and everolimus improve the survival of patients with hepatocellular carcinoma [95]. Harmine exhibited anticancer properties in breast cancer cell lines [96]. Berberine inhibited autophagy by participating in the PTEN/Akt/mTOR pathway by reversing doxorubicin resistance in breast cancer [97]. Collectively, our CMap data suggest that these FDA-approved compounds are potential therapeutic drugs for TR breast cancer patients.
Conclusion
To summarize, through comprehensive bioinformatics analyses of the transcriptome of four datasets of ER+ breast cancer patients and further wet-lab validation, our study presented GLUL as a remarkable factor that associated with ER+ breast cancer. Furthermore, we also identified the immune signaling pathways by the top co-expressed genes with GLUL in ER+ breast cancer patients developing tamoxifen resistance. Our results provide a better understanding of the molecular mechanisms leading to tamoxifen resistance in patients undergoing treatment. Finally, computational connectivity map-based production of drugs that can serve as alternative treatments for tamoxifen-resistant ER+ breast cancer.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This research was supported by grants from the Ministry of Science and Technology (MOST) of Taiwan (MOST-110-2320-B-039-068 to W.J.W., 109-2314-B-006-018-MY3 to H.P.H, and 109-2320-B-038-009-MY2 to C.Y.W.), from Taipei Medical University Hospital (109TMU-TMUH-19, and 111TMU-TMUH-08), from National Cheng Kung University Hospital (NCKUH-10902031 & NCKUH-11002013 & NCKUH-11102007 to H.P.H.). This work was financially supported of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (DP2-111-21121-01-C-01-01). The authors truly appreciate Mr. Daniel P. Chamberlin from the Office of Research and Development at Taipei Medical University for professional English editing. The authors acknowledge the statistical/computational/technical support of the Clinical Data Center, Office of Data Science, Taipei Medical University, Taiwan.
Author contributions
T.M.X.D., C.C.W., W.J.W., H.P.H., and C.Y.W. designed the study; C.C.W., and H.P.H. collected the bioinformatics and clinical data; T.M.X.D., and C.Y.W. performed the experiments; H.D.K.T., G.A., and C.C.C. provided the biostatistics analysis; T.M.X.D. and C.C.W. wrote the initial manuscript draft; H.P.H., and C.Y.W. edited the manuscript; and all authors read and approved the final manuscript.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: a cancer journal for clinicians. 2022
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209-49
3. Polyak K. Heterogeneity in breast cancer. The Journal of clinical investigation. 2011;121:3786-8
4. Baslan T, Kendall J, Volyanskyy K, McNamara K, Cox H, D'Italia S. et al. Novel insights into breast cancer copy number genetic heterogeneity revealed by single-cell genome sequencing. eLife. 2020 9
5. Yersal O, Barutca S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World journal of clinical oncology. 2014;5:412-24
6. Ma H, Lu Y, Marchbanks PA, Folger SG, Strom BL, McDonald JA. et al. Quantitative measures of estrogen receptor expression in relation to breast cancer-specific mortality risk among white women and black women. Breast cancer research: BCR. 2013;15:R90
7. Peart O. Breast intervention and breast cancer treatment options. Radiologic technology. 2015;86:535M-58M quiz 59-62
8. Fisusi FA, Akala EO. Drug Combinations in Breast Cancer Therapy. Pharmaceutical nanotechnology. 2019;7:3-23
9. Jordan VC. The role of tamoxifen in the treatment and prevention of breast cancer. Current problems in cancer. 1992;16:129-76
10. Cersosimo RJ. Tamoxifen for prevention of breast cancer. The Annals of pharmacotherapy. 2003;37:268-73
11. Droog M, Beelen K, Linn S, Zwart W. Tamoxifen resistance: from bench to bedside. European journal of pharmacology. 2013;717:47-57
12. Mirzaei M, Sheikholeslami SA, Jalili A, Bereimipour A, Sharbati S, Kaveh V. et al. Investigating the molecular mechanisms of Tamoxifen on the EMT pathway among patients with breast cancer. Journal of medicine and life. 2022;15:835-44
13. Sukocheva OA, Lukina E, Friedemann M, Menschikowski M, Hagelgans A, Aliev G. The crucial role of epigenetic regulation in breast cancer anti-estrogen resistance: Current findings and future perspectives. Seminars in cancer biology. 2022;82:35-59
14. Sun Y-S, Zhao Z, Yang Z-N, Xu F, Lu H-J, Zhu Z-Y. et al. Risk Factors and Preventions of Breast Cancer. Int J Biol Sci. 2017;13:1387-97
15. Peng L, Jiang J, Tang B, Nice EC, Zhang YY, Xie N. Managing therapeutic resistance in breast cancer: from the lncRNAs perspective. Theranostics. 2020;10:10360-77
16. Demas DM, Demo S, Fallah Y, Clarke R, Nephew KP, Althouse S. et al. Glutamine Metabolism Drives Growth in Advanced Hormone Receptor Positive Breast Cancer. Front Oncol. 2019;9:686 -
17. Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B. et al. Bidirectional Transport of Amino Acids Regulates mTOR and Autophagy. Cell. 2009;136:521-34
18. Ko Y-H, Lin Z, Flomenberg N, Pestell RG, Howell A, Sotgia F. et al. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells. Cancer Biology & Therapy. 2011;12:1085-97
19. Nardone A, Weir H, Delpuech O, Brown H, De Angelis C, Cataldo ML. et al. The oral selective oestrogen receptor degrader (SERD) AZD9496 is comparable to fulvestrant in antagonising ER and circumventing endocrine resistance. Br J Cancer. 2019;120:331-9
20. Chen S-H, Cheung CHA. Challenges in Treating Estrogen Receptor-Positive Breast Cancer. Estrogen: IntechOpen. 2018
21. Finn RS, Aleshin A, Slamon DJ. Targeting the cyclin-dependent kinases (CDK) 4/6 in estrogen receptor-positive breast cancers. Breast Cancer Res. 2016;18:17
22. Lawal B, Liu YL, Mokgautsi N, Khedkar H, Sumitra MR, Wu ATH. et al. Pharmacoinformatics and Preclinical Studies of NSC765690 and NSC765599, Potential STAT3/CDK2/4/6 Inhibitors with Antitumor Activities against NCI60 Human Tumor Cell Lines. Biomedicines. 2021 9
23. Lawal B, Lee CY, Mokgautsi N, Sumitra MR, Khedkar H, Wu ATH. et al. mTOR/EGFR/iNOS/MAP2K1/FGFR/TGFB1 Are Druggable Candidates for N-(2,4-Difluorophenyl)-2',4'-Difluoro-4-Hydroxybiphenyl-3-Carboxamide (NSC765598), With Consequent Anticancer Implications. Front Oncol. 2021;11:656738
24. Lawal B, Kuo YC, Sumitra MR, Wu ATH, Huang HS. In vivo Pharmacokinetic and Anticancer Studies of HH-N25, a Selective Inhibitor of Topoisomerase I, and Hormonal Signaling for Treating Breast Cancer. J Inflamm Res. 2021;14:4901-13
25. Lawal B, Kuo YC, Wu ATH, Huang HS. BC-N102 suppress breast cancer tumorigenesis by interfering with cell cycle regulatory proteins and hormonal signaling, and induction of time-course arrest of cell cycle at G1/G0 phase. Int J Biol Sci. 2021;17:3224-38
26. Joseph NM, Ferrell LD, Jain D, Torbenson MS, Wu T-T, Yeh MM. et al. Diagnostic utility and limitations of glutamine synthetase and serum amyloid-associated protein immunohistochemistry in the distinction of focal nodular hyperplasia and inflammatory hepatocellular adenoma. Modern Pathology. 2014;27:62-72
27. Fares HM, Lyu X, Xu X, Dong R, Ding M, Mi S. et al. Autophagy in cancer: The cornerstone during glutamine deprivation. European Journal of Pharmacology. 2022;916:174723
28. Zhou Y, Eid T, Hassel B, Danbolt NC. Novel aspects of glutamine synthetase in ammonia homeostasis. Neurochemistry International. 2020;140:104809
29. Romero-Garcia S, Lopez-Gonzalez JS, Báez-Viveros JL, Aguilar-Cazares D, Prado-Garcia H. Tumor cell metabolism: an integral view. Cancer Biol Ther. 2011;12:939-48
30. Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer. 2017;3:169-80
31. Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B. et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521-34
32. Eliasen MM, Winkler W, Jordan V, Pokar M, Marchetti M, Roth E. et al. Adaptive cellular mechanisms in response to glutamine-starvation. Frontiers in bioscience: a journal and virtual library. 2006;11:3199-211
33. DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313-24
34. Liu Y, Zhang X, Wang W, Liu T, Ren J, Chen S. et al. Ammonia promotes the proliferation of bone marrow-derived mesenchymal stem cells by regulating the Akt/mTOR/S6k pathway. Bone Research. 2022;10:57
35. Chanrion M, Negre V, Fontaine H, Salvetat N, Bibeau F, Mac Grogan G. et al. A gene expression signature that can predict the recurrence of tamoxifen-treated primary breast cancer. Clin Cancer Res. 2008;14:1744-52
36. Yau C, Benz CC. Genes responsive to both oxidant stress and loss of estrogen receptor function identify a poor prognosis group of estrogen receptor positive primary breast cancers. Breast Cancer Res. 2008;10:R61
37. Hoppe R, Achinger-Kawecka J, Winter S, Fritz P, Lo WY, Schroth W. et al. Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. Eur J Cancer. 2013;49:3598-608
38. Zhong X, Xie G, Zhang Z, Wang Z, Wang Y, Wang Y. et al. MiR-4653-3p and its target gene FRS2 are prognostic biomarkers for hormone receptor positive breast cancer patients receiving tamoxifen as adjuvant endocrine therapy. Oncotarget. 2016;7:61166-82
39. Wang CY, Chiao CC, Phan NN, Li CY, Sun ZD, Jiang JZ. et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am J Cancer Res. 2020;10:95-113
40. Wang CY, Chao YJ, Chen YL, Wang TW, Phan NN, Hsu HP. et al. Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer. Int J Med Sci. 2021;18:256-69
41. Wu YH, Yeh IJ, Phan NN, Yen MC, Liu HL, Wang CY. et al. Severe acute respiratory syndrome coronavirus (SARS-CoV)-2 infection induces dysregulation of immunity: in silico gene expression analysis. Int J Med Sci. 2021;18:1143-52
42. Hsu HP, Wang CY, Hsieh PY, Fang JH, Chen YL. Knockdown of serine/threonine-protein kinase 24 promotes tumorigenesis and myeloid-derived suppressor cell expansion in an orthotopic immunocompetent gastric cancer animal model. J Cancer. 2020;11:213-28
43. Kao TJ, Wu CC, Phan NN, Liu YH, Ta HDK, Anuraga G. et al. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging (Albany NY). 2021;13:17970
44. Vejnar CE, Zdobnov EM. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40:11673-83
45. McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM. et al. The biochemical basis of microRNA targeting efficacy. Science. 2019 366
46. Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697
47. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D13
48. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-50
49. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Research. 2017;77:e108
50. Liu HL, Yeh IJ, Phan NN, Wu YH, Yen MC, Hung JH. et al. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect Genet Evol. 2020;85:104438
51. Wu YH, Yeh IJ, Phan NN, Yen MC, Hung JH, Chiao CC. et al. Gene signatures and potential therapeutic targets of Middle East respiratory syndrome coronavirus (MERS-CoV)-infected human lung adenocarcinoma epithelial cells. J Microbiol Immunol Infect. 2021
52. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419
53. Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929-35
54. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 4
55. Sleightholm R, Neilsen BK, Elkhatib S, Flores L, Dukkipati S, Zhao R. et al. Percentage of Hormone Receptor Positivity in Breast Cancer Provides Prognostic Value: A Single-Institute Study. J Clin Med Res. 2021;13:9-19
56. Jordan VC. Tamoxifen as the first targeted long-term adjuvant therapy for breast cancer. Endocr Relat Cancer. 2014;21:R235-R46
57. Tsoi H, You C-P, Leung M-H, Man EPS, Khoo U-S. Targeting Ribosome Biogenesis to Combat Tamoxifen Resistance in ER+ve Breast Cancer. Cancers (Basel). 2022;14:1251
58. Fang F, Guo C, Zheng W, Wang Q, Zhou L. Exosome-Mediated Transfer of miR-1323 from Cancer-Associated Fibroblasts Confers Radioresistance of C33A Cells by Targeting PABPN1 and Activating Wnt/β-Catenin Signaling Pathway in Cervical Cancer. Reprod Sci. 2022;29:1809-21
59. Li L, Gao J, Li J, Wang J. MiR-711 regulates gastric cancer progression by targeting CD44. Cancer Biomark. 2022;35:71-81
60. Xia M, Lu J, Wu Y, Feng X. MicroRNA-4287 alleviates inflammatory response via targeting RIPK1 in osteoarthritis. Autoimmunity. 2022;55:301-9
61. Xu L, Yu QW, Fang SQ, Zheng YK, Qi JC. MiR-650 inhibits the progression of glioma by targeting FAM83F. Eur Rev Med Pharmacol Sci. 2018;22:8391-8
62. Huang L, Tang X, Shi X, Su L. miR-532-5p promotes breast cancer proliferation and migration by targeting RERG. Exp Ther Med. 2020;19:400-8
63. Chen EG, Zhang JS, Xu S, Zhu XJ, Hu HH. Long non-coding RNA DGCR5 is involved in the regulation of proliferation, migration and invasion of lung cancer by targeting miR-1180. Am J Cancer Res. 2017;7:1463-75
64. Tan G, Wu L, Tan J, Zhang B, Tai WC, Xiong S. et al. MiR-1180 promotes apoptotic resistance to human hepatocellular carcinoma via activation of NF-kappaB signaling pathway. Sci Rep. 2016;6:22328
65. Ning N, Liu S, Liu X, Tian Z, Jiang Y, Yu N. et al. Curcumol inhibits the proliferation and metastasis of melanoma via the miR-152-3p/PI3K/AKT and ERK/NF-kappaB signaling pathways. J Cancer. 2020;11:1679-92
66. Danza K, De Summa S, Pinto R, Pilato B, Palumbo O, Merla G. et al. MiR-578 and miR-573 as potential players in BRCA-related breast cancer angiogenesis. Oncotarget. 2015;6:471-83
67. Bacci M, Lorito N, Ippolito L, Ramazzotti M, Luti S, Romagnoli S. et al. Reprogramming of Amino Acid Transporters to Support Aspartate and Glutamate Dependency Sustains Endocrine Resistance in Breast Cancer. Cell Rep. 2019;28:104-18 e8
68. Guo Y, Yue P, Wang Y, Chen G, Li Y. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis. Biomed Pharmacother. 2019;118:109255
69. Zhang W, Xu J, Shi Y, Sun Q, Zhang Q, Guan X. The novel role of miRNAs for tamoxifen resistance in human breast cancer. Cellular and Molecular Life Sciences. 2015;72:2575-84
70. Gao Y, Zhang W, Liu C, Li G. miR-200 affects tamoxifen resistance in breast cancer cells through regulation of MYB. Scientific Reports. 2019;9:18844
71. Amiruddin A, Massi MN, Islam AA, Patellongi I, Pratama MY, Sutandyo N. et al. microRNA-221 and tamoxifen resistance in luminal-subtype breast cancer patients: A case-control study. Annals of Medicine and Surgery. 2022;73:103092
72. Loh YN, Hedditch EL, Baker LA, Jary E, Ward RL, Ford CE. The Wnt signalling pathway is upregulated in an in vitro model of acquired tamoxifen resistant breast cancer. BMC Cancer. 2013;13:174
73. Brett JO, Spring LM, Bardia A, Wander SA. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Research. 2021;23:85
74. Rytomaa M, Lehmann K, Downward J. Matrix detachment induces caspase-dependent cytochrome c release from mitochondria: inhibition by PKB/Akt but not Raf signalling. Oncogene. 2000;19:4461-8
75. Sun L, Li T, Wei Q, Zhang Y, Jia X, Wan Z. et al. Upregulation of BNIP3 mediated by ERK/HIF-1alpha pathway induces autophagy and contributes to anoikis resistance of hepatocellular carcinoma cells. Future Oncol. 2014;10:1387-98
76. Ng TL, Leprivier G, Robertson MD, Chow C, Martin MJ, Laderoute KR. et al. The AMPK stress response pathway mediates anoikis resistance through inhibition of mTOR and suppression of protein synthesis. Cell Death Differ. 2012;19:501-10
77. Wang Y, Fan S, Lu J, Zhang Z, Wu D, Wu Z. et al. GLUL Promotes Cell Proliferation in Breast Cancer. J Cell Biochem. 2017;118:2018-25
78. Muthu M, Kumar R, Syed Khaja AS, Gilthorpe JD, Persson JL, Nordstrom A. GLUL Ablation Can Confer Drug Resistance to Cancer Cells via a Malate-Aspartate Shuttle-Mediated Mechanism. Cancers (Basel). 2019 11
79. Hahn S, Jackstadt R, Siemens H, Hünten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. Embo j. 2013;32:3079-95
80. Han Q, Lv L, Wei J, Lei X, Lin H, Li G. et al. Vps4A mediates the localization and exosome release of β-catenin to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2019;457:47-59
81. Fu Q, Cash SE, Andersen JJ, Kennedy CR, Madadi AR, Raghavendra M. et al. Intracellular patterns of sialophorin expression define a new molecular classification of breast cancer and represent new targets for therapy. Br J Cancer. 2014;110:146-55
82. Hong Y, Liang H, Uzair Ur R, Wang Y, Zhang W, Zhou Y. et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci Rep. 2016;6:37421
83. Kim KY, Han W, Noh DY, Kang D, Kwack K. Impact of genetic polymorphisms in base excision repair genes on the risk of breast cancer in a Korean population. Gene. 2013;532:192-6
84. Oh JH, Lee JY, Yu S, Cho Y, Hur S, Nam KT. et al. RAE1 mediated ZEB1 expression promotes epithelial-mesenchymal transition in breast cancer. Sci Rep. 2019;9:2977
85. Xu WX, Song W, Jiang MP, Yang SJ, Zhang J, Wang DD. et al. Systematic Characterization of Expression Profiles and Prognostic Values of the Eight Subunits of the Chaperonin TRiC in Breast Cancer. Front Genet. 2021;12:637887
86. He J, Lee HJ, Saha S, Ruan D, Guo H, Chan CH. Inhibition of USP2 eliminates cancer stem cells and enhances TNBC responsiveness to chemotherapy. Cell Death Dis. 2019;10:285
87. Ak J, Lakshmanagowda PB, G CMP, Goturu J. Impact of music therapy on breast milk secretion in mothers of premature newborns. J Clin Diagn Res. 2015;9:Cc04-6
88. Di Leo A, Jerusalem G, Petruzelka L, Torres R, Bondarenko IN, Khasanov R. et al. Results of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2010;28:4594-600
89. Li Z, Yang SS, Yin PH, Chang T, Shi LX, Fang L. et al. Activated estrogen receptor-mitogen-activated protein kinases cross talk confer acquired resistance to lapatinib. Thorac Cancer. 2015;6:695-703
90. Suina K, Tsuchihashi K, Yamasaki J, Kamenori S, Shintani S, Hirata Y. et al. Epidermal growth factor receptor promotes glioma progression by regulating xCT and GluN2B-containing N-methyl-d-aspartate-sensitive glutamate receptor signaling. Cancer Sci. 2018;109:3874-82
91. North WG, Liu F, Lin LZ, Tian R, Akerman B. NMDA receptors are important regulators of pancreatic cancer and are potential targets for treatment. Clin Pharmacol. 2017;9:79-86
92. Espanol AJ, Salem A, Rojo D, Sales ME. Participation of non-neuronal muscarinic receptors in the effect of carbachol with paclitaxel on human breast adenocarcinoma cells. Roles of nitric oxide synthase and arginase. Int Immunopharmacol. 2015;29:87-92
93. Choudhary A, Zachek B, Lera RF, Zasadil LM, Lasek A, Denu RA. et al. Identification of Selective Lead Compounds for Treatment of High-Ploidy Breast Cancer. Mol Cancer Ther. 2016;15:48-59
94. Kim N, Choi JW, Song AY, Choi WS, Park HR, Park S. et al. Direct potentiation of NK cell cytotoxicity by 8-azaguanine with potential antineoplastic activity. Int Immunopharmacol. 2019;67:152-9
95. Grigg SE, Sarri GL, Gow PJ, Yeomans ND. Systematic review with meta-analysis: sirolimus- or everolimus-based immunosuppression following liver transplantation for hepatocellular carcinoma. Aliment Pharmacol Ther. 2019;49:1260-73
96. Ding Y, He J, Huang J, Yu T, Shi X, Zhang T. et al. Harmine induces anticancer activity in breast cancer cells via targeting TAZ. Int J Oncol. 2019;54:1995-2004
97. Wang Y, Liu Y, Du X, Ma H, Yao J. Berberine Reverses Doxorubicin Resistance by Inhibiting Autophagy Through the PTEN/Akt/mTOR Signaling Pathway in Breast Cancer. Onco Targets Ther. 2020;13:1909-19
Author contact
![]() Corresponding author: Chih-Yang Wang, E-mail: chihyangedu.tw.
Corresponding author: Chih-Yang Wang, E-mail: chihyangedu.tw.

 Global reach, higher impact
Global reach, higher impact