Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(14):2033-2043. doi:10.7150/ijms.74713 This issue Cite
Review
CircRNAs in hepatocellular carcinoma: characteristic, functions and clinical significance
1. Department of Hepatobiliary and Pancreatic Surgery, Huaihua First People's Hospital, Huaihua, Hunan, P. R. China.
2. Cardiovascular Center, Huizhou First Municipal People's Hospital, Huizhou, Guangdong, P. R. China.
3. Department of Hepatobiliary Surgery, Clinical Medical College, Yangzhou University, Yangzhou, Jiangsu, P. R. China.
Received 2022-5-4; Accepted 2022-9-7; Published 2022-11-14
Abstract
Hepatocellular carcinoma (HCC) is one of the most common and serious types of cancer worldwide, with high incidence and mortality rates. Circular RNAs (circRNAs) are a novel class of non-coding RNA with important biological functions. In recent years, multiple circRNAs have been found to be involved in the biological processes of tumorigenesis and tumor development. Increasing evidence has shown that circRNAs also play a crucial role in the occurrence and development of HCC. However, the specific molecular mechanism of circRNAs in HCC has not been fully elucidated. The present review systematically summarized the classification and basic characteristics of circRNAs, their biological functions and their role in the occurrence and development of HCC. By summarizing the previous studies on circRNAs in HCC, this study aimed to indicate potential approaches to improving the early diagnosis and treatment of HCC.
Keywords: hepatocellular carcinoma, circRNAs, molecular sponge, circRNA-miRNA-mRNA axis
Introduction
Liver cancer is one of the most common and serious types of cancer worldwide, with high incidence and mortality rates [1]. Hepatocellular carcinoma (HCC) accounts for 90% of all primary liver cancer cases and is the most common histological subtype [2]. Currently, treatment options for HCC include surgical resection, liver transplantation, image-guided ablation, transcatheter arterial chemoembolization and targeted therapy, such as sorafenib, lenvatinib and regorafenib [3-5]. However, despite these comprehensive treatments, the prognosis of HCC remains poor [6]. Therefore, exploring the pathogenesis of HCC, identifying new therapeutic targets and designing more effective treatments are crucial.
Circular RNAs (circRNAs) are a circular type of non-coding RNAs (ncRNAs) with important biological functions that are characterized by a covalently closed ring structure without a 5' cap and 3' polyadenylation tail [7]. It has been shown that circRNAs are involved in the pathogenesis of a variety of human diseases, including cardiovascular diseases, diabetes, neurological diseases and cancer [8]. An amount of evidence indicates that circRNAs play a key role in the occurrence and development of HCC. Some circRNAs can promote the progression of HCC. For example, circ_0008450 can promote the proliferation, invasion and migration of HCC cells and inhibit apoptosis by regulating miR-548p [9]; circRNA-104718 can also promote the proliferation, invasion, and proliferation of HCC cells by regulating the miRNA-218-5p/TXNDC5 axis. migration and inhibit the apoptosis of HCC [10]. However, some circRNAs have inhibitory effects on the progression of HCC. For example, circADAMTS14 inhibits the proliferation, invasion and migration of HCC cells and promotes HCC apoptosis by regulating miR-572/RCAN1 [11]; circRNA-5692 exerts the same inhibitory effect by regulating miR-328-5p/DAB2IP [12]. Although a small number of circRNAs have been functionally characterized, the role of circRNAs in the occurrence and development of HCC has not been fully elucidated and requires further investigation.
The role of circRNAs in the occurrence and development of HCC should be further studied. Therefore, the present study described the basic characteristics of circRNAs, elaborated on their biological functions and summarized their specific roles in HCC, to provide basic ideas for the further study of circRNAs in HCC.
Basic characteristics and classification of circRNAs
As an important member of ncRNA, circRNAs have been widely studied, and have been shown to have the following basic characteristics: 1. CircRNAs have a covalent closed continuous loop, which means that they do not have a 5'-3' polar or polyadenylated tail. This feature allows circRNAs to escape the fate of being degraded by exonuclease, and theoretically makes them have a more stable structure than linear RNAs [13]; 2. the circRNA studies that have been carried out so far have indicated that circRNAs are widespread in eukaryotic cells, and consist of several types [14]; 3. the distribution of circRNAs in cells is not uniform, as most of them are located in the cytoplasm and a few in the nucleus [15]; 4. the majority of circRNA sequences are highly conserved within a species [16]; 5. CircRNAs are often expressed in different developmental stages and various tissues with significant specificity [17]; 6. CircRNAs play a regulatory role at the level of transcription or post-transcription [15].
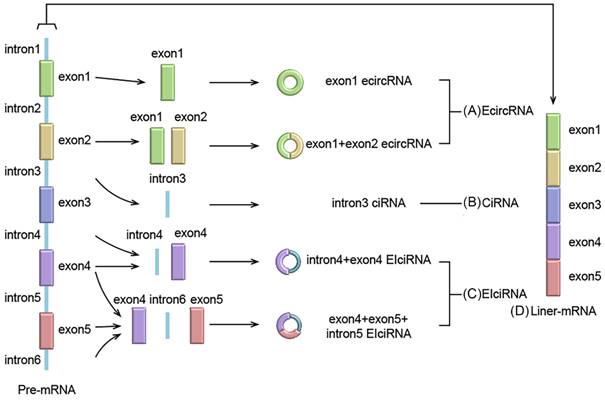
Pre-mRNA is the parent of both circRNAs and linear RNA. Unlike the classical splicing of linear RNA, most circRNAs are formed by reverse splicing of pre-mRNA [18]. circRNAs have a wide range of sources, and circRNAs may be produced in all regions of the genome, including intergenic, intronic, antisense and untranslated regions [15]. circRNAs are classified into the following three categories, based on origin: Exon circRNAs (EcircRNAs), circular intron RNAs (CiRNAs) and exon-intron circRNAs (EIciRNAs). circRNAs are divided into these three subtypes due to their different cyclization mechanisms [19]. EcircRNAs are derived from exons, and most of the circRNAs currently studied are EcircRNAs. According to the different formation methods of EcircRNAs, two different models have been proposed. The first is the lariat-driven circularization model, which contains only one exon, and is formed by reverse-splicing the 5' splice site of the same exon to the 3' splice site. The second is the circularization model driven by intron pairs, which contains multiple exons and is formed by reverse splicing of the 5' splice site to the 3' splice site of another exon. CiRNAs, unlike EcircRNAs, lack exons and are all produced from intron lariats, which can avoid the debranching and degradation of normal introns. EIciRNAs integrate the characteristics of EcircRNAs and CiRNAs, including both exons and introns that are not completely spliced. And through different combinations, various types of EIciRNAs can be formed (Figure 1).
Formation of circRNA and linear RNA. (A) Exon-circRNA (EcircRNA): EcircRNAs are only derived from exons; (B) Circular intron RNA (CiRNA): CiRNAs are derived from intron lariats; (C) Exon-intron circRNA (EIciRNA): EIciRNA are derived from both exons and intron lariats; (D) Traditional linear mRNA comes from pre-mRNA classic splicing.
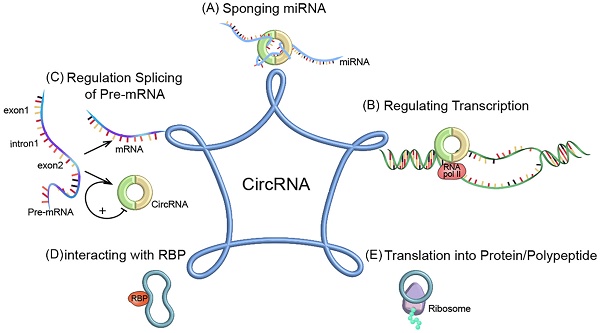
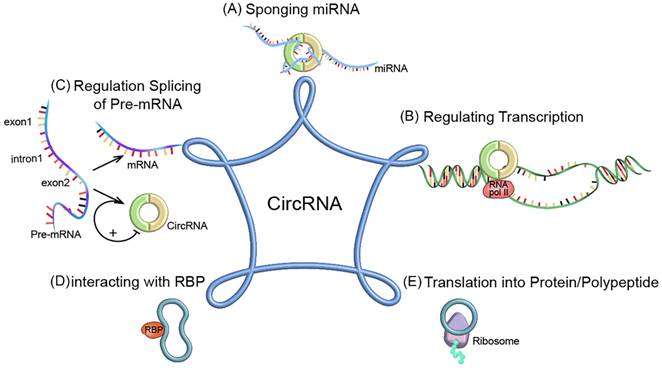
The functional mechanisms of circRNAs. (A) Acting as miRNA sponge; (B) Regulating transcription; (C) Regulating splicing of pre-mRNA; (D) Interacting with RBP; (E) Translated into protein/polypeptide.
CircRNAs have been shown to play an important role in a variety of physiological and pathological processes, including tumorigenesis and tumor progression. Understanding and clarifying these basic characteristics is important to further study the function and mechanism of circRNAs in HCC.
The biological function of circRNAs
CircRNAs play a key role in cancer growth, metastasis and treatment resistance. The biological functions of circRNAs have been extensively studied. Their functions can be divided into five categories: (1) Acting as microRNA (miRNA) sponges to inhibit miRNA-related functions [20]; (2) regulating transcription and translation; (3) regulating alternative splicing of pre-mRNAs; (4) regulating gene expression by interacting with RNA-binding proteins (RBPs); (5) some circRNAs can be translated into proteins as translation templates (Figure 2).
circRNAs can function as miRNA sponges
According to the central dogma of genetics, mRNAs can be translated into proteins, which comprises the dominant protein formation process [21]. miRNAs are a type of endogenous RNA ~22 nt in length, which can play an important regulatory role by targeting mRNAs for cleavage or translation inhibition [22]. The discovery of miRNA response elements shows that circRNAs can competitively bind to miRNAs to affect the pathophysiological process of animals and plants. This mechanism is widespread in the human body and plays a regulatory role in several biological processes. CircRNAs can be used as a miRNA molecular sponge to prevent miRNAs from performing their original biological function [23], or they can competitively inhibit miRNAs by binding to the target molecule. This results in the miRNA function being inhibited, which, in turn, affects the downstream function. One of the best examples of this circRNA mechanism is cerebellar degeneration-related protein 1 antisense (CDR1as). First, CDR1as can bind to miR-7, since it has >70 selectively conserved miR-7 binding site circular molecules [24]. CDR1as can also be used as a molecular sponge to bind to miR-1270 [25], miR-135-5p [26], miR-219a-5p [27] and miR-671-5p [28]. CDR1as has been closely associated with the occurrence and development of various cancer types, such as HCC [29-31], gastric [32], colorectal [33] and ovarian [26] cancers. Similar molecular sponging mechanisms include circ-CDYL [34], circASAP1 [35], circPVT1 [36, 37] and cSMARCA5 [38]. In conclusion, the circRNA-miRNA axis may play an important role in the development of cancer. Further research on the direct mechanism of the circRNA-miRNA axis is required to fully elucidate the code of life.
circRNAs can act as transcriptional regulators
Unlike EcircRNAs, which are mainly distributed in the cytoplasm, CiRNAs and EIciRNAs retain introns and are largely located in the nucleus. The mechanism of action of these two circRNAs is also different from that of EcircRNAs. CiRNAs and EIciRNAs cannot be used as molecular sponges to participate in the pathophysiology of organisms, but they can be combined with RNA polymerase II to affect the transcription process of the parent gene, thus also affecting protein synthesis [39]. EIciRNAs can participate in the regulation of the transcription mechanism, since its sequence retains the intron of the parent gene. The following describes several typical adjustment procedures: circ-EIF3J and circ-PAIP2 contain the sequence of the promoter region of the parent gene, so they can interact with U1 snRNPs and RNA polymerase II in the promoter region of the host gene, thereby enhancing the transcription process of the parent genes (PAIP2 and EIF3J) [40]. CiRNAs can also affect the activity of RNA polymerase II, thereby regulating the transcription process, further affecting the amount of target mRNA. This mechanism corresponds to CiRNAs such as ci-ANKRD52, ci-MCM5 and ci-SIRT7, all of which extend the activity of RNA polymerase II to promote transcription, thereby promoting the expression of parental genes. On the contrary, when the levels of these CiRNAs are reduced, the transcription levels of their parent genes are reduced [41].
CircRNAs can compete with traditional linear mRNAs for pre-mRNAs
In the source-based classification, exon-derived circRNAs constitute the majority [42]. In addition, both circRNAs and traditional linear mRNAs are derived from pre-mRNAs. Pre-mRNAs form circRNAs through exon circularization and can compete with linear mRNAs through canonical splicing. Therefore, circRNAs formed by exon circularization can compete with linear mRNAs in this splicing process [43]. Pre-mRNA exon circularization for the formation of circRNAs inevitably leads to the existence of fewer pre-mRNAs for the formation of mRNAs through canonical splicing. This suggests that the formation of circRNAs usually occurs at the expense of reduced mRNA production [44]. Similarly, the canonical spliceosome mechanism can affect the formation of circRNAs from pre-mRNAs [45]. Consequently, the production of circRNAs can block pre-mRNA splicing into mRNAs [46, 47]. As a result, the production of circRNAs leads to the reduction of its linear isoform mRNA [48], which leads to the production of circRNAs blocking the process of mRNA formation from pre-mRNAs, ultimately affecting protein production. However, this mechanism of competing for splicing sites requires further systematic research to provide data support, as it cannot fully account for the effect of this mechanism on the final product.
CircRNAs can regulate gene expression by interacting with RBPs
Post-transcriptional regulation is not limited to the splicing process; RBPs also play an important role in it. circRNAs can bind to miRNAs to act as miRNA molecular sponges, while certain circRNAs are also able to bind to RBPs to act as a protein sponge, thus affecting the post-transcriptional modification process, which all possess RBPs binding sites. For example, the human antigen R (HuR) can bind to PABPN1 mRNA and positively regulate the translation process of PABPN1. circPABPN1 acts as an RBPs sponge. It can first bind to HuR and inhibit the binding of HuR to PABPN1 mRNA, thereby inhibiting the positive effect of HuR on the translation of PABPN1 mRNA [49]. This mechanism of acting as an RBP molecular sponge has also been observed in circ-FOXO3 [50], circ-MBL [43] and circ-Amotl1 [51-53].
circRNAs possess a translational ability
Due to the lack of the polyadenylated tails and internal ribosome entry sites (IRES) required for protein translation, circRNAs are widely classified as endogenous non-coding RNAs (ncRNAs) [54, 55]. However, the latest research appears to have proven that certain circRNAs have the ability to be translated into proteins. At present, 4 such circRNAs have been discovered: circ-ZNF609 [56], circ-SHPRH [57], circ-FBXW7 [58] and circMBL [59]. When IRES is introduced into the circRNA sequence, circRNAs can be translated in vivo and in vitro. However, it remains unclear whether such a reaction can occur in an organism, or whether it can be widespread and participate in the pathophysiology of the human body. This type of biological behavior undermines our understanding of circRNAs. Therefore, it is worth studying whether circRNAs can be considered non-coding RNAs.
In recent years, there has been an increasing amount of research results on the biological functions of circRNAs, a considerable portion of which are different from our current understanding. circRNAs are an important supplement to the central dogma of genetics. They play a regulatory role in multiple steps of protein synthesis and participate in the pathophysiological process of tumor development. However, our understanding of circRNAs remains incomplete, and further research is required.
Functional circRNAs in HCC
CircRNAs are commonly aberrantly expressed in tissues from several types of cancer, including HCC. These differentially expressed circRNAs ultimately promote or inhibit the occurrence and development of cancer. These circRNAs may play a crucial role in the carcinogenesis and development of HCC.
The up-regulated circRNAs in HCC
| CircRNA | Gene Symbol | Expression Change | miRNA sponged | Competitive mRNA | Function | Reference |
|---|---|---|---|---|---|---|
| circ-CDYL | CDYL | Up | miR-892a miR-328-3p | HDGF HIF1AN | proliferation (+); T-ICs (+); spheroid growth (+); chemotherapy resistance (+); EPCAM+cells (+) | [34] |
| circ-0046600 | Up | miR-640 | HIF-1α | migration (+); | [60] | |
| CircASAP1 | ASAP1 | Up | miR-326; miR-532-5p | MAPK1; CSF-1 | proliferation (+); migration (+); invasion (+) | [35] |
| hsa_circ_0101432 | RPPH1 | Up | miR-1258/miR-622 | MAPK1 | proliferation (+); invasion (+); apoptosis (-) | [61] |
| circMAN2B2 | MAN2B2 | Up | miRNA-217 | MAPK1 | proliferation (+) | [62] |
| circPTGR1 | PTGR1 | Up | miR-449a | MET | migration (+); invasion (+) | [66] |
| circ-DB | Up | miR-34a | USP7/Cyclin A2 | [67] | ||
| circRNA Cdr1as | CDR1 | Up | miR-1270 | AFP | proliferation (+); migration (+) | [68] |
| circRNA PVT1 | PVT1 | Up | miR-203 | HOXD3 | proliferation (+); migration (+) | [36] |
| circPVT1 | PVT1 | Up | miR-3666 | SIRT7 | proliferation (+); apoptosis (-) | [37] |
| circRNA-100338 | Up | miR-141-3p | RHEB/EIF5; mTOR signaling pathway | proliferation (+) | [63] | |
| circRNA-100338 | SNX27 | Up | miR-141-3p | migration (+); invasion (+) | [64] | |
| hsa_circ_0008450 | CMTM3 | Up | miR-214-3p | EZH2 | proliferation (+); migration (+); apoptosis (-) | [65] |
| circ_0008450 | CMTM3 | Up | miR‐548p | proliferation (+); migration (+); invasion (+); apoptosis (-) | [9] | |
| hsa_circRNA_103809 | ZFR | Up | miR-377-3p | FGFR1/ERK | proliferation (+); cell cycle (+); migration (+) | [72] |
| circRNA-104718 | Up | miRNA-218-5p | TXNDC5 | proliferation (+); migration (+); invasion (+); apoptosis (-) | [10] | |
| circMYLK | MYLK | Up | miR-362-3p | Rab23 | proliferation (+); invasion (+); migration (+) | [71] |
| circ-ZNF652 | ZNF652 | Up | miR-203/ miR-502-5p | Snail | Invasion (+); migration (+) | [73] |
| circ_0000267 | Up | miR‐646 | proliferation (+); migration (+); invasion (+); apoptosis (-) | [74] | ||
| circ-FOXP1 | FOXP1 | Up | miR-875-3p/ miR-421 | SOX9 | proliferation(+);invasion(+); apoptosis (-) | [69] |
| circRNA_104075 | Up | miR-582-3p | YAP | [75] | ||
| hsa_circ_0078710 | THBS2 | Up | miR-31 | HDAC/CDK2/cyclinA/cyclin D1/CDK4/p21 | proliferation (+); migration (+); invasion (+); cell cycle (+) | [76] |
| hsa_circ_101280 | SLAIN1 | Up | miR-375 | JAK2 | proliferation (+); apoptosis (-) | [77] |
| circRNA-101368 | Up | miR-200a | HMGB1/RAGE/NF-κB/E-Cadherin | migration (+) | [78] | |
| circ-ZEB1.33 | Up | miR-200a-3p | CDK6 | [79] | ||
| circFBLIM1 | FBLIM1 | Up | miR-346 | FBLIM1 | proliferation (+); invasion (+); apoptosis (-) | [80] |
| hsa_circ_0103809 | AP4E1 | Up | miR-490-5p | SOX2 | proliferation (+); migration (+); apoptosis (-) | [81] |
| hsa_circ_0016788 | TRIM11 | Up | miR-486 | CDK4 | proliferation (+); invasion (+); apoptosis (-) | [82] |
| hsa_circRBM23 | RBM23 | Up | miR-138 | vimentin/CCND3 | viability (+); proliferation (+); migration (+); cell cycle (+) | [83] |
| hsa_circ_0005075 | EIF4G3 | Up | miR-431 | proliferation (+); migration (+); invasion (+) | [84] | |
| hsa_circ_0067934 | PRKC1 | Up | miR-1324 | FZD5/Wnt/β-catenin | proliferation (+); migration (+); invasion (+); apoptosis (-) | [85] |
| circABCC2 | ABCC2 | Up | miR-665 | ABCC2 | proliferation (+); invasion(+) | [70] |
| hsa_circ_100338 | SNX27 | Up | miR-141-3p | ZEB1 | proliferation (+) | [86] |
| CircFBXO11 | FBXO11 | Up | miR-605 | FOXO3/ABCB1 | proliferation (+); cell cycle (+); chemotherapy; resistance (+) | [87] |
| circ_0091581 | Up | miR-526b | c-MYC | proliferation (+) | [88] | |
| circPCNX | PCNX | Up | miR-506 | Snail2/YAP | proliferation (+); apoptosis (-) | [89] |
| Circ-PRMT5 | PRMT5 | Up | miR-188-5p | HK2 | proliferation (+); migration (+); glycolysis (+) | [90] |
| hsa_circ_0000092 | Up | miR-338-3p | HN1 | proliferation (+); migration (+); invasion (+); apoptosis (-) | [91] | |
| hsa_circ_0056836 | Up | miR-766-3p | FOSL2 | proliferation (+); migration (+); invasion (+) | [92] | |
| circ- HOMER1 | HOMER1 | Up | miR-1322 | CXCL6 | proliferation (+); migration (+); invasion (+); apoptosis (-) | [93] |
| CircABCB10 | ABCB10 | Up | miR-670-3p | HMG20A | proliferation (+); invasion (+) | [94] |
| circ_0091579 | Up | miRNA-490-3p | proliferation (+); migration (+) | [95] | ||
| circ_0001955 | Up | miR-516a-5p | TRAF6/MAPK11 | proliferation (+) | [96] | |
| Circ-TCF4.85 | Up | miR-486-5p | ABCF2 | proliferation (+); migration (+); invasion (+); apoptosis (-) | [97] |
CircRNAs function as oncogenes in HCC
In recent studies, several circRNAs have been found to be significantly overexpressed in HCC, as compared with adjacent tissues, and it has been suggested that they may play an oncogenic role in the development of HCC. The upregulated circRNAs in HCC are listed in Table 1.
Although different circRNAs regulate HCC in different ways, recent studies have found that different circRNAs may regulate HCC through a common pathway [34, 35, 60-62]. It has been reported that the expression of circ-CDYL in HCC tissue is higher than that in paracancerous tissue [34]. The high expression of circ-CDYL has been found to significantly promote the formation of HCC [34]. Functional experiments have shown that circ-CDYL could act as a molecular sponge of miR-892a and miR-328-3p, and inhibit their binding to HDGF and HIF1AN, thereby promoting the expression of HDGF and HIF1AN [34]. This, in turn, has been shown to lead to the activation of the PI3K-AKT-mTORC1/β-catenin and NOTCH2 pathways, promoting the expression of the effector proteins BIRC5/SURVIVIN and MYC proto-oncogene [34]. In conclusion, the expression of circ-CDYL is increased in the early stage of HCC and promotes the formation of HCC through a series of gene regulations [34]. In addition, hsa_circ_0046600 can also regulate hypoxia-inducible factor-1α (HIF-1α) through sponging miR-640. Recent studies have shown hsa_circ_0046600 to be significantly upregulated in HCC tissues, as compared with adjacent normal tissues, and the expression of hsa_circ_0046600 to be significantly correlated with clinicopathological factors [60]. In functional experiments, the downregulation of hsa_circ_0046600 was found to significantly inhibit the migration of HCC cells. Mechanistically, hsa_circ_0046600 is mainly used as a molecular sponge of miR-640 to regulate the level of HIF-1α, thereby regulating the biological behavior of HCC. In conclusion, hsa_circ_0046600 can promote the migration of HCC cells through the miR-640/node HIF-1α axis [60].
As compared with adjacent tissues, circASAP1 is highly expressed in HCC tissues. Functional experiments have shown that the overexpression of circASAP1 can promote cell proliferation, migration and invasion [35]. circASAP1 can promote the expression of mitogen-activated protein kinase (MAPK)1 and colony-stimulating factor (CSF)-1 by sponging miR-326 and miR-532-5p. In conclusion, circASAP1 can regulate the miR-326/miR-532-5p-MAPK1/CSF-1 axis, thereby regulating the biological behavior of HCC [35]. Hsa_circ_0101432 is significantly upregulated in HCC. The upregulated hsa_circ_0101432 can enhance the proliferation and invasion ability of HCC cells and inhibit apoptosis. Mechanistically, hsa_circ_0101432 has been reported to act as a molecular sponge of miR-1258 and miR-622 to enhance the expression of MAPK1 to promote tumor growth. In conclusion, hsa_circ_0101432 inhibits HCC cell apoptosis by targeting miR-1258 and miR-622 and upregulating MAPK1 mRNA expression, and promoting cell proliferation, invasion and HCC tumor growth [61]. The expression of circMAN2B2 was reported by Fu et al. [62] to be higher in HCC tissue than that in paracancerous tissue. It was also reported to act as a molecular sponge of miR-217 to regulate the expression of MAPK1, thereby promoting cell proliferation.
Although different circRNAs regulate HCC in different ways, recent studies have found that the same circRNAs can also regulate HCC in different ways [9, 36, 37, 63-65]. CircPVT1 is highly expressed in HCC, which indicates a poor prognosis. Functional experiments have shown that the downregulation of circPVT1 can reduce the proliferation and migration capacity of HCC cells, whereas the upregulation of circPVT1 can promote the growth and migration of HCC cells. In terms of mechanism, circPVT1 can directly bind with miR-203, and promote the occurrence and development of HCC by regulating the miR-203/homebox D3 (HOXD3) pathway. In conclusion, circPVT1 can regulate the progression of HCC by adjusting the miR-203/HOXD3 axis [37]. The circPVT1 expression is significantly upregulated in HCC tissues and cell lines. The downregulation of circPVT1 can significantly reduce the proliferation of HCC cells and increase apoptosis. Mechanistically, circPVT1 can be used as a molecular sponge to adsorb miR-3666 and reduce the inhibitory effect of miR-3666 on SIRT7. In conclusion, the circPVT1/miR-3666/SIRT7 axis has been shown to play an important role in the occurrence and development of HCC [36].
It was reported by Huang et al [63] that the activity of the mTOR signaling pathway can be regulated by circRNA-100338. Further research found that circRNA-100338 regulates the mTOR signaling pathway through the circRNA-100338/miR-141-3p/RHEB axis, and may be used as an important indicator to predict the prognosis of HCC patients [64]. The expression of circRNA_100338 in HCC tissues is higher than that in adjacent tissues, indicating a poor prognosis. Bioinformatics analysis and luciferase experiments have confirmed that circRNA_100338 regulates the occurrence and development of HCC through binding to miR-141-3p, suggesting that circRNA_100338 is a potentially valuable biomarker for HCC diagnosis and treatment [64].
By measuring the content of hsa_circ_0008450 in HCC tissues and cells, Lin et al found that hsa_circ_0008450 was significantly increased. Functional experiments have suggested that downregulating hsa_circ_0008450 can significantly inhibit cell proliferation, invasion and migration. Mechanistically, hsa_circ_0008450 has been found to promote the enhancer of zeste homolog 2 (EZH2) protein expression by sponging miR-214-3p. That study demonstrated that the hsa_circ_0008450/miR-214-3p/EZH2 axis plays an oncogenic role in the occurrence of HCC and may serve as a new target for the treatment of HCC [65]. In HCC tissue specimens and cell lines, Zhang et al also found that the circ_0008450 expression was upregulated, which was associated with poor prognosis [9]. Functional tests have suggested that the downregulation of circ_0008450 can inhibit the proliferation, migration and invasion of HCC cells, while increasing apoptosis. Mechanistically, circ_0008450 has been proven to be a sponge of miR-548p, and circ_0008450 to regulate the process of HCC by combining with miR-548p [9].
CircRNAs are abundant in cells and tissues, and recent studies have shown that exosomes also contain large amounts of circRNAs. circRNAs in exosomes have also been found to be key regulators in the occurrence and development of HCC. It has been reported that circPTGR1 is upregulated in serum exosomes of HCC patients and is associated with clinical stage and prognosis. circPTGR1 can be used as a molecular sponge of miR-449a and reduce the inhibitory effect of miR-499a on hepatocyte growth factor receptor (MET)-related mRNA, thereby promoting MET protein translation and HCC cell migration and invasion [66]. Exosomes also contain a large amount of circ-deubiquitination (circ-DB). Research has shown that the exosomal circ-DB level increases as the body fat proportion increases. It has been reported by functional experiments that exosomal circ-DB may not only promote HCC growth, but also reduce DNA damage. Mechanistically, exosomal circ-DB can act as a molecular sponge to adsorb miR-34a, thereby reducing the inhibitory effect of miR-34a on DB-related USP7 [67]. As compared with normal paracancerous tissues and normal liver cell lines, the expression of circRNA Cdr1as in HCC cell lines and tissues was significantly higher. In functional experiments, the upregulation of circRNA Cdr1as can promote the proliferation and migration of HCC cells. Mechanistically, circRNA Cdr1as promotes the protein expression of AFP by competitively binding to miR-1270 in HCC. Further research has shown that exosomes also contain a large amount of circRNA Cdr1as, the overexpression of which in HCC cells promotes the proliferation and migration of surrounding normal cells. In conclusion, circRNA Cdr1as can promote the protein expression of AFP by binding to miR-1270, thereby promoting the proliferation and migration of HCC cells [68].
Several other circRNAs [10, 69-97] have been identified to play an oncogenic role in the development and progression of HCC, and they are listed in Table 1. These circRNAs are highly expressed in HCC and act as oncogenes by promoting cancer cell proliferation, invasion, migration, anti-apoptosis, drug resistance and immune escape. Different circRNAs may regulate HCC through a common pathway, and the same circRNAs can also regulate HCC in different ways. HCC cells can not only regulate the occurrence and development of HCC through the differential expression of circRNAs in them, but also distant cells and tissues through exosomes containing differentially expressed circRNAs.
CircRNAs may function as tumor suppressors in HCC
In addition to the aforementioned circRNAs, some other circRNAs exhibit a significantly lower expression in HCC than adjacent tissues. It has been suggested that they may play the role of tumor suppressor genes in the development of HCC. These downregulated circRNAs in HCC are listed in Table 2.
The expression of cSMARCA5 is lower in HCC tissues, as compared with that in adjacent tissues, which indicates a poor prognosis. Functional experiments have suggested that cSMARCA5 can inhibit the proliferation and migration of HCC cells. Mechanistically, cSMARCA5 can promote the expression of TIMP3 by sponging miR-17-3p and miR-181b-5p. TIMP3 is a well-known tumor suppressor gene that can significantly inhibit the occurrence and development of tumors. In conclusion, the cSMARCA5/miR-17-3p/miR-181b-5p/TIMP3 axis plays an important role in the proliferation and migration of HCC [38].
The down-regulated circRNAs in HCC
| CircRNA | Gene Symbol | Expression Change | miRNA sponged | Competitive mRNA | Function | Reference |
|---|---|---|---|---|---|---|
| cSMARCA5 | SMARCA5 | Down | miR-17-3p; miR-181b-5p | TIMP3 | proliferation (-); migration (-) | [38] |
| circTRIM33-12 | TRIM33 | Down | miR-191 | TET1/5hmC | proliferation (-); migration (-); invasion (-); immune evasion (-) | [98] |
| circMTO1 | MTO1 | Down | miR-9 | p21 | Proliferation (-); invasion (-) | [99] |
| circADAMTS13 | ADAMTS13 | Down | miR-484 | Proliferation (-); | [100] | |
| circADAMTS14 | ADAMTS14 | Down | miR‐572 | RCAN1 | proliferation (-); migration (-); invasion (-); apoptosis (+) | [11] |
| hsa_circ_0070269 | PLAC8 | Down | miR-182 | NPTX1 | proliferation (-); invasion (-) | [101] |
| circRNA_101505 | Down | miR-103 | NOR1 | proliferation (-); apoptosis (+); cisplatin resistance (-) | [102] | |
| hsa_circ_0091570 | MBNL3 | Down | miR-1307 | ISM1 | proliferation (-); migration (-); apoptosis (+) | [103] |
| hsa_circ_0001649 | SHPRH | Down | miR-127-5p; miR-612; miR-4688 | SHPRH | proliferation (-); migration (-) | [104] |
| circSETD3 | SETD3 | Down | miR-421 | MAPK14 | proliferation (-); cell cycle (-); migration (-) | [105] |
| circSMAD2 | SMAD2 | Down | miR-629 | EMT | migration (-); invasion (-); EMT (-) | [106] |
| circC3P1 | C3P1 | Down | miR-4641 | PCK1 | proliferation (-); migration (-); invasion (-) | [107] |
| circ-ABCB10 | ABCB10 | Down | miR-340-5p; miR-452-5p | NRP1; ABL2 | proliferation (-); migration (-); invasion (-); apoptosis (+) | [108] |
| CircRNA-0072309 | Down | miR-665 | PI3K/AKT and Wnt/β-catenin pathways | proliferation (-); migration (-); invasion (-); apoptosis (+) | [109] | |
| circ-0051443 | Down | miR-331-3p | BAK1 | cell cycle (-); apoptosis (+) | [110] | |
| circRNA-5692 | Down | miR-328-5p | DAB2IP | proliferation (-); migration (-); invasion (-); apoptosis (+) | [12] |
Recent studies have shown that circTRIM33-12 is downregulated in HCC tissues and cell lines, which indicates a poor prognosis in HCC patients. Functional experiments have suggested that the downregulation of circTRIM33-12 in HCC cells increases their proliferation, migration, invasion and immune escape. Mechanistically, circTRIM33-12 can be used as a molecular sponge to bind miR-191, reducing the inhibitory effect of miR-191 on TET1. The upregulation of TET1 further leads to a marked reduction in the 5-hydroxymethylcytosine (5hmC) level. In conclusion, the circTRIM33-12/miR-191/TET1/5hmC axis plays an important role in the occurrence and development of HCC [98].
CircMTO1 has also been reported as hsa_circRNA_0007874 or hsa_circRNA_104135 and is significantly downregulated in HCC tissues. Poor prognosis in HCC patients was found to be associated with a low circMTO1 expression. Through functional experiments, the downregulation of circMTO1 can significantly enhance the proliferation and invasion ability of HCC cells. Mechanistically, circMTO1 has been shown to act as a molecular sponge that binds to miR-9 to reduce the inhibitory effect of miR-9 on p21, thereby promoting p21 expression. In conclusion, the circMTO1/miR-9/p21 axis plays an important role in the progression of HCC, suggesting that circMTO1 may be a potential target for the treatment of HCC [99].
Studies have shown that circADAMTS13 is significantly downregulated in HCC organizations. In addition, clinical pathological analysis has suggested that the upregulation of circADAMTS13 is negatively correlated with tumor size, but positively correlated with prognosis. Functional experiments have reported that the overexpression of circADAMTS13 can significantly inhibit the proliferation of HCC cells. Bioinformatics analysis and luciferase reporter gene detection revealed that circADAMTS13 can be combined with miR-484 as a molecular sponge. Therefore, these results indicated that circADAMTS13 can act as a tumor suppressor in the process of HCC through the functional pathway of sponging miR-484 [100]. As compared with normal paracancerous tissues and normal liver cell lines, the expression of circADAMTS14 in HCC cell lines and tissues was significantly lower. As shown by functional experiments, the upregulation of circADAMTS14 can induce HCC cell apoptosis and inhibit cell proliferation and invasion. Mechanistically, circADAMTS14 promotes the protein expression of regulator of calcineurin 1 (RCAN1) by competitively binding to miR-572 in HCC. In conclusion, circADAMTS14 can suppress the HCC process by adjusting the miR-572/RCAN1 axis [11].
Several other circRNAs [12, 101-110], which act as tumor suppressor genes in the development and progression of HCC, have been listed in Table 2. These circRNAs have been reported to act as tumor suppressor genes. In the occurrence and development of HCC, these circRNAs inhibit the proliferation, invasion, migration, anti-apoptosis, drug resistance and immune escape of cancer cells. Research on circRNAs in HCC remains highly insufficient; therefore, further research on the role of circRNAs in HCC is required.
Medical history, symptoms, signs, laboratory examinations and imaging examinations have markedly improved the diagnosis of HCC. However, several patients are already at an advanced stage at diagnosis, which prevents them from benefiting from surgery. Liver puncture cytology biopsy is the gold standard for diagnosing HCC; however, this invasive operation is difficult for patients to accept, and penetrating the tumor tissue may not be possible, depending on the experience of the clinician. The abnormal expression of circRNAs in HCC tissues and cells highlights their potential value as new diagnostic biomarkers. Exosomes, in particular, also contain multiple circRNAs. circRNAs may be used as biomarkers for early HCC diagnosis or prediction of prognosis. In addition, the aforementioned studies indicate that circRNAs play an important role in the proliferation, invasion, migration, anti-apoptosis and drug resistance of HCC cells; therefore, circRNAs may also serve as potential therapeutic targets in HCC.
Conclusions
HCC is one of the most common types of cancer and highly detrimental to people's health; however, its molecular mechanisms are incompletely understood. circRNAs are an important class of non-coding RNAs. Emerging evidence indicates that circRNAs are correlated with the pathogenesis of various human diseases. Furthermore, circRNAs have been shown to play important roles in the occurrence and development of HCC. First, circRNAs can affect several biological processes of HCC, including cell proliferation, migration, invasion, apoptosis and drug resistance. Second, circRNAs may be used as biomarkers to diagnose and evaluate the prognosis of patients with HCC, which may markedly improve early HCC diagnosis and treatment. Finally, circRNAs may also serve as potential targets for HCC treatment.
CircRNAs are an emerging class of non-coding RNAs that act as endogenous competitive RNAs in the human body. Although research on circRNAs remains insufficient, we believe that circRNAs may be widely used for the diagnosis, treatment and prognosis assessment of HCC, thereby improving the prognosis of patients with HCC. However, further research is required to confirm their value in the clinical setting.
Abbreviations
HCC: hepatocellular carcinoma; TACE: transcatheter arterial chemoembolization; CircRNAs: circular RNAs; EcircRNAs: exon circRNAs; CiRNAs: circular intron RNAs; EIciRNAs: exon-intron circRNAs; ncRNA: non-coding RNA; RBPs: RNA-binding proteins; miRNAs: microRNAs; MREs: miRNA response element; CDR1as: cerebellar Degeneration-Related protein 1 antisense; HuR: human antigen R; IRES: internal ribosome entry sites; ncRNAs: non-coding RNAs; EMT: epithelial-mesenchymal transition.
Acknowledgements
This project was supported by grants from The National Natural Science Foundation of China (No. 81871909), “13th five-year Plan” Science and Education strong Health Project leading personnel of Yangzhou (No. LJRC20181), Provincial-level discipline leader of the NJPH (No. DTRC201809), General Project of Hunan Provincial Health and Healthy Fertility Committee Fund (No. 20201954), and Key guiding project of Hunan Provincial Health Commission (No. 202104010743).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30
2. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018
3. Ko KL, Mak LY, Cheung KS, Yuen MF. Hepatocellular carcinoma: recent advances and emerging medical therapies. F1000Res. 2020 9
4. Ren Z, Ma X, Duan Z, Chen X. Diagnosis, Therapy, and Prognosis for Hepatocellular Carcinoma. Analytical cellular pathology (Amsterdam). 2020;2020:8157406
5. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-14
6. Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123-35
7. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94
8. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-91
9. Zhang J, Chang Y, Xu L, Qin L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J Cell Biochem. 2019;120:9487-94
10. Yu J, Yang M, Zhou B, Luo J, Zhang Z, Zhang W. et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond). 2019;133:1487-503
11. Song C, Li D, Liu H, Sun H, Liu Z, Zhang L. et al. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. J Cell Physiol. 2019;234:2460-70
12. Liu Z, Yu Y, Huang Z, Kong Y, Hu X, Xiao W. et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10:900
13. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331-42
14. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J. et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984-92
15. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
16. Chen X, Fan S, Song E. Noncoding RNAs: New Players in Cancers. Adv Exp Med Biol. 2016;927:1-47
17. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777
18. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
19. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y. et al. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90
20. Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67-79
21. Crick F. Central dogma of molecular biology. Nature. 1970;227:561-3
22. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
23. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
24. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-12
25. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as Upregulates SCAI to Suppress Cisplatin Resistance in Ovarian Cancer via miR-1270 Suppression. Mol Ther Nucleic Acids. 2019;18:24-33
26. Chen H, Mao M, Jiang J, Zhu D, Li P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. OncoTargets and therapy. 2019;12:3869-79
27. Li Y, Zhang J, Pan S, Zhou J, Diao X, Liu S. CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thorac Cancer. 2020;11:537-48
28. B. D. Dysregulated miR-671-5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2015;7:4746-59
29. Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS One. 2016;11:e0158347
30. Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17-27
31. Su Y. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. 2019; 11: 8183-203.
32. Li C, Li M, Xue Y. Downregulation of CircRNA CDR1as specifically triggered low-dose Diosbulbin-B induced gastric cancer cell death by regulating miR-7-5p/REGgamma axis. Biomed Pharmacother. 2019;120:109462
33. Tang W, Ji M, He G, Yang L, Niu Z, Jian M. et al. Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. OncoTargets and therapy. 2017;10:2045-56
34. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X. et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology. 2020;71:130-47
35. Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY. et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology. 2020;72:906-22
36. Li Y, Shi H, Yuan J, Qiao L, Dong L, Wang Y. Downregulation of circular RNA circPVT1 restricts cell growth of hepatocellular carcinoma through downregulation of Sirtuin 7 via microRNA-3666. Clinical and experimental pharmacology & physiology. 2020;47:1291-300
37. Zhu Y, Liu Y, Xiao B, Cai H, Liu M, Ma L. et al. The circular RNA PVT1/miR-203/HOXD3 pathway promotes the progression of human hepatocellular carcinoma. Biology open. 2019 8
38. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ. et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214-27
39. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-11
40. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
41. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
42. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
43. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
44. Kelly S, Greenman C, Cook PR, Papantonis A. Exon Skipping Is Correlated with Exon Circularization. J Mol Biol. 2015;427:2414-7
45. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH. et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103-11
46. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381-8
47. Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907
48. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16:58
49. K A. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361-9
50. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846-58
51. Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu. et al. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther. 2017;25:2062-74
52. Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L. et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609-20
53. Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z. et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. Journal of hematology & oncology. 2017;10:52
54. Wang X, Fang L. Advances in circular RNAs and their roles in breast Cancer. J Exp Clin Cancer Res. 2018;37:206
55. Liu D, Mewalal R, Hu R, Tuskan GA, Yang X. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic Res. 2017;4:17031
56. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37 e9
57. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805-14
58. Chen H, Liu Y, Li P, Zhu D. RE: Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2019;111:435
59. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L. et al. Translation of CircRNAs. Mol Cell. 2017;66:9-21 e7
60. Zhai Z, Fu Q, Liu C, Zhang X, Jia P, Xia P. et al. Emerging Roles Of hsa-circ-0046600 Targeting The miR-640/HIF-1alpha Signalling Pathway In The Progression Of HCC. OncoTargets and therapy. 2019;12:9291-302
61. Zou H, Xu X, Luo L, Zhang Y, Luo L, Yao Y. et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle. 2019;18:2398-413
62. Fu X, Zhang J, He X, Yan X, Wei J, Huang M. et al. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J Cancer. 2020;11:3318-26
63. Huang XY, Huang ZL, Zhang PB, Huang XY, Huang J, Wang HC. et al. CircRNA-100338 Is Associated With mTOR Signaling Pathway and Poor Prognosis in Hepatocellular Carcinoma. Front Oncol. 2019;9:392
64. Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428
65. Lin T, Dai Y, Guo X, Chen W, Zhao J, Cao L. et al. Silencing Of hsa_circ_0008450 Represses Hepatocellular Carcinoma Progression Through Regulation Of microRNA-214-3p/EZH2 Axis. Cancer Manag Res. 2019;11:9133-43
66. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J. et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432-45
67. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K. et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844-59
68. Su Y. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. AGING. 2019;11:8183-203
69. Wang W, Li Y, Li X, Liu B, Han S, Li X. et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed Pharmacother. 2020;121:109517
70. Bai N, Peng E, Xia F, Wang D, Li X, Li X. CircABCC2 Regulates Hepatocellular Cancer Progression by Decoying MiR-665. J Cancer. 2019;10:3893-8
71. Li Z, Hu Y, Zeng Q, Wang H, Yan J, Li H. et al. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. 2019;19:211
72. Zhan W, Liao X, Chen Z, Li L, Tian T, Yu L. et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 2020;235:1733-45
73. Guo J, Duan H, Li Y, Yang L, Yuan L. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem Biophys Res Commun. 2019;513:812-9
74. Pan H, Tang L, Jiang H, Li X, Wang R, Gao J. et al. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J Cell Biochem. 2019
75. Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y. et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9:1091
76. Xie B, Zhao Z, Liu Q, Wang X, Ma Z, Li H. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene. 2019;683:253-61
77. Cao S, Wang G, Wang J, Li C, Zhang L. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol Cell Biol. 2019;97:218-28
78. Li S, Gu H, Huang Y, Peng Q, Zhou R, Yi P. et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. 2018;17:2349-59
79. Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L. et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018;18:116
80. Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao Y. et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37:172
81. Cai H, Hu B, Ji L, Ruan X, Zheng Z. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am J Transl Res. 2018;10:1690-702
82. Guan Z, Tan J, Gao W, Li X, Yang Y, Li X. et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol. 2018;234:500-8
83. Wang B, Chen H, Zhang C, Yang T, Zhao Q, Yan Y. et al. Effects of hsa_circRBM23 on Hepatocellular Carcinoma Cell Viability and Migration as Produced by Regulating miR-138 Expression. Cancer Biother Radiopharm. 2018;33:194-202
84. Li MF, Li YH, He YH, Wang Q, Zhang Y, Li XF. et al. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed Pharmacother. 2018;99:848-58
85. Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D. et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497:626-32
86. Cheng X, Tian P, Zheng W, Yan X. Piplartine attenuates the proliferation of hepatocellular carcinoma cells via regulating hsa_circ_100338 expression. Cancer Med. 2020;9:4265-73
87. Li J, Qin X, Wu R, Wan L, Zhang L, Liu R. Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis. J Cell Mol Med. 2020;24:5152-61
88. Wei X, Zheng W, Tian P, He Y, Liu H, Peng M. et al. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle. 2020;19:817-24
89. Sun P, Fan X, Hu X, Fu X, Wei Q, Zang Y. circPCNX and Pecanex Promote Hepatocellular Carcinoma Cell Viability by Inhibiting miR-506. Cancer Manag Res. 2019;11:10957-67
90. Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 2020;19:269-79
91. Pu J, Wang J, Li W, Lu Y, Wu X, Long X. et al. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J Cell Mol Med. 2020
92. Li Z. Circular RNA hsa_circ_0056836 functions an oncogenic gene in hepatocellular carcinoma through modulating miR-766-3p/FOSL2 axis. AGING. 2020;12:2485-97
93. Zhao M, Dong G, Meng Q, Lin S, Li X. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J Cell Biochem. 2020;121:4440-9
94. Fu Y, Cai L, Lei X, Wang D. Circular RNA ABCB10 promotes hepatocellular carcinoma progression by increasing HMG20A expression by sponging miR-670-3p. Cancer Cell Int. 2019;19:338
95. W.-Y. Niu LC, P. Zhang, H. Zang, B. Zhu, W.-B. Shao. Circ_0091579 promotes proliferative ability and metastasis of liver cancer cells by regulating microRNA-490-3p. Eur Rev Med Pharmacol Sci. 2019;23:10264-73
96. Yao Z, Xu R, Yuan L, Xu M, Zhuang H, Li Y. et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10:945
97. Gao J, Dai C, Yu X, Yin XB, Zhou F. Circ-TCF4.85 silencing inhibits cancer progression through microRNA-486-5p-targeted inhibition of ABCF2 in hepatocellular carcinoma. Mol Oncol. 2020;14:447-61
98. Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC. et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18:105
99. Han D, Li J, Wang H, Su X, Hou J, Gu Y. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-64
100. Qiu L, Huang Y, Li Z, Dong X, Chen G, Xu H. et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13:441-55
101. Su X, Su J, He H, Zhan Y, Liu H. Hsa_circ_0070269 inhibits hepatocellular carcinoma progression through modulating miR-182/NPTX1 axis. Biomed Pharmacother. 2019;120:109497
102. Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R. et al. CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein 1 expression. Cell Death Discov. 2019;5:121
103. Wang YG, Wang T, Ding M, Xiang SH, Shi M, Zhai B. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer letters. 2019;460:128-38
104. Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging (Albany NY). 2019;11:3362-75
105. Xu L, Feng X, Hao X, Wang P, Zhang Y, Zheng X. et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38:98
106. Zhang X, Luo P, Jing W, Zhou H, Liang C, Tu J. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. OncoTargets and therapy. 2018;11:2853-63
107. Zhong L, Wang Y, Cheng Y, Wang W, Lu B, Zhu L. et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499:1044-9
108. Yang W JH, Tian XF. Circular RNA-ABCB10 suppresses hepatocellular carcinoma progression through upregulating NRP1/ABL2 via sponging miR-340-5p/miR-452-5p. Eur Rev Med Pharmacol Sci. 2020;24:2347-57
109. Yu Q, Dai J, Shu M. Circular RNA-0072309 has antitumor influences in Hep3B cell line by targeting microRNA-665. Biofactors. 2020
110. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L. et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer letters. 2020;475:119-28
Author contact
![]() Corresponding author: Dousheng Bai, Department of Hepatobiliary Surgery, Clinical Medical College, Yangzhou University, 98 West Nantong Road, Yangzhou, Jiangsu, P.R.China. E-mail: drbaidoushengedu.cn.
Corresponding author: Dousheng Bai, Department of Hepatobiliary Surgery, Clinical Medical College, Yangzhou University, 98 West Nantong Road, Yangzhou, Jiangsu, P.R.China. E-mail: drbaidoushengedu.cn.

 Global reach, higher impact
Global reach, higher impact