Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(8):1300-1306. doi:10.7150/ijms.73026 This issue Cite
Research Paper
MALAT1 Polymorphisms and Lung Cancer Susceptibility in a Chinese Northeast Han Population
1. Department of Epidemiology, School of Public Health, China Medical University, Shenyang 110122, P.R. China.
2. Department of Obstetrics and Gynecology, Liaoning Provincial Hospital for women and children, Shayang Street, Heping District, Shenyang 110122, P.R. China.
3. Clinical Laboratory, Shengjing Hospital of China Medical University, Shenyang, P.R. China.
4. Department of Clinical Epidemiology and Center of Evidence Based Medicine, The First Hospital of China Medical University, No. 155 Nanjing Bei Street, Heping District, Shenyang 110001, P.R. China.
Received 2022-3-19; Accepted 2022-6-21; Published 2022-7-11
Abstract
Background: LncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) was competitive endogenous RNA (ceRNA) involved in various molecular processes for metastasis development in lung cancer. Single nucleotide polymorphisms (SNPs) in MALAT1 gene might be predictive markers for lung cancer. In our study, we selected rs619586 and rs3200401 in MALAT1 gene to explore their effects on lung cancer susceptibility.
Methods: The case-control study included 444 lung cancer cases and 460 healthy controls. Genotyping was performed by Taqman allelic discrimination method. Logistic regression, Student t-test, and Chi-square test (χ2) were used to analyze the data.
Results: The findings of the study showed that rs3200401 was significantly associated with the risk of non-small cell lung cancer (NSCLC) and lung squamous cell carcinoma (LUSC). Compared with homozygous CC genotype, CT heterozygous genotype decreased risk of NSCLC (Pa = 0.034) and LUSC (Pa = 0.025). In addition, no statistical association was detected between rs619586 and lung cancer susceptibility. The interactions between genes and cigarette smoking were discovered via crossover analysis. However, there were no remarkable gene-environment interactions in additive and multiplicative model.
Conclusion: Rs3200401 in lncRNA MALAT1 was associated with the susceptibility of non-small-cell lung cancer and lung squamous cell carcinoma. The gene-environmental (cigarette smoking) interactions were not notable.
Keywords: Lung cancer, LncRNA, MALAT1, Single nucleotide polymorphism, Interaction.
Introduction
In recent years, a very important factor causing human death was malignant tumors. In particular, the incidence and mortality of lung cancer are always at the forefront [1-3]. The global data (GLOBOCAN 2018) predicted that the number of cases in Asia accounted for nearly half of all new malignancies (18.1 million) worldwide, and nearly 70% of cancer deaths are Asian [4]. Cigarette smoking is one of the environmental risk factors leading to lung cancer. But it is estimated that half of all new cases were non-smokers or smokers who quit many years ago [5]. Therefore, smoking cannot fully explain the cause of the disease. Other studies had shown that genetic factors also play a crucial role in the process of lung cancer [6-13].
Non-coding RNA (ncRNAs) are divided into two types by length: small ncRNAs (< 200 bp) and lncRNAs (> 200 bp)[14]. Many studies have increasingly proved that long non-coding RNAs play critical roles in biological process [15-22]. Genome-wide association studies (GWAS) have showed that numerous single nucleotide polymorphisms (SNPs) are associated with some diseases.
LncRNA MALAT1 located at 11q13, which is also known as NEAT2 (nuclear-enriched abundant transcript 2). MALAT-1 was first discovered in a study of non-small cell lung cancer [23]. Since the discovery of MALAT1, it had played an important role in the occurrence, development, metastasis, drug resistance and clinical treatment of the disease [16, 24-30], especially in lung cancer. Moreover, many papers had reported the risk of MALAT1 polymorphisms and diseases susceptibility [31-44]. However, few studies have investigated about MALAT1 polymorphisms and the risk of lung cancer. Therefore, the study recruited Chinese Northeast Han Population as the research objects and performed a case-control study to explore the association between MALAT1 polymorphisms and susceptibility of lung cancer.
Material and Methods
Study subjects and data collection
The Institutional Review Board of the China Medical University approved the case-control study which was carrying out in Shenyang, Liaoning Province. All the subjects were from the Chinese Northeast Han Population. Cases were from several hospitals in Shenyang. Inclusion and exclusion criteria of the cases were mentioned in our previous studies [39, 45-47]. And the healthy controls were from medical examination centers of hospital. Both cases and controls who smoke more than 100 cigarettes a lifetime were defined as smokers. After signing the informed consent form and filling out the personal information questionnaire, all participants were collected 5mL of venous blood and stored in the -20 ℃ environment for the next step of DNA extraction.
SNPs selection and Genotyping
The method of screening two SNPs was the same as the previous research [48]. The minimum allele frequencies (MAF) of the two SNPs (rs619586, rs3200401) were more than 0.05 in Chinese Han population (CHB). Genotyping were conducted by a Real-Time Polymerase Chain Reaction (PCR) with the TaqMan assay. 10% of the samples were randomly selected for repeated experiments, and the genotyping results were consistent.
Statistical analysis
The data were performed by SPSS software 20.0 (IBM SPSS, Inc., Chicago, IL, USA). The distributions among differences with cigarette smoking, age and gender were assessed by Chi-square test (χ2) and student t-test. Goodness-of-fit χ2 was used to compute the value of Hardy-Weinberg equilibrium (HWE) in the control group. Logistic regression analysis was performed to analysis the experimental data (Indicators: odds ratios (ORs) and 95% confidence intervals (CIs)). And the two indicators were adjusted by gender, age, and cigarette smoking. The interactions were described by additive and multiplicative models. P-value <0.05 (two-sided) was defined as statistically significant.
Results
Subject characteristics
The study included 460 healthy controls and 444 lung cancer cases with the average ages of 58.03 and 59.45, respectively. There were no significant differences in gender and age between case and control group, while there was a statistical difference in cigarette smoking (P<0.001). Case group contained 213 lung adenocarcinoma (LUAD) cases, 145 lung squamous cell carcinomas (LUSC) cases and 86 small cell lung cancers (SCLC). The detailed information was shown in Table 1. In addition, the values of HWE were more than 0.05(P = 0.543 for 619586, P = 0.061 for rs3200401).
Distribution of demographic variables in lung cancer and controls.
| Risk factor | Lung cancer (%) | Control (%) | P |
|---|---|---|---|
| (n=444) | (n=460) | ||
| Age(mean ± SD) | 59.45±10.768 | 58.03±13.995 | 0.087 |
| ≤60 | 226(50.9) | 227(49.3) | 0.641 |
| >60 | 218(49.1) | 233(50.7) | |
| Gender | 0.530 | ||
| Male | 263 (59.) | 263(57.2) | |
| Female | 181 (40.8) | 197(42.8) | |
| Smoking status | <0.001 | ||
| Ever | 227(51.1) | 161 (35.0) | |
| Never | 217(48.9) | 299(65.0) | |
| Pathological type | |||
| LUAD | 213(48.0) | ||
| LUSC | 145(32.7) | ||
| SCLC | 86(19.4) |
LUAD: lung adenocarcinoma, LUSC: lung squamous cell carcinoma, SCLC: small cell lung cancer.
Genotype distribution and Lung Cancer susceptibility
The association of the two SNPs with lung cancer, NSCLC, LUAD and LUSC were shown in Table 2 and Table 3. The results demonstrated that there was no significant difference between the rs619586 and the risk of the lung cancer and other pathological classifications. However, we found that the relationship between rs3200401 and non-small cell lung cancer risk (CT vs. CC: ORa = 0.707, 95%CI = 0.513-0.974, P = 0.034). At the same time, rs3200401 and lung squamous cell carcinoma had statistical differences in the two gene models (CT vs. CC: ORa = 0.581, 95%CI = 0.361-0.935, P = 0.025; TT+CT vs. CC: ORa = 0.634, 95%CI = 0.403-0.997, P = 0.048).
Interactions between SNPs and Cigarette Smoking
In Table 4, crossover analysis was performed to estimate the interaction between the two SNPs and cigarette smoking in lung cancer. The combined effect of the dangerous genes and cigarette smoking increased the risk of disease to a certain extent. The results showed that the interaction between two SNPs and cigarette smoking were found in lung cancer and NSCLC. Table 5 presented that rs619586 and rs3200401 had no additive interaction with cigarette smoking exposure. Similarly, there were no interactions between the two SNPs and cigarette smoking exposure in multiplicative model (Table 6).
Discussion
LncRNA MALAT1 was first reported in non-small cell lung cancer [23], but few studies have published on its polymorphisms and susceptibility to lung cancer. In view of the current high mortality and morbidity of lung cancer, many mechanism studies also had shown that MALAT1 was related to the occurrence, development and prognosis of lung cancer. Therefore, it was necessary to study its genetic polymorphisms and susceptibility to lung cancer. In our study, we selected rs619586 and rs3200401 to explore their risk with lung cancer susceptibility.
The association of the two SNPs with lung cancer and non-small cell lung cancer risk.
| Genotyping | Control (%) | Lung cancer | Non-small cell lung cancer | ||||
|---|---|---|---|---|---|---|---|
| (n=460) | Cases(%)(n=444) | ORª(95%CI) | Pª value | Cases (%)(n=358) | ORª(95%CI) | Pª value | |
| rs619586 | |||||||
| AA | 372(80.9) | 360(81.1) | 1.00(Ref) | 288(80.4) | 1.00(Ref) | ||
| GA | 82(17.8) | 82(18.5) | 1.057(0.749-1.493) | 0.751 | 68(19.0) | 1.115(0.776-1.604) | 0.555 |
| GG | 6(1.3) | 2(0.5) | 0.409(0.080-2.097) | 0.284 | 2(0.6) | 0.486(0.095-2.489) | 0.387 |
| GG+GA vs. AA | 1.016(0.724-1.424) | 0.929 | 1.074(0.752-1.532) | 0.696 | |||
| GG vs. AA+GA | 0.406(0.079-2.076) | 0.279 | 0.478(0.093-2.442) | 0.375 | |||
| A allele | 826(89.8) | 802(90.3) | 1.00(Ref) | 644(89.9) | 1.00(Ref) | ||
| G allele | 94(10.2) | 86(9.7) | 0.942(0.692-1.282) | 0.705 | 72(10.1) | 0.982(0.711-1.358) | 0.915 |
| rs3200401 | |||||||
| CC | 309(67.2) | 321(72.3) | 1.00(Ref) | 263(73.5) | 1.00(Ref) | ||
| CT | 143(31.1) | 112(25.2) | 0.755(0.560-1.017) | 0.065 | 85(23.7) | 0.707(0.513-0.974) | 0.034 |
| TT | 8(1.7) | 11(2.5) | 1.264(0.494-3.232) | 0.625 | 10(2.8) | 1.345(0.516-3.508) | 0.545 |
| TT+CT vs. CC | 0.783(0.585-1.047) | 0.099 | 0.743(0.545-1.014) | 0.061 | |||
| TT vs. CC+CT | 1.368(0.537-3.487) | 0.511 | 1.479(0.569-3.846) | 0.422 | |||
| C allele | 761(82.7) | 754(84.9) | 1.00(Ref) | 611(85.3) | 1.00(Ref) | ||
| T allele | 159((17.3) | 134(15.1) | 0.851(0.662-1.093) | 0.206 | 105(14.7) | 0.822(0.629-1.076) | 0.153 |
a Adjusted by age, gender, smoking. OR, odds ratio; CI, confidence interval.
The association of the two SNPs with lung adenocarcinoma and lung squamous cell carcinoma risk.
| Genotyping | Control (%) | Lung adenocarcinoma | Lung squamous cell carcinoma | ||||
|---|---|---|---|---|---|---|---|
| (n=460) | Cases (%)(n=213) | ORª(95%CI) | Pª value | Cases (%)(n=145) | ORª(95%CI) | Pª value | |
| rs619586 | |||||||
| AA | 372(80.9) | 171(80.3) | 1.00(Ref) | 117(80.7) | 1.00(Ref) | ||
| GA | 82(17.8) | 40(18.8) | 1.042(0.681-1.596) | 0.848 | 28(19.3) | 1.244(0.747-2.072) | 0.401 |
| GG | 6(1.3) | 2(0.9) | 0.913(0.179-4.671) | 0.913 | 0(0) | - | - |
| GG+GA vs. AA | 1.035(0.683-1.569) | 0.872 | 1.144(0.691-1.894) | 0.600 | |||
| GG vs. AA+GA | 0.907(0.178-4.634) | 0.907 | - | - | |||
| A allele | 826(89.8) | 382(89.7) | 1.00(Ref) | 262(90.3) | 1.00(Ref) | ||
| G allele | 94(10.2) | 44(10.3) | 1.012(0.694-1.477) | 0.950 | 28(9.7) | 0.939(0.602-1.464) | 0.782 |
| rs3200401 | |||||||
| CC | 309(67.2) | 153(71.8) | 1.00(Ref) | 110(75.9) | 1.00(Ref) | ||
| CT | 143(31.1) | 55(25.8) | 0.796(0.549-1.154) | 0.229 | 30(20.7) | 0.581(0.361-0.935) | 0.025 |
| TT | 8(1.7) | 5(2.3) | 1.288(0.410-4.044) | 0.665 | 5(3.4) | 1.486(0.422-5.234) | 0.538 |
| TT+CT vs. CC | 0.822(0.573-1.180) | 0.289 | 0.634(0.403-0.997) | 0.048 | |||
| TT vs. CC+CT | 1.377(0.441-4.304) | 0.582 | 1.706(0.486-5.988) | 0.404 | |||
| C allele | 761(82.7) | 361(84.7) | 1.00(Ref) | 250(86.2) | 1.00(Ref) | ||
| T allele | 159((17.3) | 65(15.3) | 0.862(0.629-1.180) | 0.354 | 40(13.8) | 0.766(0.526-1.114) | 0.162 |
a Adjusted by age, gender, smoking. OR, odds ratio; CI, confidence interval.
Crossover Analysis of Interaction Between the two SNPs and cigarette smoking with lung Cancer and non-small Cell lung cancer risk.
| Control (%) | Smoking | Lung Cancer | Non-small cell lung cancer | |||||
|---|---|---|---|---|---|---|---|---|
| (n=460) | Cases(%)(n=444) | OR(95%CI) | P value | Cases(%)(n=358) | OR(95%CI) | P value | ||
| rs619586 | ||||||||
| AA | 235(51.1) | Never | 177(39.9) | 1.00(ref) | 148(41.3) | 1.00(ref) | ||
| GA+GG | 64(13.9) | Never | 40(9.0) | 0.830(0.534-1.289) | 0.407 | 34(9.5) | 0.844(0.530-1.342) | 0.472 |
| AA | 137(29.8) | Ever | 183(41.2) | 1.773(1.320-2.382) | 0.000* | 140(39.1) | 1.623(1.187-2.218) | 0.002* |
| GA+GG | 24(5.2) | Ever | 44(9.9) | 2.434(1.427-4.153) | 0.001* | 36(10.1) | 2.382(1.366-4.153) | 0.002* |
| rs3200401 | ||||||||
| TT+CT | 94(20.4) | Never | 62(14.0) | 1.00(ref) | 48(13.4) | 1.00(ref) | ||
| CC | 205(44.6) | Never | 155(34.9) | 1.146(0.782-1.681) | 0.484 | 134(37.4) | 1.280(0.849-1.929) | 0.238 |
| TT+CT | 57(12.4) | Ever | 61(13.7) | 1.623(1.001-2.630) | 0.049* | 47(13.1) | 1.615(0.960-2.715) | 0.071 |
| CC | 104(22.6) | Ever | 166(37.4) | 2.420(1.616-3.623) | 0.000* | 129(36.0) | 2.429(1.575-3.746) | 0.000* |
a adjusted by age and gender. OR, odds ratio; CI, confidence interval, *Indicates statistical significance (P<0.05).
Addictive interaction measures between the two SNPs and cigarette smoking exposure.
| Lung cancer | Non-small cell lung cancer | |||||
|---|---|---|---|---|---|---|
| SNP | Measure | Estimate | 95%CI | Measure | Estimate | 95%CI |
| rs619586 | RERI | 0.831 | -0.484, 2.146 | RERI | 0.916 | -0.420, 2.251 |
| AP | 0.341 | -0.056, 0.739 | AP | 0.384 | -0.011, 0.779 | |
| S | 2.377 | 0.647, 8.731 | S | 2.964 | 0.597, 14.726 | |
| rs3200401 | RERI | 0.651 | -0.223, 1.526 | RERI | 0.534 | -0.407, 1.476 |
| AP | 0.269 | -0.075, 0.613 | AP | 0.220 | -0.154, 0.594 | |
| S | 1.847 | 0.611, 5.578 | S | 1.597 | 0.570, 4.472 | |
RERI, relative excess risk due to interaction; AP, attributable proportion due to interaction; S, synergy Index; 95 % CI, 95% confidence interval.
Multiplicative interaction between the two SNPs and cigarette smoking exposure
| Lung Cancer | Non-small cell lung cancer | |||||
|---|---|---|---|---|---|---|
| SNPs | Variables | ORa (95% CI) | Pa value | ORa (95% CI) | Pa value | |
| rs619586 | GA+GG vs. AA | 0.829(0.533-1.291) | 0.407 | 0.856(0.536-1.366) | 0.514 | |
| Smoking | 2.055(1.468-2.877) | 0.000 | 1.837(1.286-2.623) | 0.001 | ||
| Interaction | 1.660(0.822-3.350) | 0.157 | 1.758(0.842-3.672) | 0.133 | ||
| rs3200401 | CC vs. CT+TT | 1.142(0.777-1.679) | 0.499 | 1.261(0.834-1.908) | 0.272 | |
| Smoking | 1.892(1.130-3.169) | 0.015 | 1.853(1.063-3.230) | 0.030 | ||
| Interaction | 1.291(0.721-2.312) | 0.390 | 1.158(0.621-2.160) | 0.644 | ||
a adjusted by age, gender. CI confidence interval, OR odds ratio.
Malate1 expression and survival rate
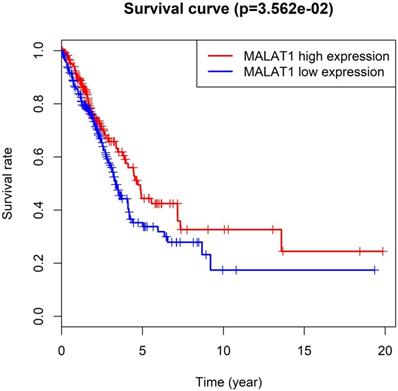
MALAT1, also known as NEAT 2, was competitive endogenous RNA (ceRNA) involved in various molecular processes [49]. Particularly, MALAT1 was a predictive marker for metastasis development in lung cancer [13]. A study discovered lncRNA-MALAT1 participates in NSCLC progression by targeting miR-202 [8]. Furthermore, increased levels of lncRNA MALAT1 could promote brain metastasis of lung cancer by inducing EMT [7]. High expression of MALAT1 promoted the progression of non-small cell lung cancer through the ERK / MAPK signaling pathway [6]. Interestingly, the results of the Cancer Genome Atlas (TCGA) analysis showed that the high expression of MALAT1 in lung adenocarcinoma has a higher survival rate than the low expression in Figure 1 (54 normal / 497 LUAD tissues). These differences required a series of experiments to verify.
Some studies have shown that lncRNA SNPs are associated with cancer risk and are potential predictive biomarkers of cancer risk [30]. The association between rs3200401 polymorphism of MALAT1 in terms of drug efficacy and toxicity has been confirmed, and the rs3200401 CT genotype can be used as a toxicity biomarker [50]. The association between rs3200401 polymorphism of MALAT1 in terms of drug efficacy and toxicity has been confirmed, and the rs3200401 CT genotype can be used as a toxicity biomarker [30]. It can be seen that rs3200401 C and T alleles can change the structural characteristics of MALAT1 and reshape the expression level of cancer-related genes, thus participating in the occurrence and development of cancer [49, 51]. There is an association between MALAT1 rs3200401 and lymph node status, Perhaps further research can reveal their potential as genetic biomarkers for Colorectal cancer (CRC) [51]. Wang et al. showed that T allele of rs3200401 was protective factor in survival outcomes of patients with advanced lung adenoma[9]. In a recent meta-analysis that did not include lung cancer, the researchers did not find a relationship between the MALAT1 rs3200401 polymorphism and overall cancer risk, rs3200401 C > T polymorphism plays different roles in cancers [52]. Our study revealed that rs3200401 reduces the risk of non-small cell lung cancer and lung squamous cell carcinoma. Similar to our results, Ding et al. found that the rs3200401 CT and TT genotypes significantly reduced the risk of developing Oral squamous cell carcinoma (OSCC) [53]. However, MALAT1 rs3200401 was significantly associated with increased disease risk in esophageal squamous cell carcinoma (ESCC), CRC and atrophic gastritis [32, 51, 54]. Moreover, the polymorphisms of rs3200401 were not statistically significant in many diseases, such as breast cancer (BC), multiple sclerosis (MS), ischemic stroke (IS) and coronary artery disease (CAD) and so on[33, 35-37, 39, 42, 44]. MALAT1 rs619586 G allele enhanced the binding of mir-214 to MALAT1 and promoted OSCC development [53]. In breast cancer (BC), Peng et al. found rs619586 AG genotype had a lower risk of BC [39]. Furthermore, patients with the G allele of the rs619586 polymorphism had a significantly increased risk of developing a high-grade Gleason grade in prostate cancer [55]. MALAT1 rs619586 A>G has A protective effect on meningioma invasion by inhibiting the activation of collagen type V alpha (COL5A1) downstream gene [56]. A previous study showed that MALAT1 rs619586 polymorphism significantly reduced the risk of lung cancer [57]. But in this study, rs619586 and lung cancer were not statistically significant difference, the differences were found in many other diseases [33, 35, 37-39, 44]. We did not find gene-environment interactions in multiplication and addition models. The reason might be that the sample size is not large enough.
To obtain more accurate results, OR and 95% CI were adjusted by age, gender, and cigarette smoking. At the same time, we strictly control the standards of each step. However, this study also had some inadequacies with all similar articles. Factors such as selection bias, sample size, and statistical power may affect the results of our study. Functional mechanism experiments would be needed to verify this result in the future.
Conclusion
The polymorphisms rs3200401 in MALAT1 was associated with the risk of non-small cell lung cancer and lung squamous cell carcinoma in Chinese Northeast Han population. The gene-environmental (cigarette smoking) interactions were not notable.
Abbreviations
BC: breast cancer; CAD: coronary artery disease; COL5A1: collagen type V alpha; CHB: Chinese Han population; Cis: confidence intervals; ceRNA: competitive endogenous RNA; ESCC: esophageal squamous cell carcinoma; GWAS: Genome-wide association studies; HWE: Hardy-Weinberg equilibrium; IS: ischemic stroke; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; MALAT1: metastasis-associated lung adenocarcinoma transcript 1; MAF: minimum allele frequencies; MS: multiple sclerosis; NcRNAs: Non coding RNA; NSCLC: non-small cell lung cancer; NEAT2: nuclear-enriched abundant transcript 2; ORs: odds ratios; OSCC: Oral squamous cell carcinoma; PCR: polymerase chain reaction; SNPs: single nucleotide polymorphisms; SCLC: small cell lung cancers.
Acknowledgements
Thanks for all the participants and funding support agencies.
Funding
Natural Science Foundation of Liaoning Province: 2021-MS-164.
Ethics Approval and Consent to Participate
The Institutional Review Board of China Medical University approved the study. Subjects in the study signed informed consent forms before participating.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108
2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90
3. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
4. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
5. Yang C, Stueve TR, Yan C, Rhie SK, Mullen DJ, Luo J. et al. Positional integration of lung adenocarcinoma susceptibility loci with primary human alveolar epithelial cell epigenomes. Epigenomics. 2018;10:1167-87
6. Liu C, Li H, Jia J, Ruan X, Liu Y, Zhang X. High Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Expression Promotes Proliferation, Migration, and Invasion of Non-Small Cell Lung Cancer via ERK/Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway. Med Sci Monit. 2019;25:5143-9
7. Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. Journal of neuro-oncology. 2015;121:101-8
8. Tiansheng G, Junming H, Xiaoyun W, Peixi C, Shaoshan D, Qianping C. lncRNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promotes Proliferation and Invasion of Non-Small Cell Lung Cancer Cells via Down-Regulating miR-202 Expression. Cell journal. 2020;22:375-85
9. Wang JZ, Xiang JJ, Wu LG, Bai YS, Chen ZW, Yin XQ. et al. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer. 2017;17:167-74
10. Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90-9
11. Wang H, Lu B, Ren S, Wu F, Wang X, Yan C. et al. Long Noncoding RNA LINC01116 Contributes to Gefitinib Resistance in Non-small Cell Lung Cancer through Regulating IFI44. Mol Ther Nucleic Acids. 2019;19:218-27
12. Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M. et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer. 2019;18:171-86
13. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180-9
14. Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705-14
15. Zhang J, Li Q, Xue B, He R. MALAT1 inhibits the Wnt/beta-catenin signaling pathway in colon cancer cells and affects cell proliferation and apoptosis. Bosn J Basic Med Sci. 2020;20:357-63
16. Liu S, Qiu J, He G, Liang Y, Wang L, Liu C. et al. LncRNA MALAT1 acts as a miR-125a-3p sponge to regulate FOXM1 expression and promote hepatocellular carcinoma progression. J Cancer. 2019;10:6649-59
17. Lin L, Li Q, Hao W, Zhang Y, Zhao L, Han W. Upregulation of LncRNA Malat1 Induced Proliferation and Migration of Airway Smooth Muscle Cells via miR-150-eIF4E/Akt Signaling. Front Physiol. 2019;10:1337-48
18. Tang R, Chen J, Tang M, Liao Z, Zhou L, Jiang J. et al. LncRNA SLCO4A1-AS1 predicts poor prognosis and promotes proliferation and metastasis via the EGFR/MAPK pathway in colorectal cancer. Int J Biol Sci. 2019;15:2885-96
19. Chen P, Zhao X, Wang H, Zheng M, Wang Q, Chang W. The Down-Regulation of lncRNA PCAT18 Promotes the Progression of Gastric Cancer via MiR-107/PTEN/PI3K/AKT Signaling Pathway. Onco Targets Ther. 2019;12:11017-31
20. Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q. et al. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16:169-82
21. Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J. et al. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol Cancer. 2017;16:171-91
22. Wang J, Cao L, Wu J, Wang Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol. 2018;52:77-88
23. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-41
24. Xu E, Liang X, Ji Z, Zhao S, Li L, Lang J. Blocking long noncoding RNA MALAT1 restrained the development of laryngeal and hypopharyngeal carcinoma. Eur Arch Otorhinolaryngol. 2020;277:611-20
25. Wang S, Yao T, Deng F, Yu W, Song Y, Chen J. et al. LncRNA MALAT1 Promotes Oxygen-Glucose Deprivation and Reoxygenation Induced Cardiomyocytes Injury Through Sponging miR-20b to Enhance beclin1-Mediated Autophagy. Cardiovasc Drugs Ther. 2019;33:675-86
26. Huang XZ, Huang J, Li WZ, Wang JJ, Song DY, Ni JD. LncRNA-MALAT1 promotes osteogenic differentiation through regulating ATF4 by sponging miR-214: Implication of steroid-induced avascular necrosis of the femoral head. Steroids. 2019;154:108533-45
27. Ji H, Qu J, Peng W, Yang L. Downregulation of lncRNA MALAT1 Inhibits Angiotensin II-induced Hypertrophic Effects of Cardiomyocytes by Regulating SIRT4 via miR-93-5p. Int Heart J. 2022;63:602-11
28. Jiang Q, Liu S, Hou L, Guan Y, Yang S, Luo Z. The implication of LncRNA MALAT1 in promoting chemo-resistance of laryngeal squamous cell carcinoma cells. J Clin Lab Anal. 2020;34:e23116
29. Wang ML, Liu JX. MALAT1 rs619586 polymorphism functions as a prognostic biomarker in the management of differentiated thyroid carcinoma. J Cell Physiol. 2020;235:1700-10
30. Lampropoulou DI, Aravantinos G, Katifelis H, Lazaris F, Laschos K, Theodosopoulos T. et al. Long non-coding RNA polymorphisms and prediction of response to chemotherapy based on irinotecan in patients with metastatic colorectal cancer. Cancer Biomark. 2019;25:213-21
31. Hu W, Ding H, Ouyang A, Zhang X, Xu Q, Han Y. et al. LncRNA MALAT1 gene polymorphisms in coronary artery disease: a case-control study in a Chinese population. Biosci Rep. 2019;39:1-13
32. Qu Y, Shao N, Yang W, Wang J, Cheng Y. Association of polymorphisms in MALAT1 with the risk of esophageal squamous cell carcinoma in a Chinese population. Onco Targets Ther. 2019;12:2495-503
33. Che D, Yang Y, Xu Y, Fang Z, Pi L, Fu L. et al. The lncRNA MALAT1 rs619586 G Variant Confers Decreased Susceptibility to Recurrent Miscarriage. Front Physiol. 2019;10:385-91
34. Motawi TMK, El-Maraghy SA, Sabry D, Mehana NA. The expression of long non coding RNA genes is associated with expression with polymorphisms of HULC rs7763881 and MALAT1 rs619586 in hepatocellular carcinoma and HBV Egyptian patients. J Cell Biochem. 2019;120:14645-56
35. Eftekharian MM, Noroozi R, Komaki A, Mazdeh M, Ghafouri-Fard S, Taheri M. MALAT1 Genomic Variants and Risk of Multiple Sclerosis. Immunol Invest. 2019;48:549-54
36. Zhu R, Liu X, He Z. Long non-coding RNA H19 and MALAT1 gene variants in patients with ischemic stroke in a northern Chinese Han population. Mol Brain. 2018;11:58-64
37. Wang G, Li Y, Peng Y, Tang J, Li H. Association of polymorphisms in MALAT1 with risk of coronary atherosclerotic heart disease in a Chinese population. Lipids Health Dis. 2018;17:75-81
38. Li Q, Zhu W, Zhang B, Wu Y, Yan S, Yuan Y. et al. The MALAT1 gene polymorphism and its relationship with the onset of congenital heart disease in Chinese. Biosci Rep. 2018;38:1-7
39. Peng R, Luo C, Guo Q, Cao J, Yang Q, Dong K. et al. Association analyses of genetic variants in long non-coding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene. 2018;642:241-8
40. Zhuo Y, Zeng Q, Zhang P, Li G, Xie Q, Cheng Y. Functional polymorphism of lncRNA MALAT1 contributes to pulmonary arterial hypertension susceptibility in Chinese people. Clin Chem Lab Med. 2017;55:38-46
41. Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J. et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2012;7:e35145
42. Li Y, Zhang D, Zhang Y, Xu X, Bi L, Zhang M. et al. Association of lncRNA polymorphisms with triglyceride and total cholesterol levels among myocardial infarction patients in Chinese population. Gene. 2020:724-35
43. Chen G, Zhang M, Liang Z, Chen S, Chen F, Zhu J, Zhao M, Xu C, He J, Hua W, Duan P. Corrigendum to: Association of polymorphisms in MALAT1 with the risk of endometriosis in Southern Chinese women. Biol Reprod. 2021;104:935-38
44. Wen J, Chen L, Tian H, Li J, Zhang M, Cao Q. et al. Effect of MALAT1 Polymorphisms on Papillary Thyroid Cancer in a Chinese Population. J Cancer. 2019;10:5714-21
45. Lv X, Cui Z, Li H, Li J, Yang Z, Bi Y. et al. Polymorphism in lncRNA AC008392.1 and its interaction with smoking on the risk of lung cancer in a Chinese population. Cancer Manag Res. 2018;10:1377-87
46. Li J, Li H, Lv X, Yang Z, Gao M, Bi Y. et al. Polymorphism in lncRNA AC016683.6 and its interaction with smoking exposure on the susceptibility of lung cancer. Cancer Cell Int. 2018;18:91-9
47. Bi Y, Cui Z, Li H, Lv X, Li J, Yang Z. et al. Polymorphisms in Long Noncoding RNA-Prostate Cancer-Associated Transcript 1 Are Associated with Lung Cancer Susceptibility in a Northeastern Chinese Population. DNA Cell Biol. 2019;38:1357-65
48. Wang S, Cui Z, Li H, Li J, Lv X, Yang Z. et al. LncRNA NEAT1 polymorphisms and lung cancer susceptibility in a Chinese Northeast Han Population: A case-control study. Pathol Res Pract. 2019;215:152723-33
49. Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757-68
50. Lv Z, Xu Q, Yuan Y. A systematic review and meta-analysis of the association between long non-coding RNA polymorphisms and cancer risk. Mutat Res Rev Mutat Res. 2017;771:1-14
51. Radwan AF, Shaker OG, El-Boghdady NA, Senousy MA. Association of MALAT1 and PVT1 Variants, Expression Profiles and Target miRNA-101 and miRNA-186 with Colorectal Cancer: Correlation with Epithelial-Mesenchymal Transition. Int J Mol Sci. 2021:22-42
52. Li K, Han Z, Wu J, Ye H, Sun G, Shi J. et al. The Relationship between MALAT1 Polymorphism rs3200401 C > T and the Risk of Overall Cancer: A Meta-Analysis. Medicina (Kaunas). 2022:58-69
53. Ding YF, Wen YC, Chuang CY, Lin CW, Yang YC, Liu YF. et al. Combined Impacts of Genetic Variants of Long Non-Coding RNA MALAT1 and the Environmental Carcinogen on the Susceptibility to and Progression of Oral Squamous Cell Carcinoma. Front Oncol. 2021;11:684941-52
54. Petkevicius V, Streleckiene G, Balciute K, Link A, Leja M, Malfertheiner P. et al. Association of Long Non-Coding RNA Polymorphisms with Gastric Cancer and Atrophic Gastritis. Genes (Basel). 2020 11
55. Hu JC, Wang SS, Chou YE, Chiu KY, Li JR, Chen CS. et al. Associations between LncRNA MALAT1 Polymorphisms and Lymph Node Metastasis in Prostate Cancer. Diagnostics (Basel). 2021;11:1505-15
56. Zheng J, Pang CH, Du W, Wang L, Sun LG, Xing ZY. An allele of rs619586 polymorphism in MALAT1 alters the invasiveness of meningioma via modulating the expression of collagen type V alpha (COL5A1). J Cell Mol Med. 2020;24:10223-32
57. Chen M, Cai D, Gu H, Yang J, Fan L. MALAT1 rs619586 A/G polymorphisms are associated with decreased risk of lung cancer. Medicine (Baltimore). 2021;100:e23716
Author contact
![]() Corresponding author: Zhihua Yin, Department of Epidemiology, School of Public Health, China Medical University. No.77 Puhe Road, Shenyang North New Area, Shenyang 110122, People's Republic of China. E-mail: zhyinedu.cn
Corresponding author: Zhihua Yin, Department of Epidemiology, School of Public Health, China Medical University. No.77 Puhe Road, Shenyang North New Area, Shenyang 110122, People's Republic of China. E-mail: zhyinedu.cn

 Global reach, higher impact
Global reach, higher impact